Abstract
Hepatocellular carcinomas (HCC) represent the third-leading cause of cancer-related deaths worldwide. The vast majority of cases arise in the context of chronic liver injury due to hepatitis B virus or hepatitis C virus infection. In order to identify genetic mechanisms of hepatocarcinogenesis, we characterized copy number alterations and gene expression profiles from the same set of tumors associated with hepatitis C virus. Most tumors harbored 1q gain, 8q gain or 8p loss, with occasional alterations in 13 additional chromosome arms. In addition to amplifications at 11q13 in 6 of 103 tumors, 4 tumors harbored focal gains at 6p21 incorporating VEGFA. Fluorescence in situ hybridization on an independent validation set of 210 tumors found 6p21 high-level gains in 14 tumors, as well as 2 tumors with 6p21 amplifications. Strikingly, this locus overlapped with copy gains in 4 of 371 lung adenocarcinomas. Overexpression of VEGFA via 6p21 gain in hepatocellular carcinomas suggested a novel, cell-nonautonomous mechanism of oncogene activation. Hierarchical clustering of gene expression among 91 of these tumors identified 5 classes, including ‘CTNNB1’, ‘proliferation’, ‘interferon-related’, and a novel class defined by polysomy of chromosome 7. These class labels were further supported by molecular data: mutations in CTNNB1 were enriched in the ‘CTNNB1’ class, while IGF1R and RPS6 phosphorylation were enriched in the ‘proliferation’ class. The enrichment of signaling pathway alterations in gene expression classes provides insights on HCC pathogenesis. Furthermore, the prevalence of VEGFA high-level gains in multiple tumor types suggests indications for clinical trials of anti-angiogenic therapies.
Keywords: Hepatocellular carcinoma, VEGFA, molecular classification
INTRODUCTION
Hepatocellular carcinoma (HCC) is the third leading cause of cancer deaths worldwide. These tumors arise in the context of chronic liver injury, inflammation and hepatocyte proliferation provoked by several etiologies. Hepatitis C virus is the most prevalent etiology in the United States and Europe, while hepatitis B virus is endemic in Asia and Africa (1, 2). The critical genomic hits at preneoplastic stages are not well defined, although aberrant methylation, genomic instability and TERT activation have been reported. In overt HCC, deregulated oncogenes (MET, CTNNB1, MYC, CCND1) and tumor suppressor genes (TP53, PTEN, CDKN2A, CDH1) due to genetic or epigenetic alterations have been characterized.
Gene expression studies have converged on major molecular classes of HCC by analyzing patients at different stages and with different etiological factors (3–8). One class is associated with high proliferation, chromosomal instability and activation of IGF or Akt/mTOR signaling. Another class features activation of Wnt signaling pathway associated with a high prevalence of mutations in CTNNB1, which is the gene with the second highest frequency of known mutations in hepatocellular carcinomas, after TP53 (7, 9). Mutations or deletions in CTNNB1 exon 3 prevent the ubiquitination of its protein product, beta-catenin, leading to its nuclear translocation and trans-activation of target genes (10). A third class is associated with interferon signaling due to leukocyte infiltration (4). Intriguingly, an interleukin gene expression signature from the adjacent liver tissue has also been linked to higher rates of venous invasion and metastasis, pointing to the importance of tumor microenvironment in the genesis and intrahepatic dissemination of HCC (11). Despite all these efforts, only two studies have been able to correlate homogeneous biological tumor patterns with survival outcomes (5, 8), although confirmatory independent validations are not yet available. In parallel, genome-wide chromosomal aberration studies (12–16) and methylation studies (17) have begun to decipher the molecular heterogeneity of HCC.
The rising worldwide incidence of hepatocellular carcinomas due to chronic hepatitis C infection underscores the need to discover biomarkers for early detection and tumor progression, as well as gene targets for primary and adjuvant therapies. We conducted extensive genomic and transcriptomic characterization of tumors from hepatitis C virus positive patients with early disease, as defined by a clinical staging system. We demonstrate that high-level copy number gain of 6p21 represents one mechanism for increasing VEGFA expression. In addition, we define 5 molecular classes that integrate copy number and gene expression alterations with signaling pathways assessment, and describe a new molecular class defined by increased dosage of chromosome 7 and overexpression of oncogenes on this region.
MATERIALS AND METHODS
Primary hepatocellular carcinoma specimens
Fresh frozen tissue specimens measuring 1 cm3 were obtained at the time of surgical resection or liver transplantation from 100 anonymous patients from the HCC Genomic Consortium (Barcelona Hospital Clinic, Milan National Cancer Institute, and Mount Sinai School of Medicine) according to institutional review board approved protocols. Two nodules were obtained from each of three transplantation patients, for a total of 103 tumor samples. For 94 of these samples, uninvolved cirrhotic tissue was obtained from the same patient at time of surgery. Clinico-pathological annotated data was analyzed in 82 samples obtained from surgical resection (Table S1). Hepatitis C virus infection was confirmed by serology in all cases, and other etiologies of HCC were ruled out. Tissue samples were treated with RNAlater (Qiagen) or liquid nitrogen and stored at −80°C until DNA and RNA extraction, as previously reported (18). Genomic DNA was extracted with the ChargeSwitch gDNA mini kit (Invitrogen), and further purified with phenol-chloroform extraction. An independent set of 210 HCC patients from all etiologies undergoing resection was included to validate high-level gains of VEGFA by fluorescence in situ hybridization of tissue microarrays (Table S1).
