Abstract
Islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP/G6PC2) is a major autoantigen in both mouse and human type 1 diabetes. IGRP is selectively expressed in islet beta cells and polymorphisms in the IGRP gene have recently been associated with variations in fasting blood glucose levels and cardiovascular-associated mortality in humans. Chromatin immunoprecipitation (ChIP) assays have shown that the IGRP promoter binds the islet-enriched transcription factors Pax-6 and BETA2. We show here, again using ChIP assays, that the IGRP promoter also binds the islet-enriched transcription factors MafA and Foxa2. Single binding sites for these factors were identified in the proximal IGRP promoter, mutation of which resulted in decreased IGRP fusion gene expression in βTC-3, HIT and Min6 cells. ChiP assays have shown that the islet-enriched transcription factor Pdx-1 also binds the IGRP promoter but mutational analysis of four Pdx-1 binding sites in the proximal IGRP promoter revealed surprisingly little effect of Pdx-1 binding on IGRP fusion gene expression in βTC-3 cells. In contrast, in both HIT and Min6 cells mutation of these four Pdx-1 binding sites resulted in an ~50% reduction in fusion gene expression. These data suggest that the same group of islet-enriched transcription factors, namely Pdx-1, Pax-6, MafA, BETA2 and Foxa2 directly or indirectly regulate expression of the two major autoantigens in type 1 diabetes.
Keywords: pancreas, transcription, promoter, diabetes
Introduction
The glucose-6-phosphatase catalytic subunit (G6Pase) is predominantly expressed in liver and kidney and plays a major role in glucose homeostasis by catalyzing the final step in the glycogenolytic and gluconeogenic pathways, namely the hydrolysis of glucose-6-phosphate to glucose and inorganic phosphate (Van Schaftingen and Gerin 2002). Two G6Pase-related proteins have recently been identified designated the islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) (Arden, et al. 1999; Martin, et al. 2001; Wang, et al. 2007b) and the ubiquitously expressed glucose-6-phosphatase catalytic subunit-related protein (UGRP), also known as G6Pase-beta. The encoded proteins exhibit around 50% sequence identity, exhibit a common 9 transmembrane domain topology and are all localized to the endoplasmic reticulum (Boustead, et al. 2004; Martin, et al. 2002).
UGRP confers low glucose-6-phosphatase activity to a broad range of tissues (Cheung, et al. 2007; Wang, et al. 2006) whereas the function of IGRP is less certain. IGRP is exclusively expressed in pancreatic islets, principally in β cells (Hutton and Eisenbarth 2003) and is a candidate for the low glucose-6-phosphatase enzyme activity detected in this tissue (Arden et al. 1999; Wang et al. 2007b). A global knockout of the IGRP gene results in a mild metabolic phenotype characterized by a ~15% decrease in fasting blood glucose (Wang et al. 2007b). This observation suggests that IGRP together with glucokinase could create a substrate cycle and therefore modulate β cell glycolytic flux and glucose-stimulated insulin secretion (Newgard, et al. 2002). Indeed, recent genetic data also suggest that variations in IGRP expression and/or activity are linked to fasting blood glucose levels and hence cardiovascular-associated mortality in humans (Bouatia-Naji, et al. 2008; Chen, et al. 2008; Fradin, et al. 2007). Unfortunately, attempts to directly demonstrate glucose-6-phosphate hydrolysis by IGRP in vitro have been mostly unsuccessful (Arden et al. 1999; Martin et al. 2001; Petrolonis, et al. 2004; Shieh, et al. 2005), raising the question as to whether IGRP requires other cellular factors to exhibit activity or whether it has an unknown catalytic function.
Although its biological function is unclear several reports have demonstrated that IGRP is an important target of cell-mediated autoimmunity in type 1 diabetes both in mice (Han, et al. 2005a; Han, et al. 2005b; Lieberman, et al. 2003; Wong, et al. 2006) and humans (Yang, et al. 2006). In the Non Obese Diabetic (NOD) mouse model of type 1 diabetes, IGRP is recognized by both CD8 (Lieberman et al. 2003) and CD4 positive (Mukherjee, et al. 2005) T cells infiltrating islets. Whether interventions directed at IGRP will be helpful in treating or preventing type 1 diabetes is unclear. In vivo administration of select IGRP epitope peptides to NOD mice appears to abrogate or delay the disease process (Han et al. 2005a). However, recent data suggests that autoimmunity toward IGRP is a secondary event with insulin being the primary autoantigen in NOD mice (Krishnamurthy, et al. 2006).
Of the relatively few islet β cell-specific gene promoters that have been identified, those sequences controlling transcription of the insulin gene have been the subject of the most detailed analyses (Hay and Docherty 2006; Melloul, et al. 2002; Sander and German 1997). Four main promoter elements in the insulin promoters of several species have been shown to be critical for activity. These elements have been referred to by numerous names, but under the current nomenclature they are known as the C1, C2, A, and E elements. The C1, C2 and A elements interact with the transcription factors, MafA, Pax-6 and Pdx-1, respectively, whereas the E elements bind the basic helix-loop-helix proteins BETA2, also known as NeuroD, E12/47 and USF (Hay and Docherty 2006; Melloul et al. 2002; Sander and German 1997). We have previously shown using a combination of gel retardation and chromatin immunoprecipitation (ChIP) assays that the IGRP promoter, like the insulin promoter, binds Pax-6, Pdx-1, BETA2, E12/47 and USF (Martin, et al. 2004; Martin, et al. 2003). To complete this comparison of the cis-acting elements between the two promoters the possibility that the equivalent of the C1 element existed in the IGRP promoter was investigated. By manually scanning the proximal IGRP promoter region it was discovered that a sequence closely resembling the C1 element, which binds MafA, is located between −186 and −157, relative to the transcription start site. We show here that MafA binds the endogenous IGRP gene and stimulates IGRP promoter activity. In addition, a binding site for a member of the Foxa family, formerly known as hepatocyte nuclear factor-3, was previously identified in the IGRP promoter located between −246 and −221 (Bischof, et al. 2001). This element corresponded to both DNA-protein contacts identified with an in situ footprinting technique and a transcriptionally active region detected by a 5′ deletion analysis (Bischof et al. 2001). We show here that Foxa2 also binds the endogenous IGRP gene and stimulates IGRP promoter activity. Foxa2 has been previously shown to be important for both Pdx-1 (Samaras, et al. 2002; Wu, et al. 1997) and MafA (Raum, et al. 2006) expression in islet β cells. These data suggest that the same group of islet-enriched transcription factors, namely Pdx-1, Pax-6, MafA, BETA2 and Foxa2 directly or indirectly regulate expression of the two major autoantigens in type 1 diabetes.
Materials and Methods
Materials
[α-32P]dATP (>3000 Ci mmol−1) and [3H] acetic acid, sodium salt (>10 Ci mmol−1) were obtained from Amersham and ICN, respectively. Antisera raised against Foxa2 (HNF-3β) (sc-65540) and c-Maf (sc-7866) were obtained from Santa Cruz Biotechnology, Inc. Antisera specific for MafA was a generous gift from Roland Stein (Matsuoka, et al. 2003).
