Abstract
Horizontal gene transfer by transposition has been widely used for transgenesis in prokaryotes. However, conjugation has been preferred for transfer of large transgenes, despite greater restrictions of host range. We examine the possibility that transposons can be used to deliver large transgenes to heterologous hosts. This possibility is particularly relevant to the expression of large secondary metabolite gene clusters in various heterologous hosts. Recently, we showed that the engineering of large gene clusters like type I polyketide/nonribosomal peptide pathways for heterologous expression is no longer a bottleneck. Here, we apply recombineering to engineer either the epothilone (epo) or myxochromide S (mchS) gene cluster for transpositional delivery and expression in heterologous hosts. The 58-kb epo gene cluster was fully reconstituted from two clones by stitching. Then, the epo promoter was exchanged for a promoter active in the heterologous host, followed by engineering into the MycoMar transposon. A similar process was applied to the mchS gene cluster. The engineered gene clusters were transferred and expressed in the heterologous hosts Myxococcus xanthus and Pseudomonas putida. We achieved the largest transposition yet reported for any system and suggest that delivery by transposon will become the method of choice for delivery of large transgenes, particularly not only for metabolic engineering but also for general transgenesis in prokaryotes and eukaryotes.
INTRODUCTION
Transposable elements were discovered by Barbara McClintock in maize (1) and subsequently in the vast majority of cells from prokaryotes to eukaryotes (2–4). The simplest transposon is a segment of DNA flanked by sequences present as inverted repeats that are recognized by a corresponding transposase, which catalyzes transposition via a cut and paste mechanism. DNA transposons have been applied for gene delivery to insert transgenes at new locations in the genome (5). Unlike homologous recombination, transposition only requires the inverted repeats and does not require homology between the transposon and its target site (6,7). Transposon technology is now widely used for several applications, including in vitro mutagenesis for DNA sequencing or protein structural–functional studies, or in vivo insertional mutagenesis for functional gene analysis and gene transfer.
Unlike the severe limitations of size for transgenes in retroviral vectors, size limits for transposon vectors remain poorly defined. Only a few studies have examined the issue (8–12). These studies all report reductions of transpositional insertion frequencies with increasing size, with suggestions that efficiency might be dramatically or even exponentially sensitive to insert size. In vitro, transposition of DNA as large as 86 kb has been reported (13), but to our knowledge, there is no report in any system of successful transposition of a transgene larger than 20 kb into a heterologous host.
In many bacterial strains, transferring and integrating large sized DNA molecules into the chromosome is difficult because both homologous and random recombination efficiencies are low. Therefore, transposition technology could be an attractive method for transgene introduction, unless inherent size limitations restrict its utility.
The mariner family of transposable elements was first discovered in Drosophila mauritiana (2) and is named because it has spread to virtually all eukaryotic and some prokaryotic organisms. Mariner transposons are usually small elements around 1300-bp long with 27 bp inverted terminal repeats, which contain a single open reading frame encoding a transposase of 345 amino acids (14). The mariner family is most closely related to the Tc1 family of transposons found in nematodes, Drosophila and fish (14,15). The MycoMar transposable element (16), a mariner transposon, has been frequently used in Gram-negative hosts for genetic modification.
DNA cloning and mutagenesis are important technologies for molecular biology and the biosciences. Conventional DNA engineering tools like restriction enzymes and DNA ligases have been successfully used for several decades (17,18). However, these technologies encounter difficulties for engineering and mutating DNA molecules >15–20 kb. We pioneered a technique for DNA manipulation, now called recombineering or Red/ET recombination, which overcomes these obstacles (19–25). Recombineering is mediated through homologous recombination, which allows the exchange of genetic information between two DNA molecules in a precise, specific and faithful manner. These qualities are optimal for DNA engineering regardless of size. Furthermore, recombineering technology requires only short regions of sequence identity (∼40–50 bp) for efficient homologous recombination. These homology sequences are short enough to be easily integrated into synthetic oligonucleotide (oligo) primers for PCR reactions (19–25).
Bioactive natural products are often synthesized by pathways of enzymes encoded in large gene clusters, such as the genes encoding megasynthetases of the polyketide synthase (PKS) and the nonribosomal peptide synthetase (NRPS) types. These pathways present enormous potential for combinatorial biosynthesis (26–28). However, complete PKS/NRPS gene clusters are often >30 kb. Modifying these large gene clusters by using conventional DNA engineering technology is difficult and time-consuming. Because recombineering has no size and site limit, it is an ideal technique for engineering large DNA molecules like these gene clusters. The myxochromide (mchS, ∼30 kb) and myxothiazol (mta, ∼60 kb) gene clusters from the myxobacterium Stigmatella aurantiaca have been engineered by using recombineering for heterologous expression in Pseudomonas putida and Myxococcus xanthus (29–31). These gene clusters were introduced into the chromosome of the heterologous host by conjugation or transformation and homologous recombination (29,31,32). Here, we extend and improve this metabolic engineering technology by showing that transposons are more efficient than other methods for achieving transgenic delivery, even for large gene clusters.
We describe engineering of two gene clusters for transposition in this article, the 30-kb myxochromide S (mchS) gene cluster (29–31) and the ∼60 kb epothilone gene cluster (33–35). Epothilones are produced by the myxobacterium Sorangium cellulosum (33) and the gene cluster (epo) has been introduced into Streptomyces coelicolor (35), M. xanthus (36) and Escherichia coli (37). In each case, the cluster was introduced in at least two smaller pieces using laborious protocols based on DNA transformation. Here, the full-length epo gene cluster was first stitched together into a single plasmid and then the promoter was exchanged, followed by addition of a transpositional cassette, conjugation origin and selection markers. Afterwards, the full size gene clusters with all the necessary components were introduced into heterologous hosts for expression.
Because many successful drugs come from prokaryotic secondary metabolites encoded in large operons, the combination of recombineering and transpositional delivery into heterologous hosts opens a new window for metabolic engineering, drug development and production.
MATERIALS AND METHODS
Bacterial strains and culturing conditions
All recombineering was performed in E. coli strain GB2005, which is a derivative of DH10B, using Luria broth (LB) medium and antibiotics (kanamycin [Km], 15 µg/ml; ampicillin [Amp], 100 µg/ml; blasticidin S [BSD], 50 µg/ml; gentamycin [Genta], 6 µg/ml; zeocin [Zeo], 15 µg/ml and tetracycline [Tet], 5 µg/ml). Heterologous hosts for PKS/NRPS gene cluster expression were M. xanthus DK1622 (38) and P. putida KT2440 (25). Myxococcus xanthus DK1622 was grown at 32°C in CTT medium (1% casitone, 8 mM MgSo4, 10 mM Tris-HCl, pH 7.6, and 1 mM potassium phosphate, pH 7.6) (38) with or without Km (50 µg/ml) before or after introduction of epothilone (epo) gene cluster. Pseudomonas putida KT2440 or FG2005 (30) was maintained in LB medium and the recombinants were selected in Pseudomonas minimum medium (PMM) medium with Km (50 µg/ml) (29) or Genta (10 µg/ml) after conjugation.
Recombineering
Recombineering, also called Red/ET recombination, was described previously (19–25,39). GB2005 cells harboring pSC101-BAD-gbaA with Tet resistance (39) were used for preparing recombineering proficient competent cells. Electrocompetent cells were electroporated with 0.3 µg of a linear DNA fragment (modification cassette), which was obtained by PCR. The selection of recombinants was carried out depending on the selection marker in the cassette. PCRs were performed with Phusion polymerase (New England Biolabs, GmbH, Frankfurt am Main, Germany) according to the manufacturer's protocol.