Single nucleotide polymorphism arrays
Genotypes and hybridization intensities for over 238,000 single nucleotide polymorphisms were measured with the StyI chip of the Affymetrix 500K Human Mapping Array set (Affymetrix). Data was analyzed with the GenePattern software package (19) according to methods previously described (20). Intensity (.CEL) files were normalized with invariant normalization and modelled using the PM-MM difference modelling method with the SNPFileCreator module (19). Copy numbers were inferred by comparing signal intensities between each tumor and a selection of the five most similar normals selected from non-tumoral cirrhotic tissue, as well as a database of multiple tumor types (21). The similarity between tumor and normal profiles was measured as the Euclidean distance between the log2-transformed signal profiles (21).
Statistical analysis of copy number alterations
Copy number data was preprocessed with GISTIC software, which includes steps for batch correction, data normalization, copy number estimation versus the average of the five closest normal samples, and copy number segmentation (refer to supplement of (21)). Copy number alterations at each probe set i are evaluated by the score Gi = fi ai, where ai represents the average amplitude of gain or loss with an overall frequency fi in the dataset. We used copy number cutoffs of 2.2 to detect broad regions of copy gain, as well as 1.8 for loss, since these cutoffs exceeded the variation observed in adjacent cirrhotic tissues obtained from the same patients. We obtained similar results by inferring copy numbers with the GEMCA algorithm (Figure S5) (22). To detect high-level gains, we raised the copy number threshold to 3.8 copies (20).
Fluorescence in situ hybridization
Four-micron tissue sections were mounted on standard glass slides and baked at 60°C for two hours. Slides were deparaffinized, dehydrated, and the tissue sections treated with Digest-All III solution (Invitrogen) according to standard protocols. Commercial alpha-satellite probes corresponding to the centromeric regions of chromosome 6 (CEP-6) or chromosome 11 (CEP-11) were purchased pre-labeled with SpectrumGreen dUTP (Abbott Molecular/Vysis, Inc.). One microgram each of BACs corresponding to the VEGFA (RP11-710L16) or CCND1 (RP11-156B3) loci were direct-labeled with SpectrumOrange dUTP using nick-translation (Abbott Molecular/Vysis, Inc.), then hybridized and washed using standard protocols. Slides were imaged using an Olympus BX51 fluorescence microscope and an Applied Imaging system running CytoVision Genus version 3.9 (Supplementary Methods).
Tissue microarrays
Tissue microarrays were constructed using the Advanced Tissue Arrayer ATA-100 (Millipore). Targets for arraying were identified by a liver pathologist (S.T.) by marking the morphologically more representative areas chosen from haematoxylin-eosin stained sections from each paraffin block. Two tissue cores with a diameter of 1.5 mm were transferred from each donor block to the recipient tissue microarray.
Quantitative real time PCR
Total RNA was extracted from 3 to 4 tissue sections, each 10 microns thick, using the TRIzol LS reagent (Invitrogen). cDNA synthesis and PCR conditions were conducted as previously described (23). Expression levels were measured with Taqman Probes® Hs00173626-m1 for VEGFA and Hs03023943-g1 for ACTB obtained from Taqman Gene Expression Assays® (Applied Biosystems). Reactions were set up as triplicates for each gene, and the median threshold cycle (Ct) value from the triplicates was used. We calculated ΔCt values between VEGFA and ACTB for each tumor, and ΔΔCt values were calculated between the ΔCt for each tumor and the median ΔCt for 5 uninvolved, non-cancerous cirrhotic controls. The Mann-Whitney test was used to evaluate differences in expression levels between groups.
Affymetrix U133 Plus 2.0 expression data
Gene expression assessment was performed in 91 tumor samples: 69 new samples obtained from surgical resection extracted as described previously and additional 22 tumor samples and 8 cirrhotic liver controls already reported (18). Affymetrix array intensity (.CEL) files were processed with the RMA algorithm (24). In order to compare expression levels across microarray platforms, we remapped probe sequences on each platform to AceView transcripts (25, 26). Array intensity data (.CEL files) have been deposited in the NCBI Gene Expression Omnibus1 (GEO) and are accessible through GEO Series accession number GSE9829.
Unsupervised classification of gene expression data
We adopted a robust hierarchical clustering approach for unsupervised classification of Affymetrix U133Plus 2.0 data (7). We averaged the results from modifying three clustering parameters: transcript lists, linkage method and number of clusters (see Supplementary Methods). The reproducibility of hierarchical clustering was assessed with the GenePattern ConsensusClustering module, version 3, using a 1 – Pearson correlation distance metric after mean-centering of genes (27).
Selection of class-specific marker genes
For each of the 5 gene expression classes obtained from consensus hierarchical clustering, we performed supervised analysis to identify genes that were significantly upregulated or downregulated among tumors. Significance analysis of microarrays with 500 permutations of class labels evaluated the significance of gene expression changes that were significantly over-expressed or under-expressed among the tumors in a single class. Up to 200 overexpressed transcripts and up to 200 underexpressed transcripts with a 2-fold or greater change and an FDR q-value < 0.01 were selected as marker genes. The list of marker genes for each class are provided in Table S6 to Table S10.
Cross-dataset prediction of gene expression classes
Affymetrix U133A array data was obtained from EBI ArrayExpress, with accession number E-TABM-36 (7). These U133A array probesets were mapped to 12,145 AceView transcripts based on exact sequence matches (25). NCI-Operon oligonucleotide microarray data were obtained from Gene Expression Omnibus, with accession numbers GSE1898 and GSE4024 (5, 6). The Prediction Analysis of Microarrays R package was used to train shrunken centroid classifiers from previously published gene expression data and class labels (28) (see Supplementary Methods).