Fusion Gene Plasmid Construction
The construction of mouse IGRP-chloramphenicol acetyltransferase (CAT) fusion genes, containing the wild type promoter sequence from −306 to +3, or the same sequence with site-directed mutations (SDMs) of four Pdx-1 binding sites, designated IGRP Quad SDM, have been previously described (Ebert, et al. 1999; Martin et al. 2004). A three step PCR strategy (Martin et al. 2004) was used to create SDMs of the Foxa2 and MafA binding sites in the IGRP promoter. The resulting constructs, designated IGRP Foxa SDM and IGRP Maf SDM, were generated within the context of the −306 to +3 IGRP promoter fragment. The sequences of the sense strand oligonucleotides used for mutagenesis were as follows (mutated nucleotides in bold lowercase): for IGRP Foxa SDM: 5′-TATGAAAAATTGCAAcgAAACACGATCCAACT-3′. For IGRP Maf SDM: 5′-AAATTGCCCCAGGTCctCATATACTGCACCTC-3′.
Cell Culture, Transient Transfection, CAT and Luciferase Assays
Hamster insulinoma tumor (HIT) and βTC-3 cells were grown in DMEM containing 2.5% (vol/vol) fetal bovine serum and 15% (vol/vol) horse serum whereas Min6 cells were grown in DMEM containing 15% (vol/vol) fetal bovine serum and HeLa cells were grown in DMEM containing 10% (vol/vol) bovine serum. Cells were transfected using lipofectamine as previously described (Martin et al. 2002). The pcDNA3-Pax6, pCMV4-Pdx-1, pcDNA3-BETA2, pcDNA3-MafA and pcDNA3-E47 expression vectors were generous gifts from Dr. Roland Stein (Qiu, et al. 2002; Zhao, et al. 2005). The pcDNA3-Foxa2 expression vector was previously described (Vander Kooi, et al. 2005). CAT, luciferase and protein assays were all performed as previously described (Martin et al. 2004; Vander Kooi et al. 2005).
Gel Retardation Assay
A) Labeled Probes: Sense and anti-sense oligonucleotides representing wild-type or mutant Foxa or MafA binding sites (see Table 1) were synthesized with BamH I compatible ends and subsequently gel purified, annealed, and labeled with [α32P]dATP using the Klenow fragment of Escherichia coli DNA Polymerase I to a specific activity of approximately 2.5 μCi/pmol (Sambrook 1989).
Table 1. Oligonucleotides used in gel retardation studies.
The sense strand sequence of the wild type (WT) and mutant (MUT) oligonucleotides used in these studies are shown. The Foxa and Maf binding motifs are boxed and the altered base pairs in the mutant IGRP sequence are shown in bold lower case letters. The consensus Foxa binding motif is taken from Ref. (Overdier, et al. 1994) and the two consensus Maf binding motifs are taken from Ref. (Kataoka, et al. 1994).

|
B) Nuclear Extract Preparation: The preparation of βTC-3 nuclear extracts through extraction of nuclei with 800 mM (high salt) or 200 mM (low salt) NaCl was as previously described (Martin et al. 2004).
C) Binding Assays: ~14 fmol of radiolabeled probe (~50,000 cpm) was incubated with the indicated nuclear extract in a final 20 μl reaction volume. Foxa2 binding reactions contained 1.5 μg high salt nuclear extract, 20 mM Hepes pH 7.9, 0.1 mM EDTA, 0.1 mM EGTA, 10% glycerol (v/v), 1 mM dithiothreitol, 1 μg poly(dI-dC)·poly(dI-dC) and 100 mM KCl. MafA binding reactions contained 3.0 μg low salt nuclear extract, 20 mM Hepes pH 7.9, 0.1 mM EDTA, 1 mM EGTA, 12.5% glycerol (v/v), 1 mM dithiothreitol, 1 μg poly(dI-dC)·poly(dI-dC), 0.375 mM spermidine, 0.075 mM spermine, 100 mM NaCl and 5 mM MgCl2. After incubation at room temperature for 20 min, samples were loaded on to a 6% polyacrylamide gel containing 1X TGE (25 mM Tris Base, 190 mM glycine, 1 mM EDTA) and 2.5% (v/v) glycerol. Samples were electrophoresed for 1.5 hrs at 150V in 1X TGE buffer before the gel was dried and exposed to Kodak XB film with intensifying screens. Competition and supershift experiments were performed as previously described (Martin et al. 2004).
Chromatin Immunoprecipitation Assay
Chromatin Immunoprecipitation (ChIP) assays were performed as previously described (Martin et al. 2004) using 10 ug of each antiserum.
Statistical Analysis
The transfection data were analyzed for differences from the control values, as specified in the Figure Legends. Statistical comparisons were calculated using an unpaired Student’s t test. The level of significance was p < 0.05 (two-sided test).
Results
MafA Binds to the IGRP Promoter In Vitro
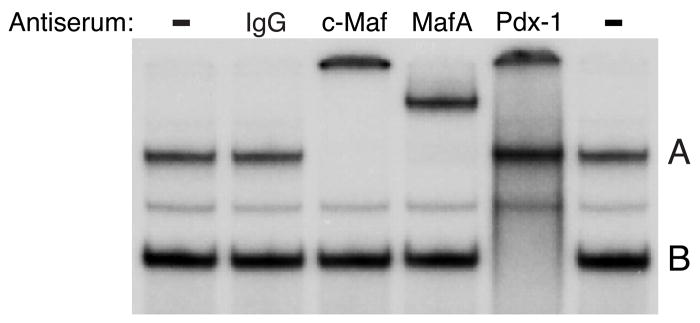
To test the possibility that a C1 element-like sequence in the IGRP promoter interacts with MafA gel retardation analyses were performed. When a labeled oligonucleotide, designated IGRP Maf WT (Table 1), representing the mouse IGRP sequence between −186 and −157, was incubated with crude βTC-3 nuclear extract two major protein-DNA complexes, designated A and B, were detected (Fig. 1). To identify the factor(s) present in complexes A and B, gel retardation assays were performed in which βTC-3 cell nuclear extract was pre-incubated with specific antisera. Previous gel retardation experiments had demonstrated Pdx-1 binding to an oligonucleotide, designated Site 2, representing the −200 to −173 IGRP promoter sequence (Martin et al. 2004). Since this region partially overlaps the −186 and −157 sequence encompassed by the IGRP Maf WT probe it was likely that either complex A or B represented Pdx-1 binding. Figure 1 shows that the addition of a control antiserum (IgG) had no effect on the formation or mobility of complexes A or B whereas addition of antiserum recognizing Pdx-1 selectively altered the mobility of complex B resulting in the appearance of a supershifted complex, while complex A was unaffected.
Figure 1. MafA and Pdx-1 bind the−186/−157 IGRP promoter region in vitro.
βTC-3 nuclear extract was incubated in the absence (−) or presence of the indicated anti-serum for 10 minutes on ice. A labeled oligonucleotide representing the wild-type −186/−157 IGRP promoter region (Maf WT; Table 1) was then added and incubation continued for 20 min at room temperature. Protein binding was then analyzed using the gel retardation assay as described in Materials and Methods. In the representative audioradiograph shown, only the retarded complexes are visible and not the free probe, which was present in excess.
