Abstract
Comparative structure models are available for two orders of magnitude more protein sequences than are experimentally determined structures. These models, however, suffer from two limitations that experimentally determined structures do not: They frequently contain significant errors, and their accuracy cannot be readily assessed. We have addressed the latter limitation by developing a protocol optimized specifically for predicting the Cα root-mean-squared deviation (RMSD) and native overlap (NO3.5Å) errors of a model in the absence of its native structure. In contrast to most traditional assessment scores that merely predict one model is more accurate than others, this approach quantifies the error in an absolute sense, thus helping to determine whether or not the model is suitable for intended applications. The assessment relies on a model-specific scoring function constructed by a support vector machine. This regression optimizes the weights of up to nine features, including various sequence similarity measures and statistical potentials, extracted from a tailored training set of models unique to the model being assessed: If possible, we use similarly sized models with the same fold; otherwise, we use similarly sized models with the same secondary structure composition. This protocol predicts the RMSD and NO3.5Å errors for a diverse set of 580,317 comparative models of 6174 sequences with correlation coefficients (r) of 0.84 and 0.86, respectively, to the actual errors. This scoring function achieves the best correlation compared to 13 other tested assessment criteria that achieved correlations ranging from 0.35 to 0.71.
The explosive growth of sequence databases has not been accompanied by commensurate growth of the protein structure database, the Protein Data Bank (PDB) (Berman et al. 2000). Of the millions of known protein sequences, well fewer than 1% of their corresponding structures have been solved experimentally. Computationally derived structure models serve to bridge this gap, owing to the prediction of two orders of magnitude more structures than are currently available (Pieper et al. 2006). In the absence of an experimentally determined structure, such computational models are often valuable for generating testable hypotheses and giving insight into existing experimental data (Baker and Sali 2001).
Computationally derived structure models, however, generally suffer two major limitations that can limit their utility: They frequently contain significant errors, and their accuracy cannot be readily assessed. Indeed, even if a method sometimes produces accurate solutions, the average precision is still low (Baker and Sali 2001; Bradley et al. 2005). There is currently no practical way to easily and robustly assess the accuracy of a predicted structure, which is problematic for the end users of the models, who cannot be certain that a model is accurate enough in the region(s) of interest to give meaningful biological insight. It is often only after performing time-consuming experiments that a model's accuracy is determined reliably.
Comparative modeling is the most widely used and generally most accurate class of protein structure prediction approaches (Marti-Renom et al. 2000; Tramontano et al. 2001; Eswar et al. 2007). The accuracy of a comparative model is weakly correlated with the sequence identity shared between the target sequence and the template structure(s) used in the modeling procedure (Sanchez et al. 2000). At high sequence identity ranges (i.e., over 50% sequence identity), comparative models can be accurate enough to be useful in virtual ligand screening or for inferring the catalytic mechanism of an enzyme (Bjelic and Aqvist 2004; Caffrey et al. 2005; Chmiel et al. 2005; Costache et al. 2005; Xu et al. 2005). At lower values of sequence identity, especially below 30%, alignment errors and differences between the target and template structures can become major sources of errors (Chothia and Lesk 1986; Rost 1999; Sauder et al. 2000; Jaroszewski et al. 2002; Ginalski et al. 2005; Madhusudhan et al. 2006; Rai and Fiser 2006). In automated comparative modeling of all known protein sequences related to at least one known structure, 76% of all models are from alignments in which the target and template share less than 30% sequence identity (Pieper et al. 2006), where the corresponding models can have a wide range of accuracies (Sanchez et al. 2000; Chakravarty and Sanchez 2004).
Because of the wide accuracy range of models, many assessment scores have been developed for tasks including (1) determining whether or not a model has the correct fold (Miyazawa and Jernigan 1996; Park and Levitt 1996; Domingues et al. 1999; Lazaridis and Karplus 1999; Gatchell et al. 2000; Melo et al. 2002; McGuffin and Jones 2003; Melo and Sali 2007), (2) discriminating between the native and near-native states (Sippl 1993; Melo and Feytmans 1997, 1998; Park et al. 1997; Fiser et al. 2000; Lazaridis and Karplus 2000; Zhou and Zhou 2002; Seok et al. 2003; Tsai et al. 2003; Shen and Sali 2006), (3) selecting the most native-like model in a set of decoys that does not contain the native structure (Shortle et al. 1998; Wallner and Elofsson 2003; Eramian et al. 2006; Qiu et al. 2007), and (4) predicting the accuracy of a model in the absence of the native structure (Wallner and Elofsson 2003, 2006; Eramian et al. 2006; McGuffin 2007). Despite the large body of work devoted to the first three tasks, however, relatively little work has been devoted to the last task, predicting the absolute accuracy of computational models. Due to the enormity of the conformational search problem, prediction methods often produce a large number of models and use a score or scores to predict which are most accurate: These approaches determine the relative accuracy of models. However, even if the selection score worked perfectly (i.e., was able to identify the most accurate model from among the many models produced), the user does not necessarily have any sense of the absolute accuracy of the selected model. Although the selected model might be more accurate than the others produced, is it accurate enough? For example, is the best model expected to have a Cα root-mean-square deviation (RMSD) of 2.0 Å, or 9.0 Å? Nearly all traditional assessment scores do not address these questions, often reporting scores in pseudo-energy units or arbitrary values that correlate poorly with accuracy measures such as RMSD. Here, we use the phrase “absolute accuracy” to mean the actual geometrical accuracy, such as RMSD and MaxSub (Siew et al. 2000), which could be calculated if the true native structure were known. In the absence of the native structure, the absolute accuracy is not known and must be predicted.
Predicting the absolute accuracy of a model is particularly difficult due to the lack of principled reasons why an individual assessment score should correlate well with accuracy measures such as RMSD, particularly if the models are not native-like (Fiser et al. 2000). Attempts to predict absolute accuracy have included methods based on neural networks (Wallner and Elofsson 2003), support vector machines (SVMs) (Eramian et al. 2006), and multivariate regression (Tondel 2004). While such approaches can perform well for small families or are able to select the most native-like model in a set of decoys that does not contain the native structure, to our knowledge no approach has demonstrated a clear ability to predict the absolute accuracy of a large, diverse set of models representative of real-world use cases.