Sequencing
PCR and sequencing were conducted by GENEWIZ (South Plainfield, NJ). PCR primers and reaction conditions are listed in Table S12 and Supplementary Information. Mutations were detected by Mutation Surveyor 2.51 (SoftGenetics), followed by manual review of candidate mutations.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were used to assess phosphorylated proteins in human tissue. Sections placed on glass were baked at 60°C for 30 min, deparaffinized in xylene, and rehydrated in a graded series of ethanol solutions. Antigens were unmasked by microwave heating the samples in 10 mM sodium citrate buffer (pH 6.0) for 5 minutes (x3). Endogenous peroxidase from the tissue was quenched using 3% hydrogen peroxide. After washing with phosphate-buffered saline, samples were incubated with anti-phosphorylated Akt antibody (1:50; Ser473, IHC specific, Cell Signaling Technology); anti-phosphorylated IGF1R (1:100; Tyr1316, Dr. Rubini, University of Ferrara); anti-CTNNB1 (1:750; Abcam) or anti-phosphorylated RPS6 (1:200; Ser240/244, Cell Signaling Technology) at 4°C overnight. DAB (3,3′-diaminobenzidine) was used as a detection system (EnVision+ System-HRP, Dako). Immunoreactivity was independently graded by two liver pathologists (S.T. and M.S.) and both agreed in the final staining score. The variables measured were as follows: (1) intensity (0=absent, 1=weak, 2=moderate, 3=strong); (2) distribution (1=very focal, 2=focal, 3=diffuse) and (3) localization of the staining (membranous, cytoplasmic, or nuclear). Samples were defined positive for Akt or IGF1R phosphorylation when intensity was 2 or higher, regardless of distribution. For RPS6 phosphorylation, only those samples with intensity and distribution of 2 or higher were considered positive. Tumors with CTNNB1 activation were defined by the presence of nuclear staining, or by more than 5% of cytoplasmic staining.
Statistical associations
Each of the 5 gene expression classes was assessed for enrichment with molecular characteristics. Fisher’s exact test was used to assess the enrichment of binary variables, such as immunostaining, mutation status, or chromosomal gain or loss. The Mann-Whitney was used to test for associations with continuous variables, such as AFP levels.
Analysis of clinical outcomes
The probability curves of recurrence and early recurrence were calculated according to Kaplan-Meier and compared by Mantel-Cox test. Median inferred copy numbers for chromosomal arms for each sample were used as covariates for univariate association with recurrence. Median copy numbers of chromosomal arms were calculated for each tumor sample, and cutoffs of 2.2 for copy gains and 1.8 for copy loss were used. For continuous variables, the cut-off level was their median value. Significant variables (p < 0.05) were included in a step-wise Cox proportional hazard regression analysis of recurrence and early recurrence. The calculations were done by the SPSS package (SPSS 15.0).
RESULTS
Copy number alterations in clinically early, HCV-related hepatocellular carcinomas
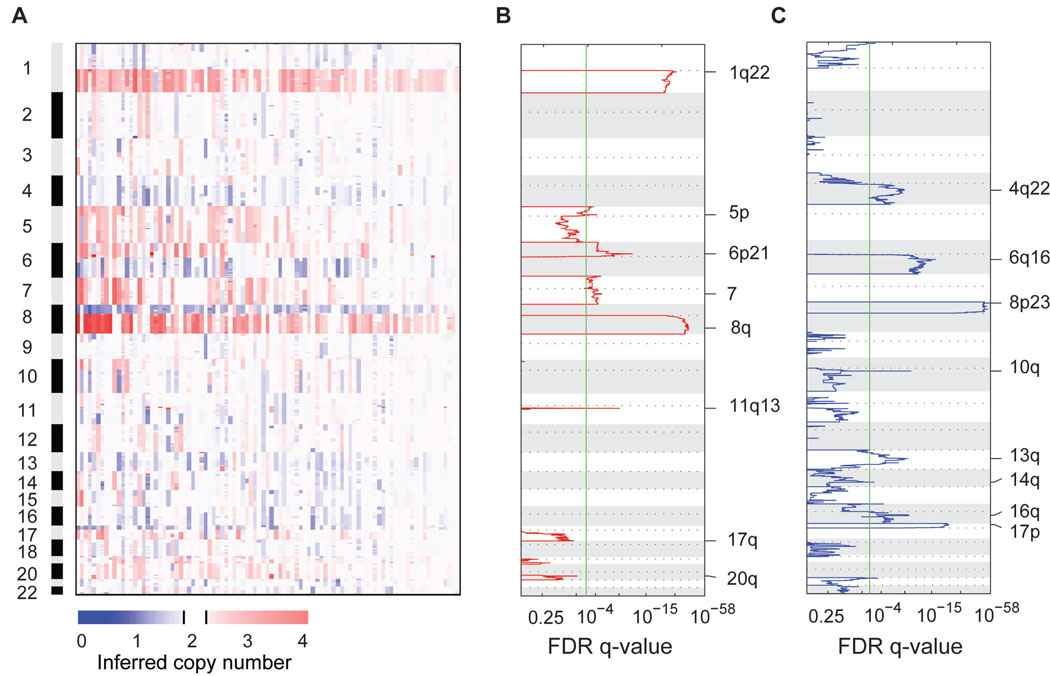
We measured copy number alterations at 238,000 genomic loci with Affymetrix Sty mapping arrays among 103 primary tumors. According to the Barcelona Clinic Liver Cancer staging system, 80% of these patients had very early or early stage disease (stage 0 or stage A), representing asymptomatic tumors with either a single nodule or up to three nodules less than 3 cm in diameter (29) (Table S1). To find evidence for driver alterations in tumor genomes, we evaluated the frequency and magnitude of copy number gains and losses with the GISTIC algorithm (21). Broad gains and losses of chromosome arms were the most prevalent changes among these tumors (Figure 1A). The most significant chromosomal gains affected chromosome arms 1q (72%) and 8q (66%), which are early events in hepatocarcinogenesis (Figure 1B) (30). Focal gains centered on 6p21 (4%) and 11q13 (6%) were the next most significant alterations, due to their high levels of copy increase. Significant copy number gains were also observed for chromosomes 5p (33%), 7 (30%), 17q (19%) and 20 (18%). Notably, gains of chromosomes 5p and 7 significantly co-occurred among these tumors (Fisher exact p < 10−4), which may indicate cooperation among multiple genes with elevated dosage on both chromosomes. Significant chromosomal losses affected chromosome arms 8p (60%), 17p (39%), 6q (27%), 4q (19%), 13q (19%), 16q (17%), 14q (13%) and 10q (11%) (Figure 1C). Aside from 1q gain, 8q gain and 8p loss in the majority of tumors, substantially lower frequencies of gains and losses on other chromosome arms suggested the presence of multiple tumor sub-types that select for alterations in distinct signaling pathways. We did not detect chromosomal gains or losses in the 94 uninvolved, non-cancerous cirrhotic tissue samples (Figure S4D).