Various antisera were tested to definitively determine whether MafA or a related protein was present in complex A. One of these antisera was raised to the c-Maf protein, which, like MafA, is a member of the large Maf family of transcription factors (Aramata, et al. 2007). This antiserum was raised to a portion of the c-Maf protein common to all the members of this family and as such is able to cross-react with other large Mafs. Figure 1 shows that the addition of c-Maf antibody specifically altered the mobility of complex A leading to the appearance of a supershifted complex. While this result indicated the binding of a large Maf protein, an antiserum that selectively recognizes MafA (Matsuoka et al. 2003) was used to address the specific presence of this protein. Similar to the effects of the c-Maf antiserum, the MafA antiserum specifically altered the mobility of complex A leading to the appearance of a supershifted complex. These results strongly suggest that MafA can interact with the −186/−157 region in the IGRP promoter in vitro.
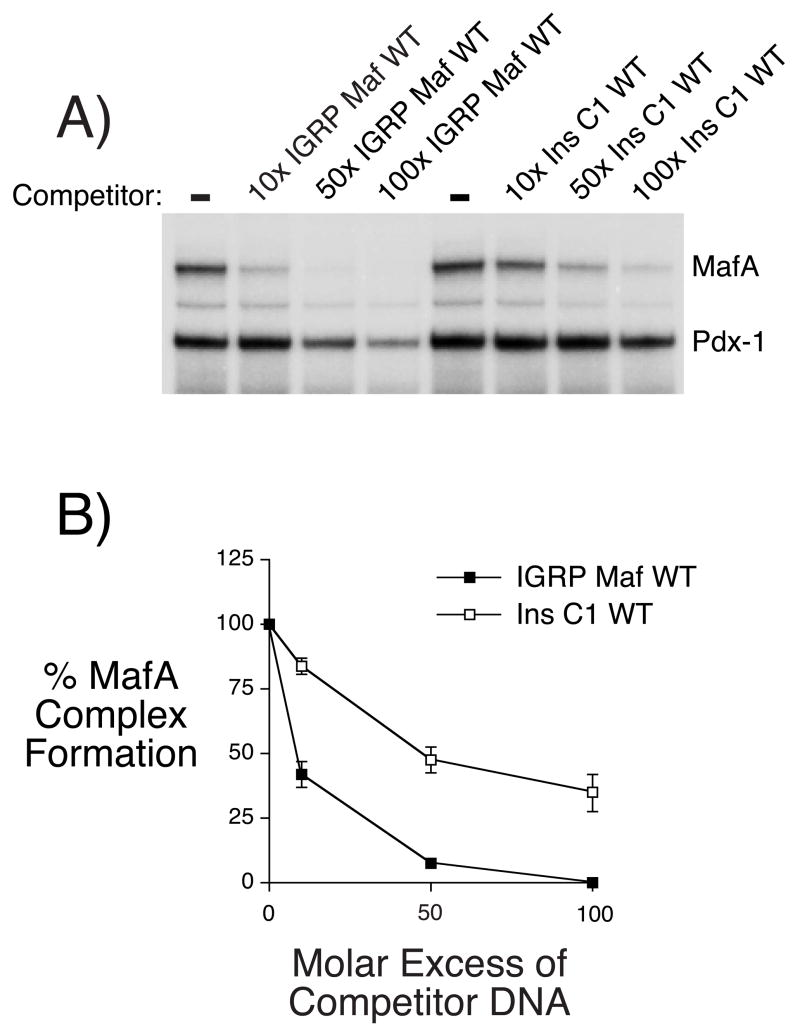
Competition experiments, in which a varying molar excess of unlabeled DNA was included with the labeled probe, were used to compare the affinity of MafA binding to the −186/−157 IGRP promoter region and the rat insulin II C1 element. Figure 2A shows that both the IGRP Maf WT oligonucleotide and an oligonucleotide representing the rat insulin II C1 element competed effectively for the formation of the MafA-DNA complex. However, quantitation of the results of several experiments showed that MafA binds the −186/−157 IGRP promoter region with approximately 5–10 fold higher affinity than the rat insulin II C1 element (Fig. 2B). In contrast, we have previously shown that Pax-6 binds the rat insulin I C2 element with an approximately 5 fold higher affinity than the −270/−246 Pax-6 binding site in the IGRP promoter (Martin et al. 2004).
Figure 2. Comparison of MafA binding to the−186/−157 IGRP promoter region and rat insulin II promoter C1 element in vitro.
Panel A: A labeled oligonucleotide representing the wild-type −186/−157 IGRP promoter region (IGRP Maf WT; Table 1) was incubated in the absence (−) or presence of the indicated molar excess of the unlabeled IGRP MafA WT or Ins C1 WT oligonucleotide (Table 1) competitors prior to the addition of βTC-3 cell nuclear extract. Protein binding was then analyzed using the gel retardation assay as described in Materials and Methods. In the representative autoradiograph shown only the retarded complexes are visible and not the free probe, which was present in excess. The MafA and Pdx-1 complexes are indicated (see Fig. 1).
Panel B: Protein binding was quantified by using a Packard Instant Imager to count 32P associated with the retarded complex. The data represents the mean + S.E.M. of three experiments.
MafA Binding is Required For Maximal IGRP Promoter Activity
Determining the contribution of MafA binding to IGRP promoter activity required the identification of a mutation that would specifically affect the binding of MafA while leaving Pdx-1 binding to the adjacent promoter sequence unaltered. Gel retardation assays were used to identify such a mutant. It was previously established that mutation of the dinucleotide AG pair at the 3′ end of the insulin C1 element disrupts MafA binding (Harrington and Sharma 2001). Figure 3 shows that while a 100-fold molar excess of the unlabeled IGRP Maf WT oligonucleotide competed effectively for the formation of the MafA-DNA complex an oligonucleotide, designated IGRP Maf MUT, that contains a mutation in the analogous dinucleotide AG pair in the IGRP Maf element (Table 1), failed to compete with the labeled probe for formation of the MafA-DNA complex. Both the IGRP Maf WT and MUT oligonucleotides compete equally effectively for formation of the Pdx-1-DNA complex (Fig. 3). This indicates that the AG mutation specifically disrupts MafA and not Pdx-1 binding.
Figure 3. Identification of a mutation that selectively disrupts MafA and not Pdx-1 binding.
A labeled oligonucleotide representing the wild-type −186/−157 IGRP promoter region (IGRP Maf WT) was incubated in the absence or presence of a 100-fold molar excess of the unlabeled IGRP Maf WT or IGRP Maf MUT (Table 1) competitors. βTC-3 nuclear extract was then added and protein binding was analyzed using the gel retardation assay as described in Materials and Methods. In the representative audioradiograph shown, only the retarded complexes are visible and not the free probe, which was present in excess. The MafA and Pdx-1 complexes are indicated (see Fig. 1).