Here, we describe a protocol for predicting absolute accuracy by which a model-specific scoring function is developed using SVM regression. For an input comparative model, a unique training set is created from an extremely large database of models of known accuracy (i.e., their native structures are known and their accuracies can thus be calculated). Two predictions are made from this training set for each query structure model: (1) the RMSD of the model and (2) the native overlap (NO3.5Å), where native overlap is defined as the fraction of Cα atoms in a model that are within 3.5 Å of the corresponding atoms in the native structure after rigid body superposition of the model to the native structure (Sanchez et al. 2000). By creating a model-specific tailored training set consisting only of models structurally similar to the assessed model, we gain the ability to predict RMSD and NO3.5Å with a high correlation to the actual RMSD and NO3.5Å values for a diverse set of 580,317 comparative models (r = 0.84 and 0.86, respectively).
We begin by describing the performance of our score at predicting absolute accuracy (Results). We then discuss the implications and application of our approach for large-scale computational prediction efforts (Discussion). Finally, we describe the test set and testing database, the metrics used to evaluate accuracy, and the process for developing the score (Methods).
Results
Test set properties
An extremely large test set of 580,317 comparative models from 6174 sequences was constructed to test our protocol. The properties of the set mirror those observed in large-scale protein structure prediction efforts (Pieper et al. 2006). Most models (461,202 models; 80%) were from alignments in which the sequence identity between the target and template was under 30%, and 94% (541,238 models) had less than 40% sequence identity (Fig. 1A). The median length of the input sequences was 181 residues, and the median model size was 111 residues (Fig. 1B). Though the median sequence length was longer than the ∼156-residue average size of protein domains found in the PDB (Berman et al. 2000; Shen et al. 2005), 78% of the models (455,347) were smaller than this size, reflecting that local, rather than global, alignments were used for modeling (Methods).
Figure 1.
Properties of the 580,317 model testing set (A–D). The y-axis on the left indicates the number of models that fall into the corresponding bin indicated by the x-axis. The line and right y-axis correspond to the cumulative percentage of total models having the appropriate feature. (A) The global sequence identity shared between target/template alignments of the test set. Approximately 80% of the models are from alignments in which the target and template share less than 30% sequence identity. (B) The length distribution of models in the test set (median = 111 amino acids). (C) The Cα RMSD distribution of the models, with a bin size equal to 2.0 Å (median = 7.0 Å). (D) The native overlap distribution, calculated using a cutoff of 3.5 Å (median = 0.46).
The accuracy distribution of the models was broad (Fig. 1C,D). The median RMSD value of the set was 7.0 Å, and the median NO3.5Å value was 0.46. Only 6% (36,063 models) had RMSD values <2.0 Å, a low number resulting from the filtering performed prior to construction of the test set, as well as the inability of the comparative modeling protocol to consistently produce models more native-like than the template structure.
Correlations between actual model accuracy and assessment scores
Correlation coefficients were calculated between the nine input features and the three geometric accuracy metrics (RMSD, NO3.5Å, and MaxSub). The accuracy of models varied widely as sequence identity decreased (Fig. 2A), making sequence identity a relatively uninformative measure for estimating model accuracy. The Pearson correlation coefficient (r) between sequence identity and native overlap was only 0.54 (Table 1).
Figure 2.
The relationships between the actual NO3.5Å and sequence identity (A; r = 0.54); the normalized DOPE score (B; r = 0.71); and the predicted native overlap (C; r = 0.86). In each plot, the diameter of a bubble represents the number of examples contained in the 2D bin indicated by the x- and y-axes. The bubble size is comparable between the different plots. Additionally, the median value for each bin is depicted by the solid line, where the upper and lower error bars indicate the third and first quartile values, respectively. (D) The relationship between the predicted and actual RMSD (r = 0.84).
Table 1.
The correlation coefficients (r) between the actual model accuracy and assessment scores on the full 580,317 model testing set
Of the nine features used for SVM training, N-DOPE had the highest correlation coefficient with RMSD, NO3.5Å, and MaxSub (Table 1). N-DOPE was particularly well suited for identifying near-native (N-DOPE scores below −1.5), or inaccurate (scores above 1.0) models. However, a majority of models (80%) had N-DOPE scores between −1.5 and 1.0, where N-DOPE was not strongly correlated with NO3.5Å (Fig. 2B) or MaxSub (Table 1). For example, the first and third quartile NO3.5Å values for models with N-DOPE values of ∼0.0 were 0.15 and 0.64, respectively, giving a wide range around the median NO3.5Å value of 0.43. The correlation coefficient between N-DOPE and NO3.5Å was 0.71.
In contrast, the correlation between the actual and predicted native overlap was 0.86 (Fig. 2C). Furthermore, the median absolute difference between the actual and predicted NO3.5Å values was only 0.07, with first and third quartile values of 0.03 and 0.16, respectively. The split between predictions that were higher and lower than the actual values was 56% and 44%, respectively. The correlation between actual and predicted RMSD was 0.84, displaying great linearity even out to high RMSD values (Fig. 2D). The median absolute difference between the actual and predicted RMSD values was 1.3 Å for all 580,317 models. Considering only those models below 5.0 Å RMSD, the median absolute difference between the actual and predicted values was only 0.71 Å. RMSD predictions were also closely split between those that were higher (48%) and lower (52%) than the actual values.
ProQ and ModFOLD were used to compare the performance of the model-specific scoring approach to approaches that do not benefit from learning from a specific training set. The correlation between the actual and predicted MaxSub scores was highest for ProQ-SS, at 0.72 (Table 1), significantly less accurate than the correlation between the predicted NO3.5 Å and MaxSub of 0.83 given by our protocol. Thus, even though our protocol was designed to predict NO3.5Å and not MaxSub, the resulting predictions were much better correlated with the actual MaxSub scores than the ProQ predictions that were designed specifically for this task.
ModFOLD was run on 36,453 randomly selected models for 225 sequences from our test set. The correlation coefficient between the ModFOLD score and RMSD for these 36,453 models was 0.51, and the correlation coefficient between NO3.5Å and the ModFOLD score was 0.63 (see, Table 3). In comparison, TSVMod's correlations were 0.85 and 0.88, respectively, which is essentially identical to the values obtained for our full test set.
Table 3.