Engineering of the mchS gene cluster
The pUC-mchS was derived from a SuperCos 1 vector that contains most of the mchS pathway. The missing gene at the 3′ end was added, along with a cassette for conjugation as described (29). Two rounds of recombineering were used for engineering of the pUC-mchS plasmid. We used our recently developed technology named triple recombination for the first round. By electroporation, PCR products of IR-Tps cassette (IR, inverted repeat; Tps, MycoMar transposase gene) and the ampicillin resistance gene (0.3 μg each in 2 μl) were cotransformed into recombineering proficient competent cells in which the pUC-mchS is resident. Recombinants were selected on LB plates containing 100 μg/ml of Amp. For second round recombineering, the IR-Tn5-kan cassette (Tn5, Tn5 promoter; kan, kanamycin resistance gene) flanked with homology arms was generated by PCR. The PCR product (0.3 μg) were used for recombineering and the recombinants were selected on LB plates with 15 μg/ml of Km (Figure 1).
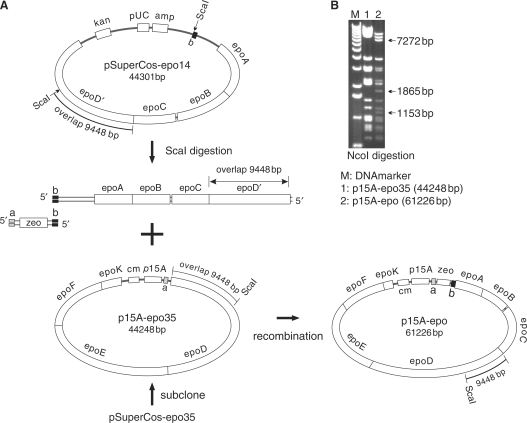
Figure 1.
Diagram of myxochromide S gene cluster engineering. In the first step, the inverted repeat and MycoMar transposase gene (IR-Tps, i) plus amp (ii) were inserted into the mchS expression plasmid (iii) backbone by triple recombination to delete zeo and kan (iv). The MycoMar Tps plus IR fragment (i) was generated by PCR from the original MycoMar transposon (12). In the second step, the oriT-tet-trpE-cm-xylS section (13) in the backbone was replaced with a right IR and Tn5-kan cassette (v), which included a ribosomal binding site for the mchS gene cluster, by selection for kanamycin resistance. In the third step, the modified plasmid (vi) was electroporated into M. xanthus and kanamycin-resistant colonies were selected. To introduce the gene cluster into P. putida, an additional cassette containing oriT and the tetracycline-resistant gene (oriT-tetR-tet, vii) was inserted between amp and the pUC origin (pUC) to form the final construct (viii) for P. putida expression. See remarks in the Materials and Methods section.
Oligos used for IR-Tps-amp insertion are listed below. Sequence as homologous arm for recombineering is in lowercase.
irtase3
5′ tcaaatccgctcccggcggatttgtcctactcaggagaTTATTCAACATAGTTCCCTTC 3′ irtase5 (the underlined sequence is the IR)
5′ tccacaggtcattcaagcgcgcgctggggaaggcaggcaggatgggatctgatcAGACCGGGGACTTATCAGCCAACCTGTTATGTGG 3′
amptase3
5′ tccagactttacgaaacacggaaaccgaagaccattcatgttgttgctcaggtcgcagaTTACCAATGCTTAATCAGTGAG 3′
amptase5
5′ tcgttataatcgttgtatcgctcttgaagggaactatgttgaataaTCTCCTGAGTAGGACAAATCCG 3′
Oligos used for insertion of IR-Tn5-kan cassette were tn5neoIR3 and tn5neoIR5.
tn5neoIR3 (the underlined sequence contains the ribosomal binding site)
5′ tggacactaccccacggctagaaacttcggtcaatacagaatttcctatcatAATCTGTACCTCCTTAAGTCAGAAGAACTCGTCAAGAAG 3′
tn5neoIR5 (the underlined sequence is the IR)
5′ tgaagttttaaatcaatctaaagtatatatgagtaaacttggtctgacagACAGGTTGGCTGATAAGTCCCCGGTCTTCACGCTGCC GCAAGCACTCAG 3′
For expression of mchS in P. putida, an oriT-tetR-tet cassette was re-inserted into the backbone to be able to conjugate the gene cluster into P. putida. A PCR product of the oriT-tetR-tet cassette with homology arms was inserted into the pTps-mchS between amp and pUC origin by recombineering (Figure 1).
Oligos used for insertion of oriT-tetR-tet cassette were oriTOtps3 and oriTOtps5.
oriTOtps3
5′ cacggaaaccgaagaccattcatgttgttgctcaggtcgcagaTCAGCGATCGGCTCGTTGCCCTG 3′
oriTOtps5
5′ tagacagatcgctgagataggtgcctcactgattaagcattggtaATAATGGTTTCTTAGAGCTTACGGCCAG 3′
Engineering the epo gene cluster
The cosmid library of S. cellulosum ce90 genomic DNA (40) was screened by using probes generated by PCR. The primers epoA-check5 (5′-TTACGCGCGCATCTTTCCTGAG-3′) and epoA-check3 (5′-TTTCAGCACGATTTCTTGGGAG-3′) were used to amplify a fragment in epoA. The primers epoK-check5 (5′-TATGACACAGGAGCAAGCGAATC-3′) and epoK-check3 (5′-TAAGGTAGATCGTGGTATCGGTG-3′) were used to amplify a fragment in epoK. The cosmid end sequencing results show that pSuperCos-epo35 contains the part of epoC and full of epoD till the end of epoK, and pSuperCos-epo14 contains epoA to epoC and the part of epoD.
To stitch the epothilone gene cluster together (Figure 3A), epoD-K genes were subcloned into a p15A-Cm minimum vector that was generated by using linearized pACYC184 (41) as template to form p15A-epo35. The zeocin (zeo) resistance gene with a 5′ end homology arm to p15A-cm-epo35 and a 3′ end homology arm to a region in front of epoA in pSuperCos-epo14 was amplified by PCR. pSuperCos-epo14 was digested with ScaI to release a 26-kb fragment which carries the short homology arm to the zeo PCR product and a long homology arm (∼9.45 kb) to p15A-cm-epo35. After mixing the zeo PCR product with the digested pSuperCos-epo14, the mixture was transformed into a recombineering proficient host containing p15A-cm-epo35. The stitched full-length epothilone gene cluster was selected by zeo and analyzed by restriction digestion (Figure 3B). The junction regions of the stitched cluster were verified by sequencing.
Figure 3.
Engineering diagram for stitching of the epothilone gene cluster. pSuperCos-epo35 and pSuperCos-epo14 were the starting clones, which include an overlap in the epoD gene. pSuperCos-epo35 was retrofitted with the p15A origin and the chloramphenicol resistance gene from pACYC184 by subcloning to remove the pSuperCos backbone, introduce the short homology arm ‘(A)’ and create p15A-epo35. pSuperCos-epo14 was digested with ScaI and the epoA-D genes were recombined into p15A-epo35 by triple recombination using a bridging zeo. In (B), the NcoI digest reveals the correct product. (the NcoI sites in p15A-epo35 and p15A-epo are shown in Appendix 1 in Supplementary Material).
To generate the conjugation/transposition cassette, by recombineering the blasticidin S-resistant gene (bsd) was inserted between the MycoMar transposase gene (Tps) and oriT in pMycoMar-hyg plasmid (32) to form the IR-Tps-bsd-oriT cassette (Figure 4A). The PCR product of this cassette with homology arms to the stitched p15A-epo-cm-zeo plasmid was used to replace the chloramphenicol (cm) gene in the backbone to form p15A-epo-IR-Tps-bsd-oriT-zeo. A PCR fragment containing the second IR plus Tn5-kan plus a ribosomal binding site (rbs) after the kan stop codon flanked with two homology arms to the p15A-epo-IR-Tps-bsd-oriT-zeo was used to replace zeo to form the final epothilone expression plasmid p15A-epo-IR-Tps-bsd-oriT-IR-kan for expressing in M. xanthus (Figure 4A). Since the Tn5 promoter is weak in P. putida (data not shown), after two rounds of recombineering the Pm promoter (toluic acid inducible) (29) plus its regulator gene (xylS) and the gentamycin-resistant gene (genta) were used to replace Tn5-kan to form p15A-epo-IR-Tps-bsd-oriT-IR-genta-xylS-Pm which is the final expression plasmid for P. putida expression. All of the engineering was done by recombineering.