Figure 1. Copy number alterations in hepatocellular carcinomas arising from hepatitis C viral infection.
(A) Overview of chromosomal gains and losses in 103 hepatocellular carcinomas. Each tumor is displayed in a separate column, and inferred copy numbers at 238,304 loci are displayed from top to bottom in genomic order. Tumors are ordered from left to right according to the number of chromosome arms with significant copy number alterations. Copy number gains are shown in red and losses are shown in blue. (B) Recurrent chromosomal gains and (C) Recurrent chromosomal losses. The GISTIC algorithm identified significant regions of copy number alterations in multiple samples. Chromosomes are displayed in ascending order along the vertical axis. The horizontal axis indicates the statistical significance of recurrent gains or losses, corresponding to the False Discovery Rate q-value obtained from GISTIC. The vertical green line indicates a significance threshold of q < 0.001.
Focal regions of copy gain or loss provide greater resolution to map oncogenes or tumor suppressors by narrowing the overlapping genomic region among multiple tumors. However, these focal lesions were rare events among clinically early hepatocellular carcinomas. Only 15% of tumors harbored at least one high-level gain, which we define as a genomic region smaller than 12 Mb with inferred copy number greater than 3.8 (Table S2). We found 9 candidate regions of homozygous deletion, all of which occurred in only one tumor (Table S3). Two of these regions affected single exons of CSMD1 or MSR1, while two regions deleted exons of GPR143, SHROOM2, WWC3, MAOA and MAOB.
To pinpoint genomic regions that may harbor key oncogenes, we repeated GISTIC analysis to find significantly recurrent high-level gains. The most frequent amplifications occurred in 6 tumors at 11q13, with a minimal common region from 68.83 to 69.30 Mb (FDR q < 10−11) (Figure S1). This region included only four known genes: CCND1, ORAOV1, FGF19 and FGF4. Fluorescence in situ hybridization with a BAC probe to these 4 genes confirmed that three of these tumors with available tissue blocks harbored amplifications. In 50% to 90% of tumor nuclei, over 10 signals corresponding to BAC RP11-710L16 were detected (Figure S1).
Overlapping high-level gains at 6p21 occur in multiple tumor types
The second most frequent high-level gains pinpointed a overlapping region that had not been previously described in hepatocellular carcinomas (FDR q < 10−11). Four tumors harbored high-level gains of 6p21, ranging in size from 2.4 Mb to 22 Mb (Figure 2A, 2B). Remarkably, a recent study of 371 lung adenocarcinomas identified this locus as the 17th most frequent region of copy gains, yet this frequency barely passed statistical significance (FDR q < 0.08) (20). Assuming that the same oncogene targets are under selection in multiple tumor types (31), we pooled regions of high-level gains from 4 hepatocellular carcinomas and 4 lung adenocarcinomas. The minimal region of overlap – between positions 43.792 Mb and 44.110 Mb – encompassed only 2 genes: VEGFA and MGC45491 (Figure 2C).
Figure 2. Overlapping high-level gains at 6p21 in hepatocellular carcinomas and lung adenocarcinomas.
(A) Copy number alterations among 4 hepatocellular carcinomas (light blue), 4 lung adenocarcinomas (light green), 4 matched cirrhotic tissues from adjacent liver (dark blue), and 3 matched normals for the lung tumors (dark green). (B) Inferred copy numbers at individual single nucleotide polymorphism probe sets. Copy numbers are shown in red for tumor samples, or in grey for the matched normal from the same patient. Vertical dashed lines delineate the boundaries of the minimal overlapping region among the 8 tumors. (C) Annotated genes found in the minimal overlapping region at 6p21. Inclusion of the SM-11XV lung tumor narrows the overlapping region to the boundaries of the shaded box. The genomic location of BAC probe RP11-710L16 used for fluorescence in situ hybridization is indicated. (D) Confirmation of VEGFA copy gains by fluorescence in situ hybridization. Red signals indicate the BAC probe RP-710L16 centered on VEGFA, and green signals indicate BAC probe CEP-6 for the centromere of chromosome 6. Representative images are displayed for the 3 of 4 hepatocellular carcinomas with available tissue blocks, and detailed counts of probe signals are provided in Table S4.
High-level gains of 6p21 were confirmed in two fluorescence in situ hybridization (FISH) studies with a BAC probe spanning the MRPS18A and VEGFA genes (Figure 2C). First, FISH analysis on thin sections from all 3 hepatocellular carcinomas with available tissue blocks validated copy gains at 6p21, with an average of over 5.5 VEGFA probe signals per tumor (Figure 2D, Figure S2, Table S4). Second, we also performed FISH analysis on an independent tissue microarray collection of 210 hepatocellular carcinomas (Table 1, Table S1, Table S4). We defined three cytogenetic categories based on the average number of VEGFA probe signals observed per nucleus. Gains of chromosome 6 were found in 58 of 210 tumors (27.6%), with 2.3 or more probe signals. An additional 14 tumors (6.7%) also had high-level gains of VEGFA, as defined by 4 or more VEGFA probe signals. Two tumors showed VEGFA amplifications (1.0%), with over 8 VEGFA probe signals.