The correlation coefficients (r) between the actual model accuracy and assessment scores on a 36,453 model testing set
Fold assessment
Fold assessment is a particularly important problem at lower values of sequence identity, when it is possible that the template used to construct a model does not have the same fold. As expected, the sequence identity of the target/template pair used to construct the model was only marginally useful for assessing whether the model had the correct fold. The GA341 and N-DOPE scores, both developed for fold assessment, were better at classifying models correctly. By use of 0.30 as the NO3.5Å threshold for defining whether or not a model has the correct fold, the calculated areas under the ROC curve (Methods) for sequence identity, GA341, N-DOPE, and the predicted NO3.5Å were 0.80, 0.86, 0.87, and 0.93, respectively (Fig. 3). With a NO3.5Å threshold of 0.50, these values were 0.81, 0.84, 0.86, and 0.93, respectively (data not shown). Thus, using the predicted NO3.5Å value to classify whether or not a model has the correct fold was significantly more accurate than the other fold assessment scores tested.
Figure 3.
Receiver operating characteristic (ROC) curves for fourfold assessment classifiers: the predicted native overlap (solid black line); the normalized DOPE score (dashed black line); the GA341 score (solid gray line); and the sequence identity shared between the target and the template (dashed gray line). For each measure, the area under the curve is noted. A model was defined as having the correct fold if NO3.5Å ≥ 0.30.
Residue neighborhood accuracy
Two structure-derived properties, the solvent exposure state and the residue neighborhood (Chakravarty and Sanchez 2004), were calculated for 25,000 models of 100–200 residues randomly selected from the test set. The accuracy of a residue's neighborhood was calculated by comparing the contacts made by a residue with its neighbors in the model, versus those made by that residue in the native structure, thereby measuring the percentage of contacts that are accurately modeled. There was a clear decrease in the median neighborhood accuracy (Fig. 4A) for models constructed from target/template pairs sharing less than 40% sequence identity (96% of the 25,000 models), with an overall correlation of r = 0.57. In contrast, the neighborhood accuracy was more correlated with the predicted native overlap value (r = 0.82) (Fig. 4B), with much tighter first and third quartile error bars.
Figure 4.
Relationship between structure-derived properties and the predicted accuracy for 25,000 randomly selected models of length 100–200 amino acids. (A) The percentage of correct neighborhood (solid line) is plotted versus the sequence identity shared between the target and the template used to construct the model (r = 0.57). The solid line indicates the median value for the bin; the upper and lower error bars indicate the third and first quartile values for the bin, respectively. The columns indicate the fraction of examples that are contained in each bin (right y-axis). (B) Relationship between the predicted native overlap and the neighborhood accuracy of a model (r = 0.82). (C) The percentage of exposed residues correctly modeled as exposed versus sequence identity (r = 0.56). (D) Percentage of exposed residues correctly modeled as exposed versus predicted NO3.5Å (r = 0.65).
Residue exposure state
The second assessed structure-derived property was the exposure state of a residue. A residue was defined as exposed if it had relative surface accessibility larger than 40%, using the method of Lee and Richards (1971) calculated by Naccess v2.1.1 (Hubbard et al. 1991). Exposure state accuracy was observed to be higher than neighborhood accuracy; with less decrease in accuracy as sequence identity fell below 40% (Fig. 4C). The overall correlation between the sequence identity and exposure state accuracy was 0.56, well below that between the predicted native overlap and correct exposure state (r = 0.65) (Fig. 4D).
Discussion
A limitation of comparative models is that their accuracy cannot be readily and robustly assessed. We have addressed this problem by developing a protocol for deriving SVM regression models optimized specifically for predicting the actual RMSD and NO3.5Å values of a model in the absence of its native structure. SVM regression was used to combine up to nine features (sequence identity, N-DOPE, Z-PAIR, Z-SURFACE, Z-COMBINED, percentage of gaps in the target/template alignment, GA341, and two PSIPRED/DSSP scores) extracted from a tailored training set unique for the query structure model being assessed. This protocol is able to predict the RMSD and NO3.5Å values for a large, diverse set of comparative models with correlation coefficients of 0.84 and 0.86, respectively, to the actual RMSD and NO3.5Å values (Table 1).
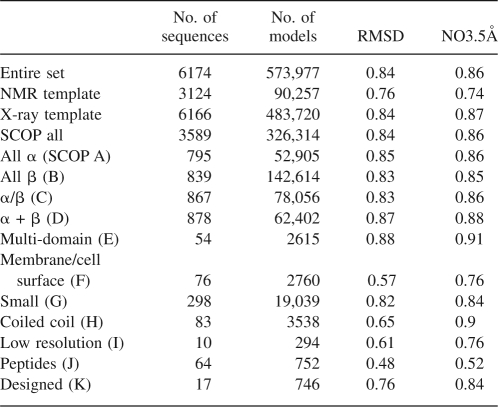
The test set used for this study consisted of 580,317 models, for 6174 sequences. This set is approximately an order of magnitude larger, and contains one to two orders of magnitude more sequences, than typical model assessment test sets (Samudrala and Levitt 2000; Tsai et al. 2003; Wallner and Elofsson 2003; Eramian et al. 2006). The properties of this set parallel those seen in large-scale comparative modeling, and the models span virtually all SCOP (Andreeva et al. 2004) fold types (Table 2), protein sizes (Fig. 1B), and accuracies (Fig. 1C,D). There are two features of this test set that reflect the difference between our goal of predicting the absolute accuracy of comparative models and traditional model assessment tests. First, the set contains no native structures, only models. A common relative accuracy test is that the native structure scores lower than all other models (Gatchell et al. 2000). While it is certainly a necessary condition that the native state is separable from decoys, it is far from sufficient, particularly in real use cases, where the best model produced is often far from native. Second, only one model, rather than many, is built per alignment, because the goal was not to determine the ability of assessment scores to identify the best model from among sets of similar models. Such relative accuracy assessments are important because they more closely replicate the real-world conditions in which assessment scores are used. However, such tests overlook that even if the model assessment scores are able to both correctly identify the native structure from among a set of decoys and identify the most accurate model from among a set of similar models, an end-user still has little information about how accurate the model actually is.
Table 2.