Figure 4.
Construction of the epo gene cluster and its expression in M. xanthus. (A), IR-Tps-bsd-oriT cassette (i) in pMycoMar-bsd-hyg plasmid was used as template to generate the PCR product of IR-Tps-bsd-oriT with homology arms. After recombineering, IR-Tps-bsd-oriT was inserted into the plasmid backbone of the stitched epo gene cluster (ii) to form p15A-epo-IR-Tps-bsd-oriT-zeo (iii). Background free template R6K-Tn5-kan was then used to generate IR-Tn5-kan PCR product (iv) with homology arms. The second round of recombineering was performed to build the final expression construct p15A-epo-IR-Tps-bsd-oriT-IR-kan (v). The verified and purified expression construct was electroporated into M. xanthus and the DNA fragment between the two IRs was integrated into the M. xanthus chromosome. (B) Extracts from the stable integrant M. xanthus DK1622-Mut8 were analyzed by HPLC-MS. Rows 1–3 show extracted ion chromatograms of HPLC–MS runs demonstrating that the different epothilones are biosynthesized. Epothilone B, C and D are the major compounds produced in this clone. Rows 4–6 show the same analysis for the wild-type M. xanthus DK1622 (WT) without the epo gene cluster. The fragmentation pattern of epothilone D produced in M. xanthus (row 7) is compared to authentic epothilone D reference (row 8).
To create the IR-Tps-bsd-oriT cassette, BSD PCR was generated by using oligonucleotides bsdINtps3 5′ tgacgccgttggatacaccaaggaaagtctacacgaaccctttggcaTTTAAATCGGATCTGATCAGCACGTGTTG 3′ and bsdINtps5 5′ ttggaaggtcgttataatcgttgtatcgctcttgaagggaactatgttgaataaTTTAAATTAGCCCTCCCACACATAACCAG 3′.
A minimum linear vector containing p15A ori and cm for subcloning of epo gene cluster from pSuperCos-epo35 was generated by using oligo 15epo35a 5′ tgcgatcaactgcgtcagatcgtcctgcgacatctcctcgACTAGTACAACTTATATCGTATGGGGCTG 3′ and 15epo35b 5′ cgaacctcattccctcatgatacagctcgcgcgcgggtgcACTAGTTAACCGTTTTTATCAGGCTCTGG 3′.
One pair of oligos epo-tri-a and epo-tri-b was used for the generation of zeo. epo-tri-a is 5′ tatctcttcaaatgtagcacctgaagtcagccccatacgatataagttgtAGCACGTGTTGACAATTAATC 3′ and epo-tri-b is 5′ gggccgcgagatcggcggcggcgaaggagtTCAGTCCTGCTCCTCGGCCAC 3′.
The bsd gene for pMycoMar-hyg was generated by using oligonucleotides 5′-tcgttataatcgttgtatcgctcttgaagggaactatgttgaataatttaaaTTAGCCCTCCCACACATAAC-3′ and 5′-tgacgccgttggatacaccaaggaaagtctacacgaaccctttggcaTTTAAATCGGATCTGATCAGCACGTGTTG-3′.
The oligonucleotides used for IR-Tps-bsd-oriT PCR were 5′-tagctcgcgggggtatcgcttcccgaacctcattccctcatgatacagAGACCGGGGACTTATCAGCCAACCTGTTATGTGGC-3′ (the underlined sequence is the IR) and 5′-tccggcggtgcttttgccgttacgcaccaccccgtcagtagctgaacaggagggacctagagTCGATCTTCGCCAGCAGGGCGAG-3′. IR-tn5-kan fragment was generated by using oligonucleotides 5′-tctcttcaaatgtagcacctgaagtcagccccatacgatataagttgtactagtACAGGTTGGCTGATAAGTCCCCGGTCTTCACGC TGCCGCAAGCACTCAGGG-3′ (the underlined sequence is the IR) and 5′-cgacgatcgcaatcggatcttcggctgcgcgctcgatgggacgatccgccatAATCTGTACCTCCTTAAGTCAGAAGAACTCGTCAAGAAGG-3′ (the underlined sequence contains the ribosomal binding site). Cm-xylS-Pm fragment was generated by using oligonucleotides 5′-tcgccgctcccgattcgcagcgcatcgccttctatcgccttcttgacgagttcttctgaTCCTGGTGTCCCTGTTGATACCG-3′ and 5′-tcgcaatcggatcttcggctgcgcgctcgatgggacgatccgccatGTTCGTGACCTCCATTATTATTGTTTCTGTTGC-3′. The gentamycin gene was generated by using oligonucleotides 5′-tgccgcaagcactcagggcgcaagggctgctaaaggaagcggaacacgtagaaagccagtccgcTGAAGGCACGAACCCAGTTGAC-3′ and 5′-tgggttggcatcgcccggctttcttagacactctccaagctctgaaatagcgttttacaaaTCGGCTTGAACGAATTGTTAGGTG-3′.
Electroporation of M. xanthus
The engineered gene clusters were introduced into the chromosome of M. xanthus DK1622 by electroporation. Briefly, M. xanthus cells from 1.7 ml of overnight culture with OD600 ∼ 0.6 were collected and electrocompetent cells were prepared after two times washing with ice-cold water. Fifty microliter of the cell suspension in cold water were mixed with 3 μg of DNA and electroporated (Electroporator 2500, Eppendorf AG, Hamburg, Germany) at 1200 V using 0.2 cm cuvette. After electroporation, 1.2 ml of CTT medium were used to resuspend cells, and the cells were incubated at 32°C in a 2 ml Eppendorf tube with a hole punched in the lid on a Thermomixer (Eppendorf) at 11 000 r.p.m. for 6 h. One milliliter of 1.5% CTT agar solution at 42°C was added to the tube and the cells were plated in soft agar for selection on CTT agar plates supplemented with Km (50 µg/ml). Km-resistant colonies appeared after 6 days and were checked by colony PCR as follows. Part of a single colony was washed once in 1 ml of H2O and resuspended in 100 μl of H2O. Then, 2 μl of the resulting suspension was used as a PCR template using Taq polymerase (Invitrogen GmbH, Karlsruhe, Germany) according to the manufacturer's protocol. The myxochromide-specific primers used to check the integration of mchS gene cluster into the M. xanthus chromosome were the same as used in previous study (29). Epothilone-specific primers were designed to detect epoA, epoC and epoK of the gene cluster to verify the integration of the whole biosynthetic gene cluster into the chromosome. For amplification of epoA fragment, epoA-check5 (5′-TTACGCGCGCATCTTTCCTGAG-3′) and epoA-check3 (5′-TTTCAGCACGATTTCTTGGGAG-3′) were used; for amplification of epoC fragment, epoC-check5 (5′-GACTGCCGAGAGAAAATCGAG-3′) and epoC-check3 (5′-ATCTGCATAGATGTCCGTCTTG-3′) were used; and for amplification of epoK fragment, epoK-check5 (5′-TATGACACAGGAGCAAGCGAATC-3′) and epoK-check3 (5′-TAAGGTAGATCGTGGTATCGGTG-3′) were used.