Table 1.
FISH analyses of 6p21 copy number alterations in 210 HCC on tissue microarrays
| Copy number alteration | Number (Frequency) | Mean VEGFA probe signals | Mean CEP6 probe signals | VEGFA:CEP6 ratios |
|---|---|---|---|---|
| VEGFA amplifications | 2 (1.0%) | 6.5 – 14.9 | 2.1 – 2.1 | 4.1 – 7.2 |
| VEGFA high-level gains | 14 (6.7%) | 4.0 – 6.5 | 2.1 – 5.9 | 1.1 – 1.8 |
| Chromosome 6 gains | 58 (27.6%) | 2.3 – 3.9 | 2.0 – 3.7 | 1.0 – 1.8 |
| Normal chromosome 6 | 136 (64.8%) |
VEGFA is a likely oncogene target of high-level gains at 6p21
Genomic regions with increased copy number are often retained in tumor cells due to selective advantages conferred by the increased expression of one or more target genes residing in the region. In order to identify candidate oncogene targets in the overlapping regions of copy gains at 6p21 and 11q13, we evaluated the association between copy numbers at these loci and the expression of each gene. A genome-wide, permutation-based t-test identified only two known genes that were significantly overexpressed among the 4 tumors with high-level gains at 6p21: VEGFA and TMEM63B (FDR q < 0.001) (Figure 3A). VEGFA was the most significantly overexpressed transcript, with an average 2.8-fold higher expression among the 4 tumors with 6p21 high-level gains (Figure 3B). TMEM63B is a transmembrane protein with unknown function, and the 4 tumors with 6p21 high-level gains had 1.9-fold higher average expression (Figure 3C). Thus, elevated levels of TMEM63B may represent a marker indicating VEGFA overexpression by increased copy dosage, rather than VEGFA overexpression due to trans-regulation by HIF1A or other regulators.
Figure 3. High-level gains at 6p21 lead to VEGFA overexpression.
(A) Significance Analysis of Microarrays identified overexpression of two genes among the 4 tumors with 6p21 amplifications. Each point represents the observed t-statistic, plotted against its expected score from permutation tests. The most significant gene is VEGFA, followed by TMEM63B (indicated in purple). The dashed lines indicate the confidence interval for the random, null distribution. (B) VEGFA or (C) TMEM63B is overexpressed in tumors with high-level gains of 6p21. Each of the 91 points represents the corresponding copy number and expression level for a single tumor. The horizontal axis indicates the median inferred copy number for 68 single nucleotide polymorphism probes in the minimal overlapping region at 6p21. The vertical axis indicates the log2 ratio between the expression level of each tumor, compared to the median of 10 normal liver samples on the Affymetrix U133 Plus 2 array. (D) Higher VEGFA expression is associated with increased copy numbers in an independent cohort. Each of the 45 points represents a single tumor chosen from the panel of 210 tumors evaluated by fluorescence in situ hybridization. The horizontal axis indicates the average number of VEGFA probes per nucleus. The vertical axis indicates the negative ΔΔCt values of VEGFA normalized to ACTB, compared to the median of 5 uninvolved, cirrhotic tissue samples. Tumors with more than 4 copies of VEGFA are shown in red (n = 10), tumors with gains of chromosome 6 are shown in black (n = 19), and diploid tumors are shown in grey (n = 16). The average negative ΔΔCt value for each of these three classes is plotted with a dashed blue line.
Notably, VEGFA expression was also associated with copy numbers measured by fluorescence in situ hybridization among 45 tumors in the tissue microarray cohort. Ten tumors with more than 4 copies of VEGFA had an average of 2.8-fold higher expression than 16 tumors that were diploid at this locus (Mann-Whitney p = 0.0007) (Figure 3D). Thus, we have validated that high-level gains of VEGFA are associated with higher VEGFA expression in two independent cohorts.
For the 6 tumors with 11q13 amplifications, we found 4 genes to be significantly overexpressed: ORAOV1, TPCN2, CCND1 and MRPL21 (FDR q < 0.001). Both ORAOV1 and CCND1 reside in the minimal common region of amplification. CCND1 is a confirmed oncogene in hepatocarcinogenesis (32), while the log expression levels of ORAOV1 showed a strikingly linear correlation with the median copy number of 11q13 (Figure S1D).
Gene expression classes are associated with signaling pathway alterations
In order to characterize the molecular heterogeneity of hepatocellular carcinomas, we measured gene expression profiles in 91 of the 103 tumors with oligonucleotide microarrays (Affymetrix U133 Plus 2 arrays). We obtained 5 gene expression classes from unsupervised classification with consensus hierarchical clustering, which considered 32 different parameter combinations (7) (Figure 4A). To validate these 5 gene expression classes with previously published studies, we trained shrunken centroid classifiers on two additional gene expression datasets (6, 7). Two classes showed extremely good concordance with the labels predicted by these external classifiers (Figure 4B). Furthermore, signaling pathway annotations could be assigned for 3 of these 5 classes by the enrichment of marker genes, immunohistochemistry and mutations (Figure 4C, Table S5 to Table S10).
Figure 4. Integrated classification of genomic and molecular alterations in hepatocellular carcinomas.
(A) Consensus hierarchical clustering of gene expression in 91 tumors. Colors in the sample dendrogram indicate the 5 gene expression classes determined by consensus hierarchical clustering over 32 different parameter set combinations. Rows display mean-centered expression levels for 1242 marker genes associated with each of the 5 gene expression classes. (B) Overlap of consensus classes with previously published classes. Class labels were assigned based on the distance to the closest shrunken centroid classifier for previous classifications (5, 7). (C) Copy number alterations among the gene expression classes. Gains and losses for the indicated chromosomes are displayed according to the color scheme in Figure 2. Mutation status (CTNNB1 exon 3 or TP53 exons 5 to 8) and immunohistochemical staining are indicated in greyscale: present (black); absent (grey); or missing data (white).