The correlation coefficients (r) between the actual model accuracy and assessment scores for different SCOP fold classes
The prediction of absolute accuracy is a difficult problem that has not been given great attention. It has been argued that there are no principled reasons why an individual assessment score should correlate with an accuracy metric, particularly if the model is not native-like (Fiser et al. 2000). Our own data support this contention, as all of the individual statistical potentials tested were relatively ill-suited for predicting absolute accuracy (Table 1). The DOPE (Discrete Optimized Protein Energy) score, for example, has been shown to be an extremely accurate model assessment score in a number of studies (Colubri et al. 2006; Eramian et al. 2006; Shen and Sali 2006; Fitzgerald et al. 2007; Marko et al. 2007; Lu et al. 2008), yet correlates poorly with accuracy measures such as RMSD and NO3.5Å when tested on our large test set (Fig. 2B). Attempts have been made to predict absolute accuracy by combining a number of assessment scores (Wallner and Elofsson 2003; Eramian et al. 2006; McGuffin 2007). Even in these studies, however, the reported correlation coefficients between the predicted and actual accuracy measures was low, ranging from 0.35 to 0.71; moreover, these results were obtained on much smaller and less diverse test sets than the set employed here. For example, when we tested the ProQ method on the 580,317 models of our test set, the correlation between the actual and predicted MaxSub was only 0.72 (Table 1). Not only was ProQ's correlation with MaxSub slightly lower than that of N-DOPE (r = 0.73), but it was far lower than the correlation between the predicted NO3.5Å and actual MaxSub obtained by the model-specific approach (r = 0.83), even though the MaxSub score was not predicted by the model-specific protocol. Had MaxSub been predicted in place of NO3.5Å, the performance gap between ProQ and the model-specific approach could only be larger. Similarly, the correlation between the accuracy measures and the ModFOLD scores were far lower than those between the actual and predicted values from TSVMod (Table 3).
These results illustrate the utility of constructing a scoring function specific for the input model. A unique feature of our approach is the optimization of the weights of the individual scores specifically for the fold and size of the model being assessed, rather than for a variety of proteins of many shapes and sizes. The difference between our approach and other composite scores is analogous to the difference between position-specific scoring matrices (PSSMs) and generalized substitution matrices (e.g., BLOSUM62) employed in alignment algorithms. The use of a tailored training set for optimizing the weights of input features is crucial, as different assessment scores perform better for different sizes and shapes of proteins, and their contributions to the overall composite score need to be adjusted accordingly. Our results show the SVM algorithm can find appropriate weights for the features: First, optimally combining the features results in scores that correlate well with RMSD and NO3.5Å (Table 1), although the overall correlation coefficients of each of the input features is low; second, there is a linear relationship between the actual and predicted accuracy (Fig. 2C,D). Most importantly, not only do the predictions correlate well with the actual accuracy of the models, but also the difference between the actual and predicted values is small (Fig. 2C,D).
The primary advantage of our approach relative to most model assessment scores is that the prediction of absolute accuracy gives users confidence in the use of models for their experiments. For comparative models, the sequence identity shared between the target sequence and template structure has historically been used to estimate the accuracy of models, as it is easy to calculate and appreciate. Sequence identity, however, is a relatively poor predictor of model accuracy, especially below 40% (Figs. 2A, 3; Table 1), and actually adds little to the performance of TSVMod: Omitting sequence identity as a feature does not change the correlation coefficient between the actual and predicted RMSD, while the correlation coefficient between the actual and predicted NO3.5Å is reduced to 0.85 (from 0.86). Given these limitations of sequence identity, many researchers have been reluctant to use comparative models based on less than 30% sequence identity in their research for fear of errors. The reason is that the accuracy of such models can vary widely (Figs. 2A, 3; Table 1) and there has been no practical way to robustly and reliably predict the absolute accuracy of these models. As a result, the utility of comparative modeling is significantly reduced, because 76% of all comparative models are based on less than 30% sequence identity (Pieper et al. 2006). Our model-specific approach would result in a dramatic increase in the number of comparative models correctly assessed as useful by helping identify those that are accurate (Fig. 2A,B) and filtering incorrect models from consideration (Fig. 3). Of the 580,317 models in the test set, 485,066 models (84%) were from alignments with less than 30% sequence identity; 173,139 of these models had actual RMSD values below 5.0Å, and were predicted as such (36% of the 580,317 total models). Figure 5 shows two of the many examples where target/template alignments shared under 12.5% sequence identity, but the models produced were accurate and assessed as such by the predicted RMSD and NO3.5Å scores.
Figure 5.
Examples of successful accuracy predictions where the sequence identity shared between the target and template was less than 12.5%, and yet very accurate models were constructed. Relying upon the individual features alone, neither model would be assessed as being very accurate, yet the weighed combination of these features using the model-specific assessment protocol leads to accurate assessments. In both images, the native structure is colored red and the model is blue. (A) Sequence from murine neuroglobin (PDB code 1q1fA) modeled using 1it2A as a template. (B) Sequence of 4-hydroxybenzoyl CoA thioesterase (1q4tA) modeled using 1s5uA as a template.
Similarly, our protocol is able to identify which models are suitable for many common experiments. Relative to purely geometric metrics like RMSD and NO3.5Å, the two structure-derived properties calculated here—the neighborhood residue accuracy and the percentage of residues correctly modeled as exposed—are informative about the utility of a model for specific tasks such as guiding mutagenesis experiments, biochemical labeling, annotation of point mutations, protein design, predicting subcellular localization, in silico ligand docking, and prediction of protein complex structures. Using the predicted accuracy of the models to estimate the accuracy of these structure-derived properties results in more precise and accurate estimates than relying upon sequence identity (Fig. 4), as has historically been done.
There are, however, a few limitations to our model assessment protocol. First, the accuracy of the protocol could be increased if we were willing to sacrifice coverage (i.e., not be able to predict the accuracy for all models). Given the current thresholds, we can predict the accuracy for 83% of the test set using tailored training sets populated by models of the same fold as the model being assessed (Methods) (Fig. 6). If we increased the minimum training set size threshold from five to 275 and did not utilize the secondary structure filtering step, predictions could be made for only 20% of the test set, but the RMSD and NO3.5Å correlation coefficients would increase from 0.84 and 0.86 to 0.90 and 0.92, respectively. A second limitation is that errors in the underlying scores can affect the accuracy of the prediction. Improvements of these scores, or the addition of other scores, will further increase the accuracy of the method.
Figure 6.
Flowchart depicting the steps to predict the RMSD and NO3.5Å of an input comparative model.