Conjugation of P. putida
Engineered gene cluster expression constructs were introduced into the chromosome of P. putida FG2005 for epo and P. putida KT2440 for mchS (30) by triparental conjugation, as described previously (42). The selection of integrants was carried out on PMM agar plates containing Km (50 µg/ml) for P. putida KT2440 for mchS and Genta (10 µg/ml) for P. putida FG2005. The obtained clones were tested by colony PCR (Taq-polymerase, Invitrogen) with the specific primers as used for checking the mchS and epo containing M. xanthus clones above.
Integration site detection
Single-primer (semi-random) PCR has been described previously (43,44). It is used to detect the integration site of transposition. Primer epo-Po-1 or epo-Po-2 was employed to amplify a PCR product containing a known region and the flanking unknown region of the epothilone gene cluster.
epo-Po-1
5′ TAGCCGAATAGCCTCTCCACC 3′
epo-Po-2
5′ GCGGGGCATCGATCAAGAAAG 3′
Primer epo-Pn-1 or epo-Pn-2 is for sequencing.
epo-Pn-1
5′ TCTTGATCCCCTGCGCCATC 3′
epo-Pn-2
5′ CTCCAGGCCAGACGTGTTTG 3′
The PCR was performed using an Eppendorf MasterCycler using either oligo epo-Po-1 or epo-Po-2. After 30 min at 94°C for denaturation, 20 cycles of 10 min at 94°C, 10 min at 55°C and 1 min at 72°C were run. Next 30 cycles with low stringency, programmed as 10 min at 94°C, 10 min at 40°C and 1 min at 72°C were employed. Finally, 30 cycles of 10 min at 94°C, 10 min at 55°C and 1 min at 72°C were used for further amplification.
Analysis of myxochromide S production in M. xanthus
The M. xanthus DK1622::pTpS-mchS mutant containing the mch S biosynthetic gene cluster was incubated in 250 ml shaking flasks containing 50 ml CTT medium (casitone 10 g/l, 1 M Tris pH 7.6 10 ml/l, 1 M K2HPO4 pH 7.6 1 ml/l, 0.8 M MgSO4 10 ml/l) amended with the adsorber resin XAD-16 (1%) and with Km (50 µg/ml). The culture was inoculated with 500 µl of a well-grown preculture and incubated for 4 days at 30°C on a rotary shaker (160 rpm). The cell mass and the Amberlite XAD-16 adsorber resin from the culture broth were harvested by centrifugation and extracted with acetone/methanol (1:1), respectively. The extract was evaporated, redissolved in 1 ml of methanol and 5 µl of the concentrated extract was analyzed using a DIONEX HPLC system with a diode-array detector (PDA-100). Chromatographic separation was carried out on a RP column (Nucleodur C18, 125 × 2 mm, 3 µm partical size; Macherey & Nagel, GmbH & Co. KG, Düren, Germany) equipped with a precolumn (8 × 3 mm/5 µm). The mobile phase gradient (solvent A: water + 0.1% formic acid and solvent B: acetonitrile + 0.1% formic acid) was linear from 50% B at 2 min to 60% B at 22 min and from 60% B at 22 min to 95% B at 26 min, followed by 3 min with 95% B at flow rate of 0.4 ml/min; detection was carried out at 400 nm. Myxochromides S were identified by comparison to the retention times and UV spectra of authentic reference standards. For quantitative analysis peak integration was carried out utilizing the Chromeleon software package (Version 6.50). A calibration curve was established from serial dilutions of myxochromides S1 and S3. Samples under investigation were diluted as required to fit the dynamic range of the method.
Analysis of Myxochromide S production in P. putida
The P. putida strain containing the myxochromide S biosynthetic gene cluster (P. putida::pTps-mchS-oriT) was incubated in 250 ml shaking flasks containing 50 ml LB medium amended with Km (50 µg/ml). The culture was inoculated with an overnight culture (1:100) and incubated for 2 days at 30°C on a rotary shaker (160 rpm). The cells were harvested by centrifugation and extracted with acetone. The extract was evaporated and redissolved in 500 µl of methanol and 5 μl of the concentrated extract was analyzed by high-pressure liquid chromatography–mass spectrometry (HPLC–MS); an Agilent 1100 series solvent delivery system coupled to Bruker HCTplus ion trap mass spectrometer was used. Chromatographic separation was carried out on an RP column Nucleodur C18 (125 by 2 mm, 3 µm particle size; Macherey and Nagel) equipped with a precolumn C18 (8 × 3 mm, 5 µm). The mobile-phase gradient (solvent A: water + 0.1% formic acid and solvent B: acetonitrile + 0.1% formic acid) was linear from 50% B at 2 min to 60% B at 22 min and from 60% B at 22 min to 95% B at 26 min, followed by 3 min with 95% B at flow rate of 0.4 ml/min. Detection was carried out in positive ionization mode. Myxochromide S1 was identified by comparison to the retention time and the MS2 pattern of the authentic reference standard (m/z [M + H]+ = 723). For quantitative analysis, samples were separated on a gradient linear from 57% B at 2 min to 90% B at 7.50 min, followed by 1.50 min at 90% B. Quantitation was carried out in manual MS2 mode. Ions of m/z [M + H]+ = 723 were collected and subjected to fragmentation. Peak integration of the characteristic fragment ions m/z 322 was carried out utilizing the Bruker Quant-Analysis v1.6 software package. A calibration curve was established from serial dilutions of myxochromide S1 down to 1 µg/ml.
Analysis of the heterologous production of epothilones
Myxococcus xanthus strains containing the gene cluster were inoculated from an overnight culture and incubated in 300-ml flasks containing 50 ml CTT medium supplemented with Km (50 µg/ml) and containing 2% XAD 16 adsorber resin (Rohm und Haas, Frankfurt, Germany) for 5 days at 30°C (200 rpm). The cells and the resin were harvested by centrifugation and extracted with acetone and methanol. Solvents were removed in vacuo, and the residue was dissolved in 1 ml methanol. An aliquot of 5 µl was analyzed by HPLC–MS as described previously (31). Detection was carried out in positive ionization mode. Epothilones were identified by comparison to the retention time and the MS2 pattern of the authentic reference standards.
The P. putida FG2005 clones carrying the epo gene cluster were cultured in LB medium with 10 μg/ml gentamycin. Induction of the culture with toluic acid was performed according to previous publications (29). Compound extraction and analysis were performed similar as described above for extraction of epothilones from M. xanthus.
RESULTS
Engineering the mchS gene cluster
The myxochromide S biosynthetic gene cluster is 29.6-kb long including three large genes. For the work here, the starting construct was the one which we previously used for heterologous expression in P. putida (29). As illustrated in Figure 1, we used triple recombination to insert the MycoMar transposase, right IR and the ampicillin resistance gene downstream of mchS gene cluster (Figure 1). The development of triple recombination for DNA engineering will be described elsewhere (J.F., A.F.S. and Y.Z.; manuscript in preparation). Briefly, we used two PCR products, with a 40 bp overlapping region to each other and one homology arm each to the targeting molecule. Further recombineering was performed to insert the left IR plus Tn5-kan in front of the mchS gene cluster. The Tn5 promoter will drive both the kan gene and the mchS gene cluster expression (Figure 1). The final construct was verified by sequencing the regions generated by PCR, as well as the recombination junctions. The MycoMar transposase gene sequence is identical to the sequences in the Gene Bank (DQ236098 or AY672108). The left IR is 5′ ACAGGTTGGCTGATAAGTCCCCGGTCT 3′ and the right IR sequence is 5′ AGACCGGGGACTTATCAGCCAACCTGT 3′.
The engineered mchS gene cluster in the transposon (pTps-mchS) for M. xanthus expression was used to electroporate P. putida KT2440 directly without success. Therefore, an oriT cassette was integrated into the construct to be able to conjugate the gene cluster into P. putida (Figure 1). As discussed below, after conjugation of this construct into P. putida, successful transposition took place.