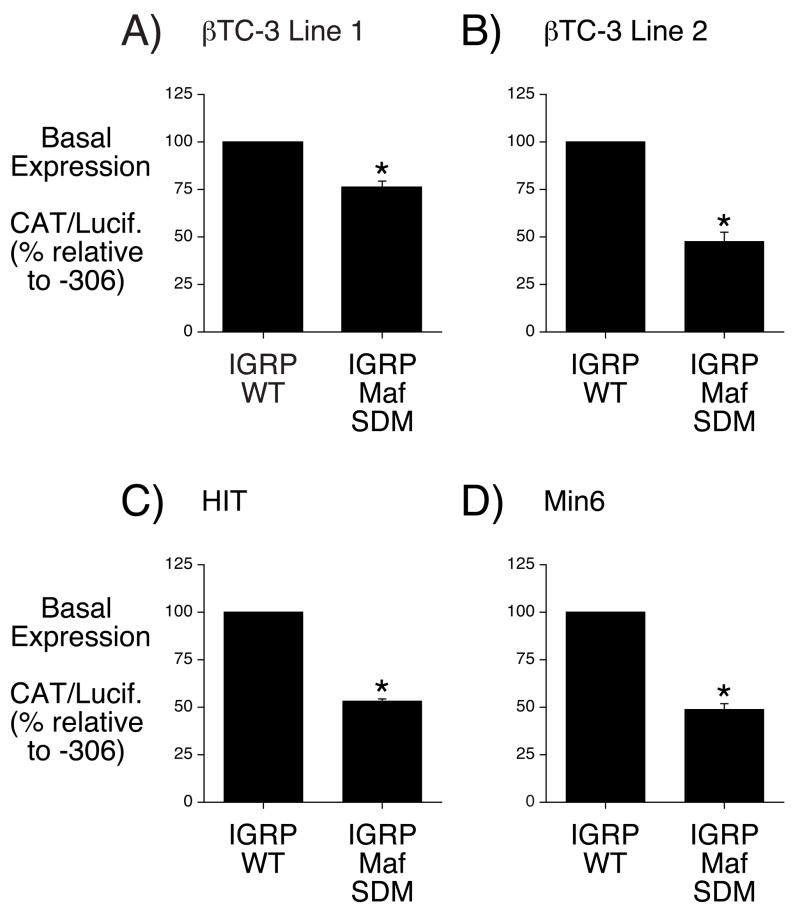
To investigate the functional significance of MafA binding, the AG dinucleotide was mutated, using site-directed mutagenesis (SDM), in the context of the −306 to +3 IGRP promoter region. A fusion gene containing this mutation, designated IGRP Maf SDM, was then analyzed by transient transfection of βTC-3, HIT and Min6 cells. Figures 4A & B show that mutation of the MafA binding site resulted in an approximately 25–50% reduction in the level of reporter gene expression in βTC-3 cells as compared to that directed by the wild type −306/+3 promoter. The magnitude of the effect of the MafA binding site mutation varied between βTC-3 cells that had been cultured for different passages (Fig. 4A & B). Figures 4C & D show that mutation of the MafA binding site resulted in an approximately 50% reduction in the level of reporter gene expression in both HIT and Min6 cells, respectively. These results demonstrate that MafA is important for IGRP promoter activity in multiple islet-derived cell lines.
Figure 4. Disruption of MafA binding reduces IGRP promoter activity.
Two separate batches of βTC-3 cells (Panels A & B), HIT cells (Panel C) and Min6 cells (Panel D) were transiently co-transfected, as described in Materials and Methods, using a lipofectamine solution containing various IGRP-CAT fusion genes (2 μg) and an expression vector encoding firefly luciferase (0.5 μg). The IGRP-CAT fusion genes represented either the wild-type promoter sequence, located between −306 and +3 (IGRP WT), or the same sequence with a site-directed mutation (SDM) in the Maf A binding site (IGRP Maf SDM). The mutation was identical to that used in the gel retardation analysis (Panel A). Following transfection, cells were incubated for 18–20 hr in serum-containing medium. The cells were then harvested and both CAT and luciferase activity was assayed as described in Materials and Methods. Results are presented as the ratio of CAT:firefly luciferase activity, expressed as a percentage relative to the value obtained with the IGRP WT fusion gene, and represent the mean of 3 experiments ± S.E.M., each using an independent preparation of each fusion gene plasmid, assayed in triplicate. *, p < 0.05 versus IGRP WT.
Foxa2 Binds to the IGRP Promoter In Vitro
We next examined Foxa2 binding to a previously identified site in the IGRP promoter (Ref. (Bischof et al. 2001); Table 1). When a labeled oligonucleotide, designated IGRP Foxa WT (Table 1), representing the mouse IGRP sequence between −246 and −221, was incubated with crude βTC-3 nuclear extract two major protein-DNA complexes, designated X and Y, were detected (Fig. 5A). To identify the factor(s) present in complexes X and Y, gel retardation assays were performed in which βTC-3 cell nuclear extract was pre-incubated with specific antisera. Figure 5A shows that the addition of a control antiserum (IgG) had no effect on the mobility of complex X or Y whereas addition of antiserum recognizing Foxa2 specifically altered the mobility of complex Y leading to the appearance of a supershifted complex, while complex X was unaffected.
Figure 5. Foxa2 binds the−246/−221 IGRP promoter region in vitro.
Panel A: βTC-3 nuclear extract was incubated in the absence (−) or presence of the indicated anti-serum for 10 minutes on ice. A labeled oligonucleotide representing the wild-type −246/−221 IGRP promoter region (IGRP Foxa WT; Table 1) was then added and incubation continued for 20 min at room temperature. Protein binding was then analyzed using the gel retardation assay as described in Materials and Methods. In the representative audioradiograph shown, only the retarded complexes are visible and not the free probe, which was present in excess. The two major complexes detected were designated X and Y. The arrow indicates a supershifted complex.
Panel B: The labeled oligonucleotide representing the wild-type −246/−221 IGRP promoter region (IGRP Foxa WT) was incubated in the absence or presence of a 100-fold molar excess of the unlabeled IGRP Foxa WT or IGRP Foxa MUT (Table 1) competitors. βTC-3 nuclear extract was then added and protein binding was analyzed using the gel retardation assay as described in Materials and Methods. In the representative audioradiograph shown, only the retarded complexes are visible and not the free probe, which was present in excess.
The specificity of this interaction was examined by testing the ability of Foxa2 to bind to an altered version of this element that contained two point mutations at positions implicated in protein binding by the in situ footprinting technique (Ref. (Bischof et al. 2001); Table 1). Figure 5B shows that while a 100-fold molar excess of the unlabeled IGRP Foxa WT oligonucleotide competed effectively for the formation of both complexes X and Y, an oligonucleotide, designated IGRP Foxa MUT, that contains a mutation in the two bases implicated in protein binding by the in situ footprinting (Ref. (Bischof et al. 2001); Table 1), failed to compete with the labeled probe for formation of the Foxa2-DNA complex (complex Y) while competition for formation of complex X was unaffected. This indicates that the mutation specifically disrupts Foxa2 and that complex X either represents a non-specific interaction or binding to another region of the probe.