There are many applications for our model assessment protocol. First, the protocol is being incorporated into our comprehensive database of comparative models, MODBASE, to increase the confidence that end-users have in using such models for their experiments (Fig. 4B,D). Second, the predicted NO3.5Å value will be used as a filter to ensure that only models assessed to have the correct fold are deposited in MODBASE (Fig. 3). Third, we suggest that the model-specific protocol may also be a good scoring function for the refinement of comparative models (D. Eramian and A. Sali, in prep.) because it displays linearity between the actual and predicted accuracy even for models that are not native-like (Fig. 2C,D). In contrast, a refinement scheme relying upon a score such as DOPE would have a more difficult time, only being able to identify the unlikely event of sampling a very near-native solution (Fig. 2B). Furthermore, any refinement scheme built upon the model-specific scoring protocol would have the benefit of giving the user an estimate of the actual accuracy of the model. Fourth, we intend to develop a version of TSVMod using different feature types that will predict per-residue accuracy, in the spirit of similar approaches such as ModFOLDclust (McGuffin 2008), Prosa (Sippl 1993), FragQA (Gao et al. 2007), and ProQres (Wallner and Elofsson 2006).
Finally, though we have principally described our protocol in the context of evaluating comparative models, we believe the construction of a tailored training set by model size and secondary structure content will ultimately be applicable to models generated by any method, including de novo predictions. Using this filtering step on the test set and using only alignment-independent scores (N-DOPE, Z-PAIR, Z-SURFACE, Z-COMBINED, and two PSIPRED/DSSP scores) as input features, we can currently predict the RMSD and NO3.5Å errors with correlation coefficients (r) of 0.80 and 0.81, respectively, to the actual errors. These numbers are lower than those of the “standard” TSVMod because: (1) The lack of a clearly defined template precludes the use of the more accurate “left” branch of tailored training set construction (Fig. 6), resulting in a suboptimal tailored training set; and (2) several informative alignment-based features (GA341, percentage of gapped residues in the alignment, sequence identity) cannot be used. In addition, an application of TSVMod to models calculated by programs other than MODELLER may also suffer from the optimization of the TSVMod features specifically for MODELLER models (e.g., DOPE can be fooled by decoys that are packed extremely tightly, as one would expect in energy-minimized models). In spite of these three significant limitations, we decided to test how well the reduced TSVMod performs relative to model quality assessment (MQAP) programs tested at CASP. The accuracy values (RMSD and native overlap) were calculated using MODELLER's superpose command for 30,186 of the CASP7 models; the remaining 10,120 coordinate files could not be read by MODELLER or the programs used to calculate features for TSVMod. TSVMod had correlations with RMSD and NO3.5Å of 0.62 and 0.73, respectively, and the global Spearman rank correlation coefficient between the actual and predicted NO3.5Å values was 0.75. Though the Pearson correlation coefficient values are below those for the “full” TSVMod on our benchmark of 0.84 and 0.86, respectively, the Spearman rank correlation coefficient is comparable to the reported values for MQAPs on the CASP7 set (McGuffin 2007), despite the fact that TSVMod was not designed for assessing CASP models. Improvement of TSVMod's performance at assessing such models would be expected if additional scores were included as features, and if the TSVMod training database was populated with models produced by the method being assessed.
In summary, we have developed a model-specific scoring protocol to predict the absolute accuracy of comparative models by optimizing the contributions of up to nine features via SVM regression. This approach has been shown to be able to predict the RMSD with a correlation to the actual RMSD of 0.84; predict the NO3.5Å with a correlation to the actual NO3.5Å of 0.86; differentiate between correct and incorrect models better than existing methods; identify models with accurate structure-derived properties better than relying upon sequence identity; and outperform the ProQ assessment score in predicting MaxSub of a model, even though our approach was not developed to predict MaxSub.
Methods
Construction of test set and training database
To create the list of target sequences, all chains in the PDB (25 April 2007 PDB release) were clustered at 40% sequence identity, resulting in 10,191 unique chains: 3926 PDB files containing chain breaks, defined as sequentially adjacent Cα atoms separated by at least 4.0 Å, were removed because chain breaks are difficult to robustly model in an automated fashion. The resulting list contained 6265 unique sequences. The template profile database was constructed by clustering the PDB at 95% sequence identity, giving 15,631 template structures (24 Feb 2007 PDB release), with each template profile built using MODELLER's profile.build command against the UniProt-90 database (Apweiler et al. 2004).
Protein structure models were calculated using MODPIPE, our automated software pipeline for large-scale protein structure modeling (Eswar et al. 2003). MODPIPE relies on MODELLER (Sali and Blundell 1993) for its functionality and calculates comparative models for large numbers of sequences using different template structures and sequence-structure alignments. Sequence-structure matches are established using a variety of fold-assignment methods, including sequence-sequence (Smith and Waterman 1981), profile-sequence (Altschul et al. 1997), and profile-profile alignments (Marti-Renom et al. 2004). Increased sensitivity of the search for known template structures is achieved by using an E-value threshold of 1.0. The main feature of the pipeline is that the validity of sequence-structure relationships is not prejudged at the fold assignment stage but rather is assessed after the construction of the model by using several model quality criteria, including the coverage of the model, sequence identity of the sequence-structure alignment, the fraction of gaps in the alignment, the compactness of the model, and statistical energy Z-scores (Melo et al. 2002; Eramian et al. 2006; Shen and Sali 2006). Using this procedure, a total of 580,317 unique target/template alignments and 5,790,889 models were produced.
The test set was constructed by taking the first model produced from each of the 580,317 unique target/template alignments. The training database consisted of all 5,790,889 models. All models in the test set are also in the training database; this redundancy is accounted for during testing so the accuracy of the method is not overestimated. The model files, alignments, and the accompanying TSVMod predictions and individual feature scores are all available for download by anonymous ftp at http://salilab.org/decoys/.
Model accuracy measures
Three geometric accuracy measures were used: the Cα RMSD value between the model and the native structure after superposition, the fraction of Cα atoms within 3.5 Å of their correct positions in the native structure (the native overlap at 3.5 Å or NO3.5Å), and the MaxSub score (Siew et al. 2000). The RMSD and NO3.5Å accuracy for each model were calculated by MODELLER's superpose command. As NO3.5Å is calculated by dividing the number of Cα atoms within 3.5 Å from their correct positions by the length of the sequence, one must choose an appropriate denominator. We chose to use the number of residues actually modeled, not the length of the input sequence, making our NO3.5Å measure a local accuracy measure. MaxSub was obtained from the Fischer laboratory and run with default parameters (Siew et al. 2000), with no correction made for the length of the input target sequence.