Production of myxochromides after introduction of the mchS pathway from S. aurantiaca into M. xanthus and P. putida
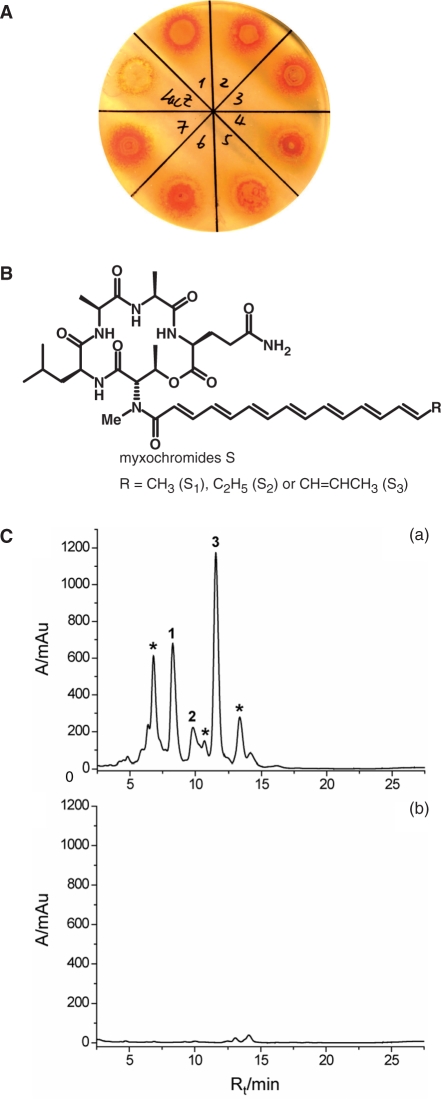
Myxochromide S compounds are characterized by their yellow–orange color and are easily observed in culture. Colonies from pTps-mchS transformation were reddish (Figure 2A) and the liquid cultures are reddish as well (data not shown). A methanol extract from M. xanthus DK1622::pTps-mchS was analyzed with HPLC and HPLC/MS for the production of myxochromides S. Myxochromides S1–3, known from S. aurantica, could be identified in extracts of the M. xanthus mutant strains via HPLC [Figure 2B, peaks 1 (S1), 2 (S2), 3 (S3)], which could also be verified via HPLC/MS analysis (data not shown). Due to the high production of myxochromides S in M. xanthus (∼500 mg/l), minor myxochromide S derivatives could also be detected (peaks marked with an asterix, Figure 2B).
Figure 2.
Detection of myxochromide S compounds in M. xanthus. (A) The color of M. xanthus colonies transformed with the mchS gene cluster is shown. Colonies from M. xanthus::pTps-mchS (1–7) and M. xanthus::pTps-lacZ (LacZ) were picked and replated on a kanamycin plate. The photo was taken after 2 days incubation. (B) the chemical structures of myxochromides S1–3. (C) HPLC profiles from extracts of the M. xanthus::pTpS-mchS mutant strain (a) in comparison to M. xanthus wild-type (b); diode array detection at 400 nm. Numbers correspond to substances as follows: 1, Myxochromide S1; 2, Myxochromide S2; 3, Myxochromide S3. Peaks marked with an asterisk are assumed to be Myxochromide S derivatives as well.
After integration of an additional oriT cassette, the transposon was conjugated and subsequently also integrated into P. putida (Figure 1). By using tri-parental conjugation, we detected more than 106 Km-resistant colonies. Twelve of these were analyzed by PCR and found to be correct genotypically. These clones were cultured and compounds were extracted from cells. The engineered mchS gene cluster was indeed expressed in P. putida but the production level was found to be low (∼100 μg/ml; data not shown).
Stitching of the epothilone gene cluster and engineering for transposition
Cosmid pSuperCos-epo35 is 53 kb in size and contains 45.3 kb of the epothilone gene cluster, whereas cosmid pSuperCos-epo14 is 42-kb large and contains 34.5 kb of the epothilone gene cluster. They share a 24-kb overlap. The stitching procedure to reconstitute the entire cluster is diagrammed in Figure 3A, and was based on two rounds of recombineering and the generation of two intermediates (zeo PCR product and linear epoA-D fragment). The first recombineering step to subclone the epoD-K region into the p15A-cm minimal vector was accomplished with high efficiency and >5 × 103 cm-resistant colonies were obtained. When 24 colonies were analyzed by restriction digestion, 21 of 24 were correct (the other 3 were found to be pACYC without inserts; data not shown). In the second step, a zeo PCR product was used in a triple recombineering exercise to bring the 26-kb long linear epoA-D fragment into p15A-cm-epo35. To facilitate this step and reduce the recombineering background due to carryover of the PCR template, the zeocin resistance gene was amplified from an R6Kγ plasmid, because replication of R6Kγ plasmid needs the π protein encoded by the pir gene, however the pir gene is not present in the E. coli strain GB2005. We expected the second round of recombineering to be difficult because: (i) long linear fragments, here 26 kb, only inefficiently enter E. coli cells; (ii) after entry into E. coli, the identical repeats in the epoA-D region can promote intramolecular rearrangements; and (iii) in triple recombination, three molecules (p15A-cm-epo35, the zeo PCR product and the 26-kb long linear epoA-D fragment) must meet each other in the same cell for productive recombination. After triple recombination, we found 100 colonies on zeocin plates. By checking with restriction digestion, we found that the majority of the recombinants (21 out of 24 clones) contained zeo plus only a short part of the epoA-D fragment because of intramolecular rearrangement. However, the other three were correct recombinants. After retransforming the correct plasmids to separate them from the unrecombined original plasmid, one plasmid was rechecked by NcoI digestion (Figure 3B) and the junction regions were verified by sequencing.
The transpositional cassette, IR-Tps-bsd-oriT, was constructed by inserting the bsd gene between the Tps and oriT in pMycoMar-hyg to form pMycoMar-bsd-hyg. A PCR product of IR-Tps-bsd-oriT (2.6 kb) flanked with homology arms was generated using high-fidelity Phusion polymerase and linearized pMycoMar-bsd-hyg as the template (Figure 4A). After electroporation into E. coli containing the stitched epo cluster, recombinants were selected by plating on BSD plus Zeo plates and incubated at 30°C overnight. This replaced the chloramphenicol (cm) resistance gene in p15A-cm-zeo-epo with the IR-Tps-bsd-oriT cassette to form the intermediate p15A-epo-IR-Tps-bsd-oriT-zeo. In the next round of recombineering, an IR-Tn5-kan-rbs PCR product flanked with homology arms was used to remove zeo and also place a ribosomal binding site in front of epoA to generate p15A-epo-IR-Tps-bsd-oriT-IR-kan. These final recombinants were selected by plating on BSD plus Km plates. More than 50 colonies were pooled from the plate and plasmid DNA was prepared and retransformed into empty E. coli cells to separate unrecombined and recombined plasmids. Three pure recombinants were verified by sequencing of IR-Tps-bsd-oriT cassette and IR-Tn5-kan-rbs-epoA region and two were found without any mutation in the functional regions.
Production of epothilones after introduction of the epo pathway into M. xanthus
Plasmid DNA from verified engineered clones was used to transform M. xanthus (Figure 4A). After entering M. xanthus, transposase expression will integrate the engineered epo cluster randomly into the M. xanthus genome. Stable integrants were selected on Km plates. For p15A-epo-IR-Tps-bsd-oriT-IR-kan, which is 60.5 kb in size, we obtained 90 colonies after 6–8 days incubation at 30°C. For pTps-oriT-mchS, which is 35 kb in size, we obtained >500 Km-resistant colonies using the same protocol (Table 2). Hence, as expected, transposition efficiency is reduced for larger transposons. Nevertheless, we report transposition of a 60-kb transgene at a readily obtained frequency that was well beyond the rate required for successful transgenesis.
Table 2.