A similar competition analysis previously indicated that these mutations only partially disrupt Foxa2 binding (Bischof et al. 2001). Subsequently, however, it was discovered that the formation of a non-specific complex that co-migrates with the Foxa2 complex had complicated the interpretation of this initial result. In the experiment shown in Figure 5 modified gel retardations conditions were used that did not allow the formation of this non-specific complex making it possible to demonstrate that the described mutations are more disruptive than previously thought (Bischof et al. 2001).
Foxa2 Binding is Required For Maximal IGRP Promoter Activity
To investigate the functional significance of these observations the Foxa2 binding site was mutated using site-directed mutagenesis (SDM) in the context of the −306 to +3 IGRP promoter region. A fusion gene containing this mutation, designated IGRP Foxa2 SDM, that contains a mutation identical to that described in the Foxa MUT oligonucleotide, was then analyzed by transient transfection of βTC-3, HIT and Min6 cells. Figures 6A & B show that mutation of the Foxa2 binding site resulted in an approximately 25–75% reduction in the level of reporter gene expression in βTC-3 cells as compared to that directed by the wild type −306/+3 promoter. As seen with the effect of the MafA binding site mutation, the magnitude of the effect of the Foxa2 binding site mutation varied between βTC-3 cells that had been cultured for different passages (Fig. 6A & B). Figures 6C & D show that mutation of the Foxa2 binding site resulted in an approximately 75% reduction in the level of reporter gene expression in both HIT and Min6 cells, respectively. These results demonstrate that Foxa2 is important for IGRP promoter activity in multiple islet-derived cell lines.
Figure 6. Disruption of Foxa2 binding reduces IGRP promoter activity.
Two separate batches of βTC-3 cells (Panels A & B), HIT cells (Panel C) and Min6 cells (Panel D) were transiently co-transfected, as described in Materials and Methods, using a lipofectamine solution containing various IGRP-CAT fusion genes (2 μg) and an expression vector encoding firefly luciferase (0.5 μg). The IGRP-CAT fusion genes represented either the wild-type promoter sequence, located between −306 and +3 (IGRP WT), or the same sequence with a site-directed mutation (SDM) in the Foxa2 binding site (IGRP Foxa SDM). The mutation was identical to that used in the gel retardation analysis (Fig. 4B). Following transfection, cells were incubated for 18–20 hr in serum-containing medium. The cells were then harvested and both CAT and luciferase activity was assayed as described in Materials and Methods. Results are presented as the ratio of CAT:firefly luciferase activity, expressed as a percentage relative to the value obtained with the −306 WT fusion gene, and represent the mean of 3 experiments ± S.E.M., each using an independent preparation of each fusion gene plasmid, assayed in triplicate. *, p < 0.05 versus IGRP WT.
Foxa2 and a Maf Family Member Interact with the IGRP Promoter In Situ
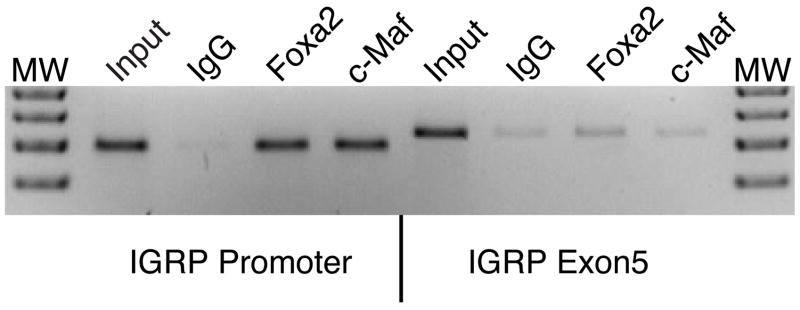
The gel retardation studies demonstrate that MafA and Foxa2 are capable of specifically binding to the IGRP promoter in vitro. To determine if these interactions occur on the IGRP promoter inside intact cells, a chromatin immunoprecipitation assay (ChIP) was performed. βTC-3 cells were treated with formaldehyde to preserve DNA-protein interactions and chromatin was subsequently isolated from these cells. The isolated chromatin was subjected to sonication to produce genomic fragments that were approximately 500 bp’s in size and this material served as the input for immunoprecipitation experiments. Antisera raised to c-Maf and Foxa2, the same antibodies described in the gel retardation experiments, were incubated with the chromatin and the resulting immune complexes were precipitated by centrifugation. The presence of the IGRP promoter was then assessed by isolating genomic DNA from the immunopellets and performing a PCR reaction with primers that specifically recognize the IGRP promoter. Figure 7 shows that the IGRP promoter is clearly enriched in the c-Maf and Foxa2 immunopellets compared to the IgG control. A similar result was seen using the MafA-specific antibody (data not shown). To test the specificity of this interaction, the immunopellets were also analyzed for the presence of a target not predicted to associate with either of these transcription factors. In this instance the control target chosen was IGRP exon 5, which is approximately 6 kilobase pairs downstream from the transcription start site. As expected, the PCR analyses show that this genomic region is not enriched in the various immunopellets (Fig. 7), confirming that c-Maf and Foxa2 associate specifically with the IGRP promoter. In summary, the ChIP experiments extend the in vitro binding studies by demonstrating that a Maf family member and Foxa2 associate with the IGRP promoter inside intact cells.
Figure 7. The IGRP promoter binds Foxa2 and a Maf family member in situ.
Foxa2 and Maf binding to the IGRP promoter were analyzed in situ using the chromatin immunoprecipitation (ChIP) assay. Chromatin from formaldehyde-treated βTC-3 cells was immunoprecipitated using anti-Foxa2 or anti-c-Maf antibodies or, as a control, using IgG. The presence of the IGRP promoter and exon 5 in the chromatin preparation prior to immunoprecipitation (1:100 input) and in the immunoprecipitates was then assayed using PCR as described in Materials and Methods. The autoradiograph shown is representative of 3 different experiments. Both the IGRP promoter and exon 5 are amplified with similar efficiencies in the chromatin preparation (Martin et al. 2003). MW; molecular weight.
Pdx-1 Binding is Required For Maximal IGRP Promoter Activity in HIT and Min6 cells
Previously published data showed that mutation of four Pdx-1 binding sites in the IGRP promoter had little effect on IGRP fusion gene expression in βTC-3 cells (Martin et al. 2004). However, ChIP assays show that this factor associates with the IGRP promoter in both βTC-3 (Martin et al. 2004) and NIT-1 (Keller, et al. 2007) cells. While MafA and Foxa2 binding are important for IGRP promoter activity in βTC-3, HIT and Min6 cells we investigated whether Pdx-1 binding was selectively important for IGRP promoter activity in HIT and Min6 cells. A fusion gene containing a mutation of all four Pdx-1 binding sites, designated IGRP Quad SDM, was analyzed by transient transfection. Figure 8 shows that mutation of the Pdx-1 binding sites resulted in an approximately 50% reduction in the level of reporter gene expression as compared to that directed by the wild type −306/+3 promoter in HIT and Min6 cells, respectively, whereas this mutation has little effect in βTC-3 cells.
Figure 8. Disruption of Foxa2 binding reduces IGRP promoter activity in HIT and Min6 but not βTC-3 cells.