The high prevalence of CTNNB1 mutations indicated alterations to canonical Wnt signaling in a subset of hepatocellular carcinomas. The CTNNB1-associated class of 24 tumors was significantly enriched for CTNNB1 exon 3 mutations (62%; Fisher p < 6 × 10−4) and nuclear localization (71%; Fisher p < 3 × 10−5) (Table S5). Further evidence for CTNNB1 activation among tumors in this class came from the significant overexpression of several liver-specific marker genes, including GLUL, LGR5, TBX3 and REG3A (33–35) (Table S6). This class was also associated with tumor diameters greater than 3 cm (Fisher p=0.002).
Another molecular sub-type of hepatocellular carcinomas featured increased proliferation, high levels of serum AFP and chromosomal instability (5, 7). We found a corresponding proliferation class of 23 tumors that overexpressed AFP (median serum level, 472 ng/mL; Mann-Whitney p=0.001), along with several genes corresponding to a proliferation gene expression signature (36) (Table S5 and Table S7). Since these tumors were enriched for IGF1R phosphorylation (48%; Fisher p < 1 × 10−4), RPS6 phosphorylation (83%; Fisher p=0.003) and Akt phosphorylation (55%; Fisher p=0.01), it is likely that tyrosine kinase activation drives the proliferation of these tumors. Reduced frequencies of CTNNB1 exon 3 mutations among these tumors (13%; Fisher p=0.02) confirmed the model that tyrosine kinase activation and CTNNB1 activation represent distinct routes for tumor progression (7, 37). Tumors in this class also had higher frequencies of 4q loss (38%; Fisher p=0.03) and 13q loss (33%; Fisher p=0.04), as well as lower frequencies of 6q loss (12%; Fisher p=0.04). A significant correlation with macrovascular invasion was observed for tumors in this proliferation class (Fisher p=0.007).
A third class of 18 tumors also harbored significantly lower rates of CTNNB1 exon 3 mutation (6%; Fisher p=0.004). Lower expression of IGF2, as well as CTNNB1 target genes, suggested that these tumors represent a distinct class. Strikingly, 4 of the 28 significantly overexpressed marker genes corresponded to interferon-stimulated genes: STAT1, ISG15, IFI6 and IFI27 (38) (Table S8). STAT1 is a transcription factor that mediates the response to hepatitis C viral infection, while elevated expression of ISG15, IFI6 and IFI27 are predictive markers of hepatitis C virus patients who failed to respond to pegylated interferon and ribavirin therapy (39). Notably, tumors about this interferon-related class were more likely to be less than 3 cm in diameter (Fisher p=0.005).
Polysomy of chromosome 7 defines a novel gene expression class
We discovered a new class of 9 tumors that was significantly associated with a lack of gains of chromosome 8q (78%; Fisher p=0.007), as well as polysomy of chromosome 7 above a median copy number of 2.7 (89%; Fisher p < 10−8). Increased dosage led to significantly higher expression levels of multiple genes on chromosome 7 (Table S9), as confirmed by gene set enrichment analysis based on cytobands (FDR q < 0.01). The most significantly overexpressed genes on chromosome 7 included COBL, CLDN15, MAD1L1, POLD2 and EPHA1, although oncogene targets of these high-level gains remain to be identified. Due to the small number of tumors in this class, we lacked the power to detect significant clinico-pathological correlations.
Prognostic significance of genomic alterations
We evaluated associations between copy number alterations and clinical outcomes among 82 patients who underwent surgical resection. Recurrence was associated with 7 gain and 13q loss by univariate analysis (Table S11). On multivariate analysis, chromosome 7 gain was retained as an independent predictor of recurrence [hazard ratio: 3.4 (95% CI: 1.7–6.8), p < 0.001], along with BCLC staging [hazard ratio: 7.1 (95% CI: 3.8–13.5), p < 0.001]. Notably, gains of chromosome 7 were associated with a significantly higher risk of recurrence within 2 years (early recurrence) after surgical resection (Table 2, Figure S3).
Table 2.
Variables with independent predictive value for clinical outcomes in multivariate analysis
| EARLY RECURRENCE | OVERALL RECURRENCE | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p-value | Hazard ratio | 95% CI | p-value | |
| Barcelona-Clinic Liver Cancer staging classification | 8.8 | 4.3 – 17.6 | < 0.001 | 7.1 | 3.8 – 13.5 | < 0.001 |
| Chromosome 7 median copy number > 2.2 | 5.8 | 2.4 – 13.8 | < 0.001 | 3.4 | 1.7 – 6.8 | < 0.001 |
DISCUSSION
To date, this study represents the largest characterization effort of paired copy number and gene expression alterations in clinically early hepatocellular carcinomas with hepatitis C virus etiology. Combined annotations from marker gene lists, mutation analyses and immunohistochemistry could associate 3 of 5 gene expression classes with previously described signaling pathway alterations (4, 5, 7). First, a CTNNB1-activated class was enriched for CTNNB1 exon 3 mutations and immunohistochemical staining. Second, a proliferation class was enriched for IGF1R and RPS6 phosphorylation, thus pointing to potential targets for combined therapeutic strategies. Small molecule inhibitors and monoclonal antibodies against IGF1R, such as A12 (Imclone) and mTOR, such as rapamycin or everolimus (Novartis), are still at pre-clinical or early clinical stage of drug development for HCC (40). Third, an interferon-related class that overexpressed several interferon-stimulated genes was associated with smaller tumor size. Notably, tumors with upregulated interferon response in another study also had higher leukocyte infiltration and tumor apoptosis (4).