Transformation efficiency in M. xanthus
| p15A-epo-IR-Tps- bsd-oriT-IR-kan | pTps-mchS | pTps-lacZ | pOPB18 | |
|---|---|---|---|---|
| Size (kb) | 61 | 35 | 15.6 | 6.7 |
| Colonies | 90 | 635 | 1020 | 9 |
Twelve Km-resistant colonies were checked by colony PCR reactions to verify full integration of the biosynthetic gene cluster. Ten were correct and eight of these were fermented for compound extraction. All eight produced epothilones B, C and D and detectable amounts of epothilone A (Figure 4B and C). Hence, we show that multiple rounds of recombineering to stitch and engineer the epo cluster, followed by transpositional integration, resulted in functional expression.
In contrast to myxochromide formation, epothilone production was low (average ∼100 µg/l), but the difference between each clones was small (data not shown) indicating that the integration site in the chromosome had little effect on epo gene cluster expression driven by the Tn5 promoter.
Conjugation and transposition to introduce the epo pathway into P. putida
The epo gene cluster is derived from myxobacterium S. cellulosum and it is not surprising that it can be used to produce epothilone compounds in M. xanthus. To test the gene cluster expression in a different bacterial species, P. putida was chosen. Wild-type P. putida has successfully been used for mchS gene cluster expression (29) and genetically engineered P. putida FG2005 producing methylmalonyl-CoA (mm-CoA) has been applied for myxothiazol gene cluster expression (30). During the P. putida engineering process, the Tn5 promoter was used to drive the lacZ gene expression and it was found to be weak in this strain (unpublished data). Therefore, the construct used for M. xanthus is most likely not suitable for epo gene cluster expression in P. putida. When introducing the mutase-epimerase-meaB genes for production of methylmalonyl-CoA from S. cellulosum So ce56 into P. putida, the Km resistance gene has been used for selection (30). Thus, a new selection marker gene had to be used for introducing the epo gene cluster into FG2005. Analyzing natural resistances of P. putida, we found this strain to be sensitive to gentamycin at 5 µg/ml. To test if a gentamycin resistance gene (genta, aacC1) functions in P. putida, genta was inserted into the RK2 ori-based plasmid pJB866 (45) to build pJB866-genta which conferred gentamycin resistance after transformation of P. putida.
We next aimed to generate a derivative of the epo gene cluster for expression in P. putida by inserting the Pm promoter. To achieve this, a cm-xylS-Pm cassette with homology arms was amplified from the template plasmid pJB866-cm (in which cm was inserted behind the Pm regulator gene xylS in pJB866). The cm-xylS-Pm PCR product was inserted in front of the epoA gene in p15A-epo-IR-Tps-bsd-oriT-IR-kan to form the construct p15A-epo-IR-Tps-bsd-oriT-IR-kan-cm-xylS-Pm (Figure 5). Afterwards, genta with homology arms was used to remove Tn5-kan and cm to form the final epo construct p15A-epo-IR-Tps-bsd-oriT-IR-genta-xylS-Pm for P. putida FG2005 expression.
Figure 5.
Engineering of epo gene cluster for P. putida FG2005 expression. PCR product of Cm resistance gene, Pm promoter and its regulator xylS gene (i) were inserted in front of epoA in the epo gene cluster (ii) to drive expression of the whole epo gene cluster (iii). PCR product of gentamycin resistance gene (iv) was used to replace Tn5-kan and cm to generate the final construct (v) for P. putida FG2005 expression. The verified final construct was integrated into the FG2005 genome by conjugation and transposition.
Sequencing verified the correctness of the construct, which was conjugated into FG2005 by tri-parental conjugation and stable integrants of epo clones were selected by gentamycin on PMM plates. Conjugation/transposition is an efficient process and more than 104 Genta-resistant colonies were obtained. The conjugation/transposition efficiency of this large sized DNA molecule was found to be similar to small sized DNA molecules like mchS gene cluster (data not shown). The same primers used for colony PCR in M. xanthus were used again to verify the P. putida clones. Verified clones were induced by toluic acid during fermentation and compounds were extracted from culture medium and cells. No epothilone could be detected in any of these extracts.
Verification of gene integration by transposition
To verify that the plasmid, p15A-epo-IR-Tps-bsd-oriT-IR-kan, had integrated into P. putida FG2005 by transposition, we used single primer PCR (43,44) to determine the sequence of the integration site of three clones (Table 1). All three sites include a duplicated TA dinucleotide at the insertion site, which is diagnostic of transposition. The three insertion sites are in the 619th gene, 4266th gene and 5136th genes of P. putida KT2440 (GenBank AE015451). The flanking sequences of the insertion sites in the three genes were found to be exactly the right IR plus the left IR. This validates the successful transposition by the designed construct with the mariner transposable element.
Table 1.
Integration site of epothilone in P. putida KT2440
| Clone | Insertion locus and orientation (IS = IR-tn5-kan-epoA-epoB- epoC-epoD-epoE-epoF-epoK-IR) | Genome location | Name of the gene |
|---|---|---|---|
| 1 | AGTTGCTCGGCCACCCGCTCAAGTA(IS) TACAAAGTCATCGACACCGAAGGCA | 725684 | Branched-chain amino acid ABC transporter, periplasmic amino acid-binding protein |
| 2 | TGTCGCGCAACGAAATCGGCAATTA (IS) TACCTTGGCCTGGCGGTAGAAACCG | 4850861 | Transcriptional regulator Anr |
| 3 | CCTGGTACTGGGCCTGGGCTACGTA (IS) TATTCTTCTTCAACCTGAACGGCAAC | 5860472 | ABC transporter, permease protein |
Transformation versus transposition efficiency in M. xanthus
Myxococcus xanthus can be transformed by electroporation and transgenes can be integrated into the chromosome via homologous recombination. However, the efficiency of integration of large DNA molecules into the chromosome is low and candidate clones must be screened carefully to exclude spontaneous mutants which give Km resistance. Transposition has been used frequently in myxobacteria for insertional mutagenesis and the efficiency of stable integration is much higher than homologous recombination (32).
To compare homologous and transpositional integration in M. xanthus, we used the validated epo and mchS expression constructs described above in a comparison with a homologous integration plasmid pOPB18 (total size 6.7 kb containing a 1.7-kb homology region to the myxovirescin gene cluster in M. xanthus) (31) and a small transposition plasmid, pTps-lacZ (derivative of pMycoMar-hyg, containing a 5.5-kb lacZ-kan-R6K cassette inside of IRs). Table 2 shows the number of transformants which were obtained. The numbers are averages of three transformations done for each plasmid on a molar basis. Interestingly, we found that the colonies which appeared on Km plates before 7 days were all correct but the colonies which appeared after 8 days were spontaneous mutants containing no inserts.
pTps-mchS is 35 kb in size and the integration fragment inside of the two IRs is 31 kb large. p15A-epo-IR-Tps-bsd-oriT-IR-kan is 61 kb in size and the transposable element is 57.5 kb. Although the large constructs have lower efficiency than small integration fragments (pTps-lacZ), their integration process is much more efficient than the small homologous integration plasmid (pOPB18), regardless of the fact that, in general, the transformation efficiency drops dramatically with increasing size of the DNA (46).
DISCUSSION
Transposons for large transgenes and metabolic engineering
Transposons have been employed as efficient mutagens in both prokaryotes and eukaryotes. They have also been used as vectors for efficient and stable introduction of transgenes into recipient genomes. These applications have been invariably based on relatively small (<10 kb) transposons. As far as we can determine, the potential to use transposons as vectors for the integration of large transgenes has not been explored in any system, whether prokaryotic or eukaryotic. Although we do observe decreased efficiencies with increased transgene size as expected, transposon-mediated transgenesis with a 57.5-kb transgene was more efficient than transgenesis via homologous recombination using a small construct. Hence, we suggest that transposition could be a valuable tool for delivery of large transgenes in various prokaryotic and eukaryotic systems. The size of the transgene is particularly relevant for metabolic engineering where the new challenges lie with manipulation of complete pathways, not just single gene products.