βTC-3 cells (Panel A), HIT cells (Panel B) and Min6 cells (Panel C) were transiently co-transfected, as described in Materials and Methods, using a lipofectamine solution containing various IGRP-CAT fusion genes (2 μg) and an expression vector encoding firefly luciferase (0.5 μg). The IGRP-CAT fusion genes represented either the wild-type promoter sequence, located between −306 and +3 (IGRP WT), or the same sequence with a site-directed mutation (SDM) in the four Pdx-1 binding sites (IGRP Quad SDM). Following transfection, cells were incubated for 18–20 hr in serum-containing medium. The cells were then harvested and both CAT and luciferase activity was assayed as described in Materials and Methods. Results are presented as the ratio of CAT:firefly luciferase activity, expressed as a percentage relative to the value obtained with the −306 WT fusion gene, and represent the mean of 3 experiments ± S.E.M., each using an independent preparation of each fusion gene plasmid, assayed in triplicate. *, p < 0.05 versus IGRP WT.
Islet-Enriched Factors Synergistically Induce IGRP Fusion Gene Expression
Previously published data and that shown in Figures 1–8 demonstrate that Pax-6, BETA2, Foxa2, MafA and Pdx-1 are important for IGRP gene expression. Co-transfection experiments were performed in cervix-derived HeLa cells, in which neither these islet-enriched transcription factors nor the IGRP gene are likely to be endogenously expressed. Several other groups have previously used this strategy to investigate the mechanism by which islet-enriched factors activate gene expression (Docherty, et al. 2005; Qiu et al. 2002). Expression vectors for Pax-6, Pdx-1, Foxa2, MafA, BETA2 and E47, a ubiquitously expressed binding partner for BETA2 (Qiu et al. 2002), were co-transfected with various −306/+3 IGRP fusion genes. Since cryptic transcription factor binding sites have been reported in the commonly used pGL2 and pGL3 luciferase vectors (Thirunavukkarasu, et al. 2000) we first compared fusion genes in which the −306/+3 IGRP promoter fragment was ligated into either the pGL2, pGL3 or pCAT(An) vectors. Supplemental Figure 1 shows that co-transfection with the transcription factor expression vectors strongly induced pGL2 and pGL3 expression, regardless of the presence of the IGRP promoter. In contrast, there was no effect on CAT expression in the absence of the IGRP promoter. The −306/+3 IGRP-CAT fusion gene was therefore used in subsequent experiments.
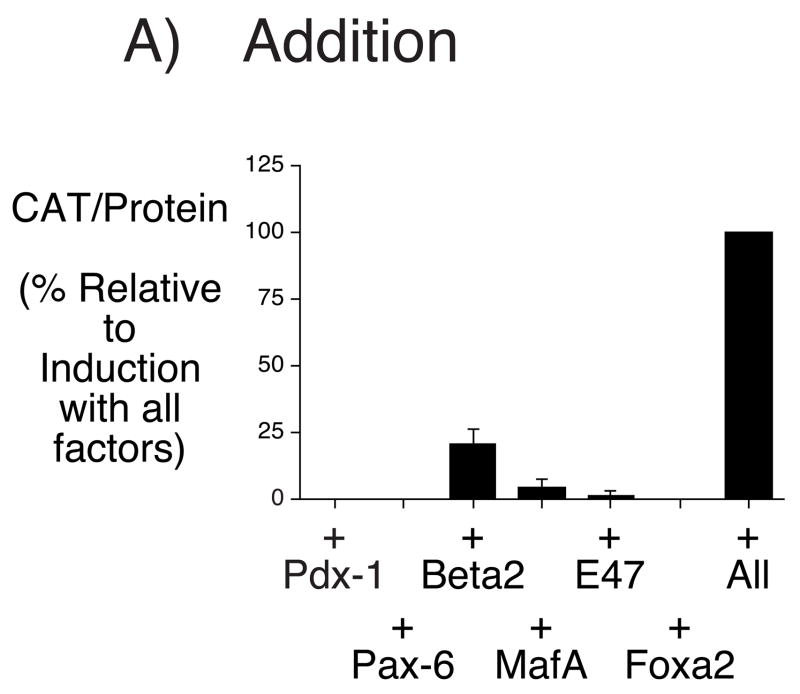
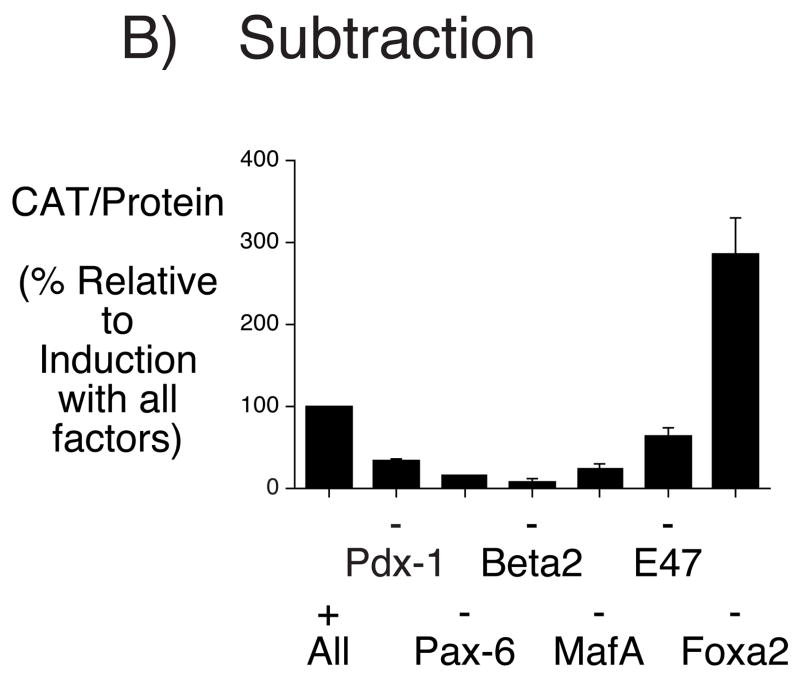
Figure 9A shows that, individually, with the exception of BETA2, none of these transcription factors markedly induced IGRP fusion gene expression whereas together they acted synergistically to strongly induce expression. We next investigated the individual contributions of each factor to this synergistic induction by assessing the effect of subtracting each one on the induction of IGRP fusion gene expression. The individual absence of Pdx-1, Pax-6, BETA2 and MafA resulted in a marked reduction in IGRP fusion gene expression (Fig. 9B). This result confirms the synergistic nature of the interactions between Pax-6, BETA2, MafA and Pdx-1. Surprisingly, the absence of Foxa2 resulted in increased expression (Fig. 9B). This suggests that, for reasons that are unclear, Foxa2 appears to repress IGRP fusion expression in HeLa cells even though mutation of the Foxa2 binding site in the IGRP promoter clearly results in reduced expression in βTC-3, Min6 and HIT cells (Fig. 6).
Figure 9. Islet-enriched transcription factors synergistically activate IGRP fusion gene transcription in HeLa cells.