Model assessment scores
MODPIPE produces a number of alignment-based and model-based assessment scores that can be used to analyze the quality of the models. For each target/template alignment, MODPIPE calculated the sequence identity and the percentage of unaligned (gapped) positions. MODPIPE also calculated five model-based assessment scores: a Cα- and Cβ-based distance-dependent statistical potential score (PAIR) (Melo et al. 2002), a Cβ-based accessible surface statistical potential score (SURFACE) (Melo et al. 2002), a combined distance and surface potential score (COMBINED) (Melo et al. 2002), a fold assessment composite score derived by a genetic algorithm (GA341) (Melo and Sali 2007), and an atomic distance-dependent statistical potential score (Discrete Optimized Protein Energy or DOPE) (Shen and Sali 2006). Next, we outline each of these scores.
For each of the PAIR, SURFACE, and COMBINED scores, a Z-score is calculated using the mean and standard deviation of the statistical potential score of 200 random sequences with the same amino acid residue type composition and structure as the model. These three scores were developed and implemented as described elsewhere (Melo et al. 2002; Eswar et al. 2003).
The GA341 score was designed to discriminate between models with the correct and incorrect fold. GA341 is a nonlinear combination of the percentage sequence identity of the alignment used to build the model, the model compactness, and the Z-score for the COMBINED statistical potential.
The DOPE score is an atomic distance-dependent statistical potential based on a physical reference state that accounts for the finite size and spherical shape of proteins by assuming a protein chain consists of noninteracting atoms in a uniform sphere of radius equivalent to that of the corresponding protein. The normalized version (N-DOPE) was used instead of the raw score; it is a standard score (Z-score) derived from the statistics of raw DOPE scores. The mean and standard deviation of the DOPE score of a given protein is estimated from its sequence. The mean score of a random protein conformation is estimated by a weighted sum of protein composition over the 20 standard amino acid residue types, where each weight corresponds to the expected change in the score by inserting a specific type of amino acid residue. The weights are estimated from a separate training set of 1,686,320 models generated by MODPIPE.
Two PSIPRED (Jones 1999) and DSSP (Kabsch and Sander 1983) agreement scores were also calculated: the percentage of amino acid residues that had different Q3 states for both the model and the target sequence (PSIPREDPRCT), and a weighted score that takes into account the PSIPRED prediction confidence (PSIPREDWEIGHT). These scores were implemented as described elsewhere (Eramian et al. 2006).
Flowchart for predicting RMSD and NO3.5Å
A flowchart outlining the steps taken to create a tailored training set and make SVM predictions is presented in Figure 6. Once a model is built, ∼20 sec of CPU are needed to calculate the nine individual assessment criteria, followed by an additional 10 sec for the filtering and SVM stages; additional time is required if PSIPRED predictions have not been precalculated.
The first step is to determine whether or not the aligned target and template sequences share more than 85% sequence identity (Fig. 6). If so, the RMSD and native overlap are predicted to be 0.5 Å and 1.0, respectively, and no further steps are taken because nearly all comparative models built on templates sharing such high sequence identity are native-like; only 0.9% of the test models surpass this threshold. The second step is to store the PDB identification code as well as starting and ending residue indices of the template used to produce each model.
Next, in the filtering step, the 5,790,889 model training database is first scanned to find all examples where the same region of the template was used either as a template or was itself the target sequence, modeled using a different template. A region is considered equivalent if the starting and ending points are each within 10 residues of the modeled region and its overall length is within 10% of the length of the model. If an entry in the training database used a chain from the same PDB file as the query target sequence, the entry was omitted to ensure that the tailored training set does not result in overestimating the accuracy of the method. Next, potential matches are filtered by the statistical potential scores and are included in the tailored training set only if the Z-PAIR, Z-SURFACE, and Z-COMBINED scores are each within 2 units of the score for the input model, and the N-DOPE score is within 0.5 units of that of the model.
If the tailored training set contained fewer than five examples, a separate filtering procedure is employed to populate the tailored training set; this occurred for 17% (99,947) of the test set. The secondary structure content of the input model is calculated using MODELLER's model.write_data command. The training database was then scanned to find all entries whose size is within 10% of the length of the model, and the helical and strand content are each within x ± 10%, where x are the values for the model. Entries from the same PDB file as the query target sequence are again omitted. Potential matches are then filtered by the statistical potential scores as described.
Finally, two SVMs are trained to predict the RMSD and NO3.5Å of the model. The SVMlight software package was used in regression mode, with a linear kernel, for all SVM training (Joachims 1999). The nine training features used are the nine aforementioned assessment scores. Tested values of the epsilon width of tube for regression training for RMSD varied from 0.01 to 0.2, and values attempted for NO3.5Å ranged from 0.01to 0.1. The final values selected for the epsilon width of tube for regression for the RMSD and NO3.5Å predictions were 0.1 and 0.05, respectively. All other SVMlight parameters were kept at their default values.
Fold assessment
The ability of individual scores to differentiate between correct and incorrect folds was assessed using receiver operating characteristic (ROC) plots (Albeck and Borgesen 1990; Metz et al. 1998), calculated with MODPIPE's ROC module. This module plots the true positive rate of classification against the false positive rate on the x-axis. A model was defined as having the correct fold if its NO3.5Å value exceeded a threshold; the two thresholds used were 0.30 and 0.50. If the model is correct, the prediction is a true positive (TP) if it is classified as correct, and a true negative (FN) if it is classified as incorrect. If instead the model is incorrect, the prediction is a true negative (TN) if the model is classified as incorrect, and a false positive (FP) if it is classified as correct. The true positive and false positive rates displayed on the ROC plot are calculated by tp = TP/P and fp = FP/P, where TP is the count of true positives, P is the sum of true positives and false negatives, FP is the count of false positives, and N is the sum of true negatives and false positives.
Comparison to other MQAP programs
To compare the model-specific approach to another approach for assessing absolute accuracy, the stand-alone version of ProQ v1.2 (Wallner and Elofsson 2003) was run for all models of the test set. ProQ is a neural network that predicts the LGScore and MaxSub of an input model, using a general, rather than a model-specific, training set. ProQ was run both with (ProQ-SS) and without (ProQ) PsiPred v2.5 secondary structure predictions (Jones 1999). We also ran the residue-based score ProQres v1.0 (Wallner and Elofsson 2006) for all models of the test set. ProQres predicts the accuracy for each residue of an input model. To obtain a single score for an input model, the ProQres scores for each residue were summed and divided by the number of residues in the model.