For prokaryotes, there are two limiting factors for integrating transgenes into the host chromosome by conventional methods: transformation efficiency and homologous recombination efficiency. Many bacterial hosts, especially those which are known as prominent secondary metabolite producers (e.g. actinomycetes, cyanobacteria, myxobacteria), appear to possess limited capacity for homologous recombination. Consequently, chromosomal integration appears difficult even with small transgenes. Transposition in vivo is an active process and often thousands of colonies can be obtained after each transformation with small constructs. To evaluate transposition for delivery of large transgenes in M. xanthus, we compared a small (∼6 kb), medium (∼31 kb) and a large sized insert (over 57 kb) in transpositional constructs. As expected, the transformation and transpositional integration efficiencies depended on the size of the insert. When a medium size construct with a 1-kb homology arm for homologous integration was used to transform M. xanthus, very few (<10) colonies appeared. But more than 500 colonies were obtained when the same construct was introduced by transposition (Table 2). With the larger epo gene cluster (57.5 kb), transposition delivered fewer colonies. However, this number (90 clones) is still considerably larger than our experience with integration via homologous recombination of smaller constructs. Hence, we suggest that transpositional delivery can be a useful tool for stable transgenesis, which is particularly relevant for large constructs.
Aspects of secondary metabolite expression in heterologous hosts
Other observations from this report are discussed below.
The production of mchS gene cluster in M. xanthus was higher than 500 mg/l and the total myxochromide compounds in some isolates was >1 g/l. Interestingly, the same (Tn5) promoter-driven mchS gene cluster in P. putida produced only 100 μg/l of myxochromides at 30°C (and no compound was detected at 16°C). However, mchS production in P. putida from the Pm promoter reached 40 mg/l at 16°C (29). Together with other observations (unpublished data), we conclude that the Tn5 promoter in P. putida is weak, or under unexpected repression.
More than 80% of the clones carrying the epo or mchS gene clusters were correct as ascertained by PCR. Together with other data presented here, this indicates that the transformation/transpositional integration strategy delivers a high proportion of full length, unmutated, large transgenes into heterologous hosts like M. xanthus. The transposition is mediated by a transient expression of MycoMar transposase gene. Therefore, after integration, the insert is stable in the chromosome. Since the type I PKS/NRPS gene clusters are composed by many repetitive sequences, we have tested the clones carrying the gene clusters by Southern blot and PCR. There was no rearrangement in six tested clones (data not shown). In our experience with various strains, we have not seen production or growth changes over many generations. We conclude that transpositional integration of type I PKS/NRPS gene clusters into heterologous host is stable, with few rearrangements or position effects.
Electroporation of large transgenes into P. putida is difficult. Small plasmids can be easily electroporated into P. putida but plasmids >30 kb construct can hardly be transformed (data not shown). Conjugation has been applied in P. putida to deliver large chromosomal sections (47,48). The combination of conjugation and transposition with small constructs has been reported some time ago (49–51). We integrated the ∼60 kb epo gene cluster into the P. putida genome with similar efficiencies compared to a small construct and the majority of the recombinants were correct as confirmed by PCR tests. Hence, previous size problems with P. putida transformation appear to have been solved.
The epothilone gene cluster was engineered and introduced into the heterologous hosts, M. xanthus and P. putida. Epothilone production was low or undetectable, respectively. There are several possible reasons. First, the Tn5 promoter may be too weak to drive substantial expression in these heterologous hosts. Second, culture conditions like temperature or media components may not be optimal. Third, in P. putida FG2005, endogenous mm-CoA levels may be the limiting factor for epothilone production.
Advanced recombineering for metabolic engineering
Recombineering is a DNA engineering methodology based on homologous recombination in E. coli (19–25). Conventional DNA engineering technology relies on DNA restriction and ligation. However, it is very hard to find convenient restriction sites in the large DNA regions which encode secondary metabolite pathways. DNA ligation relies on purified linear DNA molecules, which are difficult to obtain in time-consuming processes that are inefficient especially for large sizes. Because there are no size limits for recombineering, and in vitro DNA handling steps are eliminated or reduced, it is well suited for the challenges presented by secondary metabolite pathways.
In addition to several routine applications, here we present an advanced recombineering exercise involving DNA stitching by triple recombination. DNA stitching mediated by recombineering has been previously applied for bacterial artificial chromosome (BAC) stitching (52,53) and a similar approach has been applied in yeast for more than a decade (54–56). Here, we were working with cosmids, so the genomic DNA insert average is 40 kb in size. Because most type I PKS/NRPS gene clusters are rarely contained within one cosmid and usually present in two or three, a convenient method for stitching cosmids together will be useful. We developed a strategy to stitch using a selection marker (zeo) as a bridge to bring two large molecules, which share a region of sequence overlap at one end, together in a triple recombination step. Triple recombination presents two advantages for stitching. First, it avoids PCR amplification of the cloned DNA, so it can be applied to large molecules and without fear of mutagenesis in functional regions. Second, it represents a simple and easy way to introduce the second region of homology. The first region is the overlap between the two parental cosmids. However for productive recombination, another region is required. This can be easily introduced into the reaction via the oligonucleotides used to PCR the zeocin cassette. Because the PCR product is small and encodes the zeocin resistance gene, the disadvantages of PCR mutagenesis do not compromise the recombination product.
Triple recombination for stitching presents the further advantage of speed and simplicity because it can save several DNA engineering steps. However, it is much less efficient than standard recombineering applications based on recombination of a single linear DNA substrate into a circular target. Furthermore, we observed a high unspecific recombination rate when stitching the epo gene cluster due to intramolecular rearrangement.
This work further extends our technology platform for the engineering of full-length gene clusters for heterologous expression. This platform includes DNA stitching by recombineering, transpositional cassette insertion, conjugation cassette insertion and inducible promoter insertion. Several technologies including recombineering, electro-transformation, conjugation and transposition have been explored for this platform. We aim to make further technical advances as well as to venture into the unknown territory represented by silent secondary metabolite clusters discovered in genome sequencing and metagenomic programs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank Zoltan Ivics for discussions. This work was partly funded by a BioMet, BMBF grant (Project No: 03I4005), by a grant from EFRE (Europäischen Fonds für Regionale Entwicklung) and the Saxonian Ministry for Science and Art to AFS. Research in the laboratory of R.M. was funded by the Bundesministerium für Bildung und Forschung (BMB + F). Funding to pay the Open Access publication charges for this article was provided by Gene Bridges GmbH, Germany.
Conflict of interest statement. None declared.