A novel gene expression class was defined by polysomy of chromosome 7 and the concomitant overexpression of multiple genes along this chromosome. Intriguingly, most of these tumors lacked 8q gains, which are the second most frequent chromosomal alterations in hepatocellular carcinomas and include the known oncogenes MYC, PTK2 and COPS5 (12, 41). This observation suggests that unknown target oncogenes on chromosome 7 may contribute to a distinct mechanism of tumor progression. Indeed, we found that even low-level gains of chromosome 7 were independent predictors of recurrence within 2 years of surgical resection. Although EGFR and MET are frequently cited oncogene candidates on chromosome 7, we only observed a modest overexpression of EGFR (1.39-fold versus normals) and MET (1.82-fold versus normals) among the 9 tumors in this class. Surprisingly, we did not observe amplifications of MET among these hepatitis C virus positive tumors, even though previous studies have reported a few instances of focal MET amplification, mostly in tumors arising in the context of hepatitis B virus infection (12, 13, 31). Thus, this discrepancy in MET amplification or overexpression could be explained by differences in viral etiologies or accompanying cirrhosis.
Due to the limited number of patients and events in each class, the size of this cohort was underpowered to detect robust differences in overall survival, and only correlation with metastatic potential was captured. Nevertheless, we speculate that these gene expression classes may represent distinct combinations of signaling pathway alterations, and would thus demonstrate different responses to molecular targeted therapies. Thus, biomarkers for HCC molecular classes should be assayed in future clinical trials in order to provide evidence of treatment response in a genetically defined subset of patients. In addition, animal models that recapitulate these molecular classes should be instrumental for testing novel anti-tumoral agents in pre-clinical experiments. For instance, a double transgenic MYC/TGFA mouse model of HCC has similar gene expression patterns with human tumors in the proliferation class (42). Additional mouse models that recapitulate the remaining classes need to be characterized.
We propose that high-level gains of VEGFA may represent a genetic dependency of tumors arising in multiple tissue types, and that these high-level gains may predict response to a variety of inhibitors against VEGFA or its receptors. While chromosome 6p gains have been previously reported in HCC (30, 43), the higher resolution of SNP arrays in this study has localized a minimal common region of copy gains that included VEGFA. We also found that 3 of the 4 tumors with focal gains of 6p21 were assigned to an unannotated gene expression class.
Although the prognostic significance of elevated transcript and serum VEGFA levels in early hepatocellular carcinomas have been recognized (44, 45), this study is the first report of focal copy gains as a mechanism for elevated VEGFA expression. Intriguingly, KDR/VEGFR-2 and FLT4/VEGFR-3 are among the putative targets of sorafenib (IC50 of 20–90 nM), a multi-kinase inhibitor that has been recently demonstrated to extend survival of patients with advanced HCC (46, 47). In contrast to the cell autonomous effects of most oncogenes, high-level gains of VEGFA suggest that tumors may also select for genetic alterations that mediate tumor-stromal interactions. VEGFA can mediate at least two signals among epithelial and endothelial cells. Proliferating hepatocytes stimulate angiogenesis in a partial hepatectomy model by secreting VEGFA, which binds to FLT1 or KDR on sinusoidal endothelial cells (48). Conversely, VEGFA-activated endothelial cells can also produce HGF in response to increased VEGFA levels, and this elevation stimulates hepatocyte proliferation (49). As further evidence for the paracrine effects of VEGFA, RNA interference against VEGFA did not affect proliferation of the SMMC-7221 hepatocellular carcinoma cell line in vitro, but inhibited tumor growth and induced apoptosis in nude mice xenografts (50).
In conclusion, our integrated molecular classification encapsulates key signaling pathway alterations of HCV-related hepatocellular carcinomas. With their high concordance with other gene expression studies, these classes should be assayed for stratified analyses of future clinical trials. In addition, focal copy number gains and overexpression of VEGFA, as well as multiple genes on chromosome 7, suggest potential targets for molecular therapies.
Supplementary Material
ACKNOWLEDGEMENTS
We dedicate this work to the memory of our friend and colleague, Eric Lemmer. We thank Rameen Beroukhim and Craig Mermel for assistance with the GISTIC algorithm, as well as Barbara Weir and Roel Verhaak for a critical reading of the manuscript.
Financial Support
This work was supported by grants from the National Institutes of Health (DK076986 to J.M.L., CA109038 to M.M., DK037340 to S.L.F.); Samuel Waxman Cancer Research Foundation (J.M.L.); Spanish National Health Institute (SAF-2007-61898 to J.M.L.); Instituto de Salud Carlos III (PI 05/645 to J.B., FIS CM04/00044 to B.M.); Italian Association for Cancer Research (V.M.); Davies Charitable Foundation (D.Y.C.); Fundación Pedro Barrié de la Maza (A.V.); National Cancer Center (A.V.); European Association for the Study of the Liver (A.V.); Charles A. King Trust (Y.H.); Departament d’Educació i Universitats de la Generalitat de Catalunya (J.B.). Ciberehd is funded by the Instituto de Salud Carlos III.