ModFOLD (McGuffin 2007) is a neural network that combines data from ModSSEA (Pettitt et al. 2005), MODCHECK (McGuffin and Jones 2003), and ProQ to predict the accuracy of an input model. ModFOLD was trained using TM-scores (Zhang and Skolnick 2004) and is available as a web server. The ModFOLD server (McGuffin 2008) allows a user to upload .tar.gz files of up to 1000 models; the user must also upload the sequence of the model(s) being assessed. Because of this manual task and the high computational demands our 580,317 model set would place on the ModFOLD hardware, we instead tested the ModFOLD server (v1.1) by 36,453 randomly selected models for 225 sequences from our test.
Acknowledgments
We acknowledge funds from Sandler Family Supporting Foundation, U.S. National Institutes of Health (Grants R01-GM54762, R01-GM083960, U54-RR022220, U54-GM074945, P01-GM71790, U54-GM074929), U.S. National Science Foundation (Grant IIS-0705196), as well as Hewlett-Packard, Sun Microsystems, IBM, NetApp Inc., and Intel Corporation for hardware gifts.
Footnotes
Reprint requests to: Andrej Sali, University of California at San Francisco, Byers Hall, Suite 503B, 1700 4th Street, San Francisco, CA 94158, USA; e-mail: sali@salilab.org; fax: (415) 514-4231.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.036061.108.
References
- Albeck, M.J., Borgesen, S.E. ROC-curve analysis. A statistical method for the evaluation of diagnostic tests. Ugeskr. Laeger. 1990;152:1650–1653. [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva, A., Howorth, D., Brenner, S.E., Hubbard, T.J., Chothia, C., Murzin, A.G. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Bairoch, A., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D., Sali, A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelic, S., Aqvist, J. Computational prediction of structure, substrate binding mode, mechanism, and rate for a malaria protease with a novel type of active site. Biochemistry. 2004;43:14521–14528. doi: 10.1021/bi048252q. [DOI] [PubMed] [Google Scholar]
- Bradley, P., Misura, K.M., Baker, D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- Caffrey, C.R., Placha, L., Barinka, C., Hradilek, M., Dostal, J., Sajid, M., McKerrow, J.H., Majer, P., Konvalinka, J., Vondrasek, J. Homology modeling and SAR analysis of Schistosoma japonicum cathepsin D (SjCD) with statin inhibitors identify a unique active site steric barrier with potential for the design of specific inhibitors. Biol. Chem. 2005;386:339–349. doi: 10.1515/BC.2005.041. [DOI] [PubMed] [Google Scholar]
- Chakravarty, S., Sanchez, R. Systematic analysis of added-value in simple comparative models of protein structure. Structure. 2004;12:1461–1470. doi: 10.1016/j.str.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Chmiel, A.A., Radlinska, M., Pawlak, S.D., Krowarsch, D., Bujnicki, J.M., Skowronek, K.J. A theoretical model of restriction endonuclease NlaIV in complex with DNA, predicted by fold recognition and validated by site-directed mutagenesis and circular dichroism spectroscopy. Protein Eng. Des. Sel. 2005;18:181–189. doi: 10.1093/protein/gzi019. [DOI] [PubMed] [Google Scholar]
- Chothia, C., Lesk, A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colubri, A., Jha, A.K., Shen, M.Y., Sali, A., Berry, R.S., Sosnick, T.R., Freed, K.F. Minimalist representations and the importance of nearest neighbor effects in protein folding simulations. J. Mol. Biol. 2006;363:835–857. doi: 10.1016/j.jmb.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Costache, A.D., Pullela, P.K., Kasha, P., Tomasiewicz, H., Sem, D.S. Homology-modeled ligand-binding domains of zebra fish estrogen receptors α, β1, and β2: From in silico to in vivo studies of estrogen interactions in Danio rerio as a model system. Mol. Endocrinol. 2005;19:2979–2990. doi: 10.1210/me.2004-0435. [DOI] [PubMed] [Google Scholar]
- Domingues, F.S., Koppensteiner, W.A., Jaritz, M., Prlic, A., Weichenberger, C., Wiederstein, M., Floeckner, H., Lackner, P., Sippl, M.J.1999Sustained performance of knowledge-based potentials in fold recognition Proteins Suppl 3: 112–120. [PubMed] [Google Scholar]
- Eramian, D., Shen, M.Y., Devos, D., Melo, F., Sali, A., Marti-Renom, M.A. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar, N., John, B., Mirkovic, N., Fiser, A., Ilyin, V.A., Pieper, U., Stuart, A.C., Marti-Renom, M.A., Madhusudhan, M.S., Yerkovich, B., et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar, N., Webb, B.M., Marti-Renom, M., Madhusudhan, M.S., Eramian, D., Shen, M.Y., Pieper, U., Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007;Chapter 2:Unit 2.9. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- Fiser, A., Do, R.K., Sali, A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, J.E., Jha, A.K., Colubri, A., Sosnick, T.R., Freed, K.F. Reduced Cβ statistical potentials can outperform all-atom potentials in decoy identification. Protein Sci. 2007;16:2123–2139. doi: 10.1110/ps.072939707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X., Bu, D., Li, S.C., Xu, J., Li, M. FragQA: Predicting local fragment quality of a sequence-structure alignment. Genome Inform. 2007;19:27–39. [PubMed] [Google Scholar]
- Gatchell, D.W., Dennis, S., Vajda, S. Discrimination of near-native protein structures from misfolded models by empirical free energy functions. Proteins. 2000;41:518–534. [PubMed] [Google Scholar]
- Ginalski, K., Grishin, N.V., Godzik, A., Rychlewski, L. Practical lessons from protein structure prediction. Nucleic Acids Res. 2005;33:1874–1891. doi: 10.1093/nar/gki327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, S.J., Campbell, S.F., Thornton, J.M. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- Jaroszewski, L., Li, W., Godzik, A. In search for more accurate alignments in the twilight zone. Protein Sci. 2002;11:1702–1713. doi: 10.1110/ps.4820102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims, T. Making large-scale SVM learning practical. In: Schölkopf B., et al., editors. Advances in kernel methods: Support vector learning. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Lazaridis, T., Karplus, M. Discrimination of the native from misfolded protein models with an energy function including implicit solvation. J. Mol. Biol. 1999;288:477–487. doi: 10.1006/jmbi.1999.2685. [DOI] [PubMed] [Google Scholar]
- Lazaridis, T., Karplus, M. Effective energy functions for protein structure prediction. Curr. Opin. Struct. Biol. 2000;10:139–145. doi: 10.1016/s0959-440x(00)00063-4. [DOI] [PubMed] [Google Scholar]