REFERENCES
- 1.Comfort NC. From controlling elements to transposons: Barbara McClintock and the Nobel Prize. Trends Biochem. Sci. 2001;26:454–457. doi: 10.1016/s0968-0004(01)01898-9. [DOI] [PubMed] [Google Scholar]
- 2.Berg DE, Howe MM. Mobile DNA. Washington, DC: ASM Press; 1989. [Google Scholar]
- 3.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 4.Merlin C, Mahillon J, Nesvera J, Toussaint A. Gene recruiters and transporters: the modular structure of bacterial moblile elements. In: Thomas CM, editor. The Horizontal Gene Pool. Harwood Academic: Amsterdam; 2000. pp. 363–409. [Google Scholar]
- 5.Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 6.Nevers P, Saedler H. Transposable genetic elements as agents of gene instability and chromosome rearrangements. Nature. 1977;268:109–115. doi: 10.1038/268109a0. [DOI] [PubMed] [Google Scholar]
- 7.Roth JR, Schmid MB. Arrangement and rearrangement of the bacterial chromosome. Stadler Genet. Symp. 1981;13:53–70. [Google Scholar]
- 8.Way JC, Kleckner N. Transposition of plasmid-borne Tn10 elements does not exhibit simple length-dependence. Genetics. 1985;111:705–713. doi: 10.1093/genetics/111.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohe AR, Hartl DL. Reduced germline mobility of a mariner vector containing exogenous DNA: effect of size or site? Genetics. 1996;143:1299–306. doi: 10.1093/genetics/143.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izsvák Z, Ivics Z, Plasterk RH. Sleeping beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 12.Karsi A, Moav B, Hackett P, Liu Z. Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar. Biotechnol. 2001;3:241–245. doi: 10.1007/s101260000072. [DOI] [PubMed] [Google Scholar]
- 13.Basu J, Stromberg G, Compitello G, Willard HF, Van Bokkelen G. Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res. 2005;33:587–596. doi: 10.1093/nar/gki207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson HH, Lampe DJ. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol. Evol. 1995;12:850–862. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- 15.Robertson HM. The mariner element is widespread in insects. Nature. 1993;362:24l–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 16.Rubin E, Akerley B, Novik V, Lampe D, Husson R, Mekalanos J. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl Acad. Sci. USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen SN, Chang AC, Boyer HW, Heiling RB. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl Acad. Sci. USA. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. 51th Cold Spring Harb. Symp. Quant. Biol. 1986:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 20.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotech. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 22.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. RecE/RecT and Reda/Redb initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 23.Muyrers JP, Zhang Y, Stewart AF. Recombinogenic engineering–new options for cloning and manipulating DNA. Trends Biochem. Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 26.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 27.Grünewald J, Marahiel MA. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 2006;70:121–146. doi: 10.1128/MMBR.70.1.121-146.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 29.Wenzel S, Gross F, Zhang Y, Fu J, Stewart F, Müller R. Heterologous expression of a myxobacterial natural products assembly line in Pseudomonads via Red/ET recombineering. Chem. Biol. 2005;12:349–356. doi: 10.1016/j.chembiol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Gross F, Ring MW, Perlova O, Fu J, Schneider S, Gerth K, Kuhlmann S, Stewart AF, Zhang Y, Müller R. Metabolic engineering of Pseudomonas putida for methylmalonyl-CoA biosynthesis to enable complex heterologous secondary metabolite formation. Chem. Biol. 2006;13:1–13. doi: 10.1016/j.chembiol.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Perlova O, Fu J, Kuhlmann S, Krug D, Stewart AF, Zhang Y, Müller R. Reconstitution of the myxothiazol biosynthetic gene cluster by Red/ET recombination and heterologous expression in Myxococcus xanthus. Appl. Environ. Microbiol. 2006;72:7485–7494. doi: 10.1128/AEM.01503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp M, Irschik H, Gross F, Perlova O, Sandmann A, Gerth K, Müller R. Critical variations of conjugational DNA transfer into secondary metabolite multiproducing Sorangium cellulosum strains So ce12 and So ce56: development of a mariner-based transposon mutagenesis system. J. Biotechnol. 2004;107:29–40. doi: 10.1016/j.jbiotec.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Gerth K, Bedorf N, Höfle G, Irschik H, Reichenbach H. Epothilones A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria) J. Antibiot. 1996;49:560–563. doi: 10.7164/antibiotics.49.560. [DOI] [PubMed] [Google Scholar]
- 34.Molnár I, Schupp T, Ono M, Zirkle RE, Milnamow M, Nowak-Thompson B, Engel N, Toupet C, Stratmann A, Cyr DD, et al. The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90. Chem. Biol. 2000;7:97–109. doi: 10.1016/s1074-5521(00)00075-2. [DOI] [PubMed] [Google Scholar]
- 35.Tang L, Sanjay S, Loleta C, John C, Leonard K, Chaitan K, Bryan J. Cloning and heterologous expression of the epothilone gene cluster. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 36.Bryan J, Sanjay S. Heterologous expression of epothilone biosynthetic genes in. Myxococcus xanthus. Antimicrob. Agents. Chemother. 2002;46:2772–2778. doi: 10.1128/AAC.46.9.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mutka SC, Carney JR, Liu Y, Kennedy J. Heterologous production of epothilone C and D in Escherichia coli. Biochemistry. 2006;45:1321–1330. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- 38.Meiser P, Bode HB, Müller R. The unique DKxanthene secondary metabolite family from the myxobacterium Myxococcus xanthus is required for developmental sporulation. Proc. Natl Acad. Sci. USA. 2006;103:19128–19133. doi: 10.1073/pnas.0606039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Sarov M, Rientjes J, Fu J, Hollak H, Kranz H, Xie W, Stewart AF, Zhang Y. An improved recombineering approach by adding RecA to lambda Red recombination. Mol. Biotechnol. 2006;32:43–54. doi: 10.1385/mb:32:1:043. [DOI] [PubMed] [Google Scholar]
- 40.Beyer S, Kunze B, Silakowski B, Müller R. Metabolic diversity in myxobacteria: identification of the myxalamid and the stigmatellin biosynthetic gene cluster of Stigmatella aurantiaca Sg a15 and a combined polyketide-(poly)peptide gene cluster from the epothilone producing strain Sorangium cellulosum So ce90. Biochim. Biophys. Acta. 1999;1445:185–195. doi: 10.1016/s0167-4781(99)00041-x. [DOI] [PubMed] [Google Scholar]
- 41.Rose RE. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill D, Stein J, Torkewitz N, Morse A, Howell C, Pachlatko J, Becker J, Ligon J. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 1994;60:78–85. doi: 10.1128/aem.60.1.78-85.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermann SRJAM, O’Neill S, Tsao TT, Harding RM, Dale JL. Single primer amplification of flanking sequences. Biotechniques. 2000;29:1176–1180. doi: 10.2144/00296bm04. [DOI] [PubMed] [Google Scholar]
- 44.Kofoid EC, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 46.Sheng Y, Mancino V, Birren B. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res. 1995;23:1990–1996. doi: 10.1093/nar/23.11.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishi A, Tominaga K, Furukawa K. A 90-kilobase conjugative chromosomal element coding for biphenyl and salicylate catabolism in Pseudomonas putida KF715. J. Bacteriol. 2000;182:1949–1955. doi: 10.1128/jb.182.7.1949-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravatn R, Studer S, Springael D, Zehnder AJ, van der Meer JR. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. Strain B13. J. Bacteriol. 1998;180:4360–4369. doi: 10.1128/jb.180.17.4360-4369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott JR, Churchward GG. Conjugative transposition. Annu. Rev. Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 50.Salyers AA, Shoemaker NB, Stevens AM, Li LY. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pembroke JT, MacMahon C, McGrath B. The role of conjugative transposons in the Enterobacteriaceae. Cell. Mol. Life Sci. 2002;59:2055–2064. doi: 10.1007/s000180200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotzamanis G, Huxley C. Recombining overlapping BACs into a single larger BAC. BMC Biotechnol. 2004;4:1. doi: 10.1186/1472-6750-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang XM, Huang JD. Combination of overlapping bacterial artificial chromosomes by a two-step recombinogenic engineering method. Nucleic Acids Res. 2003;31:e81. doi: 10.1093/nar/gng081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raymond CK, Sims EH, Olson MV. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 2002;12:190–197. doi: 10.1101/gr.205201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitagawa K, Abdulle R. In vivo site-directed mutagenesis of yeast plasmids using a three-fragment homologous recombination system. BioTechniques. 2002;33:288–294. doi: 10.2144/02332bm07. [DOI] [PubMed] [Google Scholar]
- 56.Raymond CK, Pownder TA, Sexson SL. General method for plasmid construction using homologous recombination. BioTechniques. 1999;26:134–141. doi: 10.2144/99261rr02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.