Footnotes
Conflicts of Interest
Please see Author Conflict of Interest forms for complete list. None of these companies had any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 2.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breuhahn K, Vreden S, Haddad R, et al. Molecular profiling of human hepatocellular carcinoma defines mutually exclusive interferon regulation and insulin-like growth factor II overexpression. Cancer Res. 2004;64:6058–6064. doi: 10.1158/0008-5472.CAN-04-0292. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 6.Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 7.Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 9.Laurent-Puig P, Legoix P, Bluteau O, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763–1773. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]
- 10.Provost E, McCabe A, Stern J, Lizardi I, D'Aquila TG, Rimm DL. Functional correlates of mutation of the Asp32 and Gly34 residues of beta-catenin. Oncogene. 2005;24:2667–2676. doi: 10.1038/sj.onc.1208346. [DOI] [PubMed] [Google Scholar]
- 11.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Patil MA, Gutgemann I, Zhang J, et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis. 2005;26:2050–2057. doi: 10.1093/carcin/bgi178. [DOI] [PubMed] [Google Scholar]
- 13.Midorikawa Y, Yamamoto S, Ishikawa S, et al. Molecular karyotyping of human hepatocellular carcinoma using single-nucleotide polymorphism arrays. Oncogene. 2006;25:5581–5590. doi: 10.1038/sj.onc.1209537. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Shen HH, Shen T, et al. Correlation between genomic DNA copy number alterations and transcriptional expression in hepatitis B virus-associated hepatocellular carcinoma. FEBS Lett. 2006;580:3571–3581. doi: 10.1016/j.febslet.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 15.Poon TCW, Wong N, Lai PBS, Rattray M, Johnson PJ, Sung JJY. A tumor progression model for hepatocellular carcinoma: Bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262–1270. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Katoh H, Ojima H, Kokubu A, et al. Genetically distinct and clinically relevant classification of hepatocellular carcinoma: putative therapeutic targets. Gastroenterology. 2007;133:1475–1486. doi: 10.1053/j.gastro.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 17.Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurmbach E, Chen YB, Khirtrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 19.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nature Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 20.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komura D, Shen F, Ishikawa S, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llovet JM, Chen YB, Wurmbach E, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–1767. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter SL, Eklund AC, Mecham BH, Kohane IS, Szallasi Z. Redefinition of Affymetrix probe sets by sequence overlap with cDNA microarray probes reduces cross-platform inconsistencies in cancer-associated gene expression measurements. BMC Bioinformatics. 2005;6:107. doi: 10.1186/1471-2105-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monti S, Tamayo P, Mesirov JP, Golub TR. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 28.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 30.Moinzadeh P, Breuhahn K, Stutzer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade - results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–941. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zender L, Spector MS, Xue W, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida N, Fukuda Y, Komeda T, et al. Amplification and overexpression of the cyclin D1 gene in aggressive human hepatocellular carcinoma. Cancer Res. 1994;54:3107–3110. [PubMed] [Google Scholar]
- 33.Cadoret A, Ovejero C, Terris B, et al. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- 34.Cavard C, Terris B, Grimber G, et al. Overexpression of regenerating islet-derived 1 alpha and 3 alpha genes in human primary liver tumors with beta-catenin mutations. Oncogene. 2006;25:599–608. doi: 10.1038/sj.onc.1208860. [DOI] [PubMed] [Google Scholar]
- 35.Renard CA, Labalette C, Armengol C, et al. Tbx3 is a downstream target of the Wnt/beta-catenin pathway and a critical mediator of beta-catenin survival functions in liver cancer. Cancer Res. 2007;67:901–910. doi: 10.1158/0008-5472.CAN-06-2344. [DOI] [PubMed] [Google Scholar]
- 36.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 37.Calvisi DF, Factor VM, Ladu S, Conner EA, Thorgeirsson SS. Disruption of beta-catenin pathway or genomic instability define two distinct categories of liver cancer in transgenic mice. Gastroenterology. 2004;126:1374–1386. doi: 10.1053/j.gastro.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Taylor MW, Tsukahara T, Brodsky L, et al. Changes in gene expression during pegylated interferon and ribavirin therapy of chronic hepatitis C virus distinguish responders from nonresponders to antiviral therapy. J Virol. 2007;81:3391–3401. doi: 10.1128/JVI.02640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asselah T, Bieche I, Narguet S, et al. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516–524. doi: 10.1136/gut.2007.128611. [DOI] [PubMed] [Google Scholar]
- 40.Llovet JM. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008 doi: 10.1002/hep.22506. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okamoto H, Yasui K, Zhao C, Arii S, Inazawa A. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology. 2003;38:1242–1249. doi: 10.1053/jhep.2003.50457. [DOI] [PubMed] [Google Scholar]
- 42.Lee JS, Chu IS, Mikaelyan A, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 43.Chen YJ, Yeh SH, Chen JT, et al. Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology. 2000;119:431–440. doi: 10.1053/gast.2000.9373. [DOI] [PubMed] [Google Scholar]
- 44.Mise M, Arii S, Higashituji H, et al. Clinical significance of vascular endothelial growth factor and basic fibroblast growth factor gene expression in liver tumors. Hepatology. 1996;23:455–464. doi: 10.1053/jhep.1996.v23.pm0008617424. [DOI] [PubMed] [Google Scholar]
- 45.Poon RT, Ho JW, Tong CS, Lau C, Ng IO, Fan ST. Prognostic significance of serum vascular endothelial growth factor and endostatin in patients with hepatocellular carcinoma. Br J Surg. 2004;91:1354–1360. doi: 10.1002/bjs.4594. [DOI] [PubMed] [Google Scholar]
- 46.Wilhelm SM, Carter C, Tang LY, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 47.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular cancer; ASCO Proceedings 2007; 2007. p. LBA1. [Google Scholar]
- 48.Shimizu H, Miyazaki M, Wakabayashi Y, et al. Vascular endothelial growth factor secreted by replicating hepatocytes induces sinusoidal endothelial cell proliferation during regeneration after partial hepatectomy in rats. J Hepatol. 2001;34:683–689. doi: 10.1016/s0168-8278(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 49.LeCouter J, Moritz DR, Li B, et al. Angiogenesis-independent endothelial protection of liver: Role of VEGFR-1. Science. 2003;299:890–893. doi: 10.1126/science.1079562. [DOI] [PubMed] [Google Scholar]
- 50.Hao JH, Yu M, Li HK, Shi YR, Li Q, Hao XS. Inhibitory effect of antisense vascular endothelial growth factor RNA on the profile of hepatocellular carcinoma cell line in vitro and in vivo. World J Gastroenterol. 2006;12(7):1140–1143. doi: 10.3748/wjg.v12.i7.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.