HeLa cells were transiently co-transfected, as described in Materials and Methods, using a lipofectamine solution containing the −306/+3 IGRP-CAT fusion gene (2 μg) and expression vectors (0.1 μg each) encoding the indicated transcription factors or the matching empty vector pCMV4/pcDNA3 controls. Panel A shows the effect of adding individual factors whereas Panel B shows the effect of removing individual factors. Following transfection, cells were incubated for 18–20 hr in serum-free medium. The cells were then harvested and both CAT activity and the protein concentration of the cell lysate were assayed as described in Materials and Methods. Results are presented as the ratio of CAT activities, corrected for the protein concentration in the cell lysate, relative to that obtained in the presence of all factors, expressed as a percentage, and represent the mean ± S.E.M. of three experiments, each using an independent preparation of the fusion gene plasmid with each condition assayed in triplicate.
Pdx-1 does not directly regulate IGRP fusion gene expression in HeLa cells
While Pdx-1 acts synergistically with Pax-6, MafA, BETA2 and E47 to induce expression of the wild type −306/+3 IGRP fusion gene (Fig. 10A) surprisingly this induction was not impaired comparing the wild type fusion gene with the −306/+3 IGRP Quad SDM fusion gene, that contains combined mutations of all four Pdx-1 binding sites (Fig. 10B). This result suggested that perhaps Pdx-1 stimulated IGRP fusion gene expression through a DNA binding-independent mechanism in HeLa cells. However, Figure 10C shows that this is also not the case because the induction of −306/+3 IGRP Quad SDM fusion gene expression is not dependent on Pdx-1. Taken together, these data indicate that Pdx-1 must be stimulating wild type −306/+3 IGRP fusion gene expression in HeLa cells by displacing an endogenous repressor rather than directly stimulating expression itself. This observation reveals a limitation in the analysis of islet-enriched transcription factors in non-islet derived cell lines and highlights the importance of control experiments to demonstrate the involvement of DNA binding.
Figure 10. Pdx-1 does not directly regulate IGRP fusion gene expression in HeLa cells.
HeLa cells were transiently co-transfected, as described in Fig. 8, with various IGRP-CAT fusion genes (2 μg) and expression vectors (0.1 μg) encoding Pax-6, MafA, BETA2 and E47 and either pCMV4-Pdx-1 (+ All) or pCMV4 alone (−Pdx-1). The IGRP-CAT fusion genes represented either the wild-type (WT) promoter sequence, located between −306 and +3, or the same sequence with site-directed mutation (SDM) in the four Pdx-1 binding sites (Quad SDM). Following transfection, cells were incubated for 18–20 hr in serum-free medium. The cells were then harvested and both CAT activity and the protein concentration of the cell lysate were assayed as described in Materials and Methods. Results are presented as the ratio of CAT activities, corrected for the protein concentration in the cell lysate, relative to that obtained with the WT fusion gene in the presence of all factors, expressed as a percentage, and represent the mean ± S.E.M. of three experiments, each using an independent preparation of each fusion gene plasmids, with each condition assayed in triplicate.
Discussion
We have been interested in understanding the molecular mechanisms that determine the islet-specific expression of the IGRP gene (Wang, et al. 2008). A 306 base pair IGRP promoter region upstream of the transcription start site is sufficient to drive expression of reporter genes specifically in β cell-derived cell lines in situ and newborn pancreatic islets in vivo (Frigeri, et al. 2004). The results of a 5′ deletion analysis, coupled with in situ footprinting, indicated that multiple cis-acting elements within this 306 base pair region contribute to IGRP promoter activity (Bischof et al. 2001). Subsequent studies demonstrated the importance of two conserved E-Box motifs (Martin et al. 2003) and two non-consensus Pax-6 binding sites (Martin et al. 2004) for IGRP promoter activity. The results presented here show that the IGRP promoter also binds the islet-enriched transcription factors Foxa2 and MafA in intact cells (Fig. 7) and that mutation of the binding sites for MafA (Figs. 1–3) and Foxa2 (Fig. 5) identified in the proximal IGRP promoter results in decreased IGRP fusion gene expression in βTC-3, HIT and Min6 cells (Figs. 4 and 6). In addition, while mutation of the four Pdx-1 binding sites in the IGRP promoter had little effect on fusion gene expression in βTC-3 cells, it resulted in reduced expression in both HIT and Min6 cells (Fig. 8). Why Pdx-1 binding would be selectively important for IGRP fusion gene expression in HIT and Min6 but not βTC-3 cells is currently unknown. However, these results suggest that the same islet-enriched transcription factors regulate, directly or indirectly, the promoter activity of the two major autoantigens in type 1 diabetes, namely insulin and IGRP.
The Foxa proteins, of which there are three isoforms Foxa1–3, previously designated HNF-3α, β, and γ, have previously been studied in the context of the pancreas. Promoter analyses, for example, have implicated Foxa1 in the regulation of glucagon gene expression and, accordingly, mice that are deficient in this factor exhibit impaired glucagon synthesis and are hypoglycemic (Gauthier, et al. 2002; Kaestner, et al. 1999). Similar types of analyses have shown that Foxa2 regulates the expression of Pdx-1 (Samaras et al. 2002; Wu et al. 1997). Because Foxa2 is required for the formation of the foregut endoderm, a global knockout approach could not be used to assess the role of this transcription factor in pancreas function (Ang and Rossant 1994). To circumvent these early embryonic defects a conditional allele was introduced into mice that allowed the inactivation of Foxa2 later in development and specifically in pancreatic β-cells (Sund, et al. 2001). Interestingly, like Foxa1 knockout mice (Kaestner et al. 1999), the conditional Foxa2 knockout mice are severely hypoglycemic (Sund et al. 2001). In this case however, the mice exhibited hyperinsulinemia owing to dysregulation of the insulin secretory pathway (Sund et al. 2001). Further investigations revealed this phenotype could be at least partly explained by decreased expression of the KATP channel subunits, Sur1 and Kir6.2 (Lantz, et al. 2004; Sund et al. 2001), Pdx-1 (Lee, et al. 2002) and altered insulin secretory vesicle docking (Gao, et al. 2007). These results are generally consistent with what was observed when a dominant negative form of Foxa2 was overexpressed in INS-1 cells (Wang, et al. 2002). In these experiments the expression of various genes involved in the insulin secretory pathway, as well as other previously identified targets of Foxa2 were measured under conditions where Foxa2 was functionally impaired. Consistent with the in vivo experiments, the KATP channel subunits were down regulated, various genes that play a positive role in insulin secretion were upregulated and there was a leftward shift in glucose-stimulated insulin secretion (Wang et al. 2002). However, in contrast to the in vivo experiments (Lee et al. 2002), Pdx-1 expression was unaltered (Wang et al. 2002). Overall these data suggest that Foxa2 has a negative impact on insulin secretion. Although these studies did not report whether IGRP expression was altered, a positive role for Foxa2 in IGRP gene expression would be consistent with this concept given that IGRP appears to negatively effect stimulus-secretion coupling (Wang et al. 2007b).