- Lee, B., Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lu, M., Dousis, A.D., Ma, J. OPUS-PSP: An orientation-dependent statistical all-atom potential derived from side-chain packing. J. Mol. Biol. 2008;376:288–301. doi: 10.1016/j.jmb.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudhan, M.S., Marti-Renom, M.A., Sanchez, R., Sali, A. Variable gap penalty for protein sequence-structure alignment. Protein Eng. Des. Sel. 2006;19:129–133. doi: 10.1093/protein/gzj005. [DOI] [PubMed] [Google Scholar]
- Marko, A.C., Stafford, K., Wymore, T. Stochastic pairwise alignments and scoring methods for comparative protein structure modeling. J. Chem. Inf. Model. 2007;47:1263–1270. doi: 10.1021/ci600485s. [DOI] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A.C., Fiser, A., Sanchez, R., Melo, F., Sali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Madhusudhan, M.S., Sali, A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin, L.J. Benchmarking consensus model quality assessment for protein fold recognition. BMC Bioinformatics. 2007;8:345. doi: 10.1186/1471-2105-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin, L.J. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24:586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- McGuffin, L.J., Jones, D.T. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- Melo, F., Feytmans, E. Novel knowledge-based mean force potential at atomic level. J. Mol. Biol. 1997;267:207–222. doi: 10.1006/jmbi.1996.0868. [DOI] [PubMed] [Google Scholar]
- Melo, F., Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- Melo, F., Sali, A. Fold assessment for comparative protein structure modeling. Protein Sci. 2007;16:2412–2426. doi: 10.1110/ps.072895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, F., Sanchez, R., Sali, A. Statistical potentials for fold assessment. Protein Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz, C.E., Herman, B.A., Roe, C.A. Statistical comparison of two ROC-curve estimates obtained from partially paired datasets. Med. Decis. Making. 1998;18:110–121. doi: 10.1177/0272989X9801800118. [DOI] [PubMed] [Google Scholar]
- Miyazawa, S., Jernigan, R.L. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- Park, B., Levitt, M. Energy functions that discriminate X-ray and near-native folds from well-constructed decoys. J. Mol. Biol. 1996;258:367–392. doi: 10.1006/jmbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- Park, B.H., Huang, E.S., Levitt, M. Factors affecting the ability of energy functions to discriminate correct from incorrect folds. J. Mol. Biol. 1997;266:831–846. doi: 10.1006/jmbi.1996.0809. [DOI] [PubMed] [Google Scholar]
- Pettitt, C.S., McGuffin, L.J., Jones, D.T. Improving sequence-based fold recognition by using 3D model quality assessment. Bioinformatics. 2005;21:3509–3515. doi: 10.1093/bioinformatics/bti540. [DOI] [PubMed] [Google Scholar]
- Pieper, U., Eswar, N., Davis, F.P., Braberg, H., Madhusudhan, M.S., Rossi, A., Marti-Renom, M., Karchin, R., Webb, B.M., Eramian, D., et al. MODBASE: A database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J., Sheffler, W., Baker, D., Noble, W.S. Ranking predicted protein structures with support vector regression. Proteins. 2007;71:1175–1182. doi: 10.1002/prot.21809. [DOI] [PubMed] [Google Scholar]
- Rai, B.K., Fiser, A. Multiple mapping method: A novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins. 2006;63:644–661. doi: 10.1002/prot.20835. [DOI] [PubMed] [Google Scholar]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Sali, A., Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Samudrala, R., Levitt, M. Decoys ‘R’ Us: A database of incorrect conformations to improve protein structure prediction. Protein Sci. 2000;9:1399–1401. doi: 10.1110/ps.9.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, R., Pieper, U., Melo, F., Eswar, N., Marti-Renom, M.A., Madhusudhan, M.S., Mirkovic, N., Sali, A. Protein structure modeling for structural genomics. Nat. Struct. Biol. 2000;7:986–990. doi: 10.1038/80776. [DOI] [PubMed] [Google Scholar]
- Sauder, J.M., Arthur, J.W., Dunbrack R.L., Jr Large-scale comparison of protein sequence alignment algorithms with structure alignments. Proteins. 2000;40:6–22. doi: 10.1002/(sici)1097-0134(20000701)40:1<6::aid-prot30>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Seok, C., Rosen, J.B., Chodera, J.D., Dill, K.A. MOPED: Method for optimizing physical energy parameters using decoys. J. Comput. Chem. 2003;24:89–97. doi: 10.1002/jcc.10124. [DOI] [PubMed] [Google Scholar]
- Shen, M.Y., Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M.Y., Davis, F.P., Sali, A. The optimal size of a globular protein domain: A simple sphere-packing model. Chem. Phys. Lett. 2005;405:224–228. [Google Scholar]
- Shortle, D., Simons, K.T., Baker, D. Clustering of low-energy conformations near the native structures of small proteins. Proc. Natl. Acad. Sci. 1998;95:11158–11162. doi: 10.1073/pnas.95.19.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siew, N., Elofsson, A., Rychlewski, L., Fischer, D. MaxSub: An automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16:776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., Waterman, M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Tondel, K. Prediction of homology model quality with multivariate regression. J. Chem. Inf. Comput. Sci. 2004;44:1540–1551. doi: 10.1021/ci049924m. [DOI] [PubMed] [Google Scholar]
- Tramontano, A., Leplae, R., Morea, V. Analysis and assessment of comparative modeling predictions in CASP4. Proteins. 2001;(Suppl 5):22–38. doi: 10.1002/prot.10015. [DOI] [PubMed] [Google Scholar]
- Tsai, J., Bonneau, R., Morozov, A.V., Kuhlman, B., Rohl, C.A., Baker, D. An improved protein decoy set for testing energy functions for protein structure prediction. Proteins. 2003;53:76–87. doi: 10.1002/prot.10454. [DOI] [PubMed] [Google Scholar]
- Wallner, B., Elofsson, A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, B., Elofsson, A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2006;15:900–913. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Yuan, X., Xiang, Z., Mimnaugh, E., Marcu, M., Neckers, L. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat. Struct. Mol. Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- Zhou, H., Zhou, Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]