While Maf family proteins have long been known to play a role in lens development (Reza and Yasuda 2004), the role of MafA in the context of the pancreas has only recently been investigated. The impetus for these studies was the identification of MafA as the factor binding the C1 element, previously referred to as Ripe3b1, in the insulin promoter (Kataoka, et al. 2002; Matsuoka et al. 2003; Olbrot, et al. 2002). Using the C1 element in conjunction with DNA affinity chromatography, two different groups independently purified the basic-leucine zipper protein MafA (Matsuoka et al. 2003; Olbrot et al. 2002). In addition, a third group identified the C1 element-binding protein as MafA by recognizing that this element bears similarity to a MARE (Maf Recognition Element), a consensus sequence recognized by members of the Maf family (Kataoka et al. 2002). The C1 element contributes to basal insulin gene transcription and is also believed to mediate a response to glucose, an effect that is coincident with an increase in binding activity at this element (Sharma and Stein 1994). The mechanism by which glucose regulates gene transcription through MafA is unclear. Glucose does induce MafA mRNA expression (Kataoka et al. 2002) and protein stability (Han, et al. 2007), however, complex serine, threonine (Rocques, et al. 2007) and tyrosine (Matsuoka, et al. 2001) phosphorylation mechanisms are also potentially involved. Interestingly, like insulin, IGRP mRNA levels increase in response to high glucose (Petrolonis et al. 2004) and the work presented here suggests that MafA is a potential candidate for the mediator of this response.
Although MafA is expressed during pancreas development (Matsuoka, et al. 2004; Nishimura, et al. 2006) global deletion of the MafA gene does not appear to affect this process (Zhang, et al. 2005), possibly due to compensation by MafB (Artner, et al. 2007). Nevertheless, adult MafA knockout mice do exhibit glucose intolerance, consistent with the exclusive expression of MafA in pancreatic islet beta cells (Zhang et al. 2005). Interestingly, in contrast to the correlation seen with Foxa2, the stimulatory action of MafA on IGRP fusion gene expression appears inconsistent with this phenotype given that IGRP appears to negatively effect stimulus-secretion coupling (Wang et al. 2007b). However, the expression of multiple genes are likely altered in these animals (Wang, et al. 2007a) such that the contribution of altered expression of individual factors could be negated.
Supplementary Material
Acknowledgments
We thank Roland Stein, Shimon Efrat and Jun-ichi Miyazaki for providing the HIT, βTC-3 and Min6 cell lines, respectively. Research in the laboratory of R.O’B. was supported by NIH grant DK61645 and by NIH grant P60 DK20593, which funds the Vanderbilt Diabetes Center Core Laboratory. Cyrus Martin was supported by the Vanderbilt Viruses, Nucleic Acids and Cancer Training Program (5T32 CA09385).
Footnotes
Disclosure Statement: The authors have nothing to disclose
Publisher's Disclaimer: This is not the definitive version of record of this article. This manuscript has been accepted for publication in Journal of Molecular Endocrinology, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the Society for Endocrinology accepts no responsibility for any errors or omissions it may contain. The definitive version is now freely available at http://dx.doi.org/10.1677/JME-08-0062.R1.
References
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Aramata S, Han SI, Kataoka K. Roles and regulation of transcription factor MafA in islet beta-cells. Endocr J. 2007;54:659–666. doi: 10.1507/endocrj.kr-101. [DOI] [PubMed] [Google Scholar]
- Arden SD, Zahn T, Steegers S, Webb S, Bergman B, O’Brien RM, Hutton JC. Molecular cloning of a pancreatic islet-specific glucose-6-phosphatase catalytic subunit-related protein. Diabetes. 1999;48:531–542. doi: 10.2337/diabetes.48.3.531. [DOI] [PubMed] [Google Scholar]
- Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet beta cell maturation. Proc Natl Acad Sci U S A. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof LJ, Martin CC, Svitek CA, Stadelmaier BT, Hornbuckle LA, Goldman JK, Oeser JK, Hutton JC, O’Brien RM. Characterization of the Mouse Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein Gene Promoter by In Situ Footprinting. Correlation with Fusion Gene Expression in the Islet Derived bTC-3 and Hamster Insulinoma Tumor Cell Lines. Diabetes. 2001;50:502–514. doi: 10.2337/diabetes.50.3.502. [DOI] [PubMed] [Google Scholar]
- Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proenca C, Marchand M, Hartikainen AL, Sovio U, De Graeve F, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- Boustead JN, Martin CC, Oeser JK, Svitek CA, Hunter SI, Hutton JC, O’Brien RM. Identification and characterization of a cDNA and the gene encoding the mouse ubiquitously expressed glucose-6-phosphatase catalytic subunit-related protein. J Mol Endocrinol. 2004;32:33–53. doi: 10.1677/jme.0.0320033. [DOI] [PubMed] [Google Scholar]
- Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, Silver KD, Timpson NJ, Hansen T, Orru M, Grazia Piras M, et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest. 2008;118:2620–2628. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YY, Kim SY, Yiu WH, Pan CJ, Jun HS, Ruef RA, Lee EJ, Westphal H, Mansfield BC, Chou JY. Impaired neutrophil activity and increased susceptibility to bacterial infection in mice lacking glucose-6-phosphatase-beta. J Clin Invest. 2007;117:784–793. doi: 10.1172/JCI30443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty HM, Hay CW, Ferguson LA, Barrow J, Durward E, Docherty K. Relative contribution of PDX-1, MafA and E47/beta2 to the regulation of the human insulin promoter. Biochem J. 2005;389:813–820. doi: 10.1042/BJ20041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert DH, Bischof LJ, Streeper RS, Chapman SC, Svitek CA, Goldman JK, Mathews CE, Leiter EH, Hutton JC, O’Brien RM. Structure and promoter activity of an islet-specific glucose-6- phosphatase catalytic subunit-related gene. Diabetes. 1999;48:543–551. doi: 10.2337/diabetes.48.3.543. [DOI] [PubMed] [Google Scholar]
- Fradin D, Heath S, Lathrop M, Bougneres P. Quantitative trait loci for fasting glucose in young Europeans replicate previous findings for type 2 diabetes in 2q23–24 and other locations. Diabetes. 2007;56:1742–1745. doi: 10.2337/db06-1329. [DOI] [PubMed] [Google Scholar]
- Frigeri C, Martin CC, Svitek CA, Oeser JK, Hutton JC, Gannon M, O’Brien RM. The Proximal Islet-Specific Glucose-6-Phosphatase Catalytic Subunit Related Protein (IGRP) Autoantigen Promoter is Sufficient to Initiate but not Maintain Transgene Expression in Mouse Islets In Vivo. Diabetes. 2004;53:1754–1764. doi: 10.2337/diabetes.53.7.1754. [DOI] [PubMed] [Google Scholar]
- Gao N, White P, Doliba N, Golson ML, Matschinsky FM, Kaestner KH. Foxa2 controls vesicle docking and insulin secretion in mature Beta cells. Cell Metab. 2007;6:267–279. doi: 10.1016/j.cmet.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Gauthier BR, Schwitzgebel VM, Zaiko M, Mamin A, Ritz-Laser B, Philippe J. Hepatic nuclear factor-3 (HNF-3 or Foxa2) regulates glucagon gene transcription by binding to the G1 and G2 promoter elements. Mol Endocrinol. 2002;16:170–183. doi: 10.1210/mend.16.1.0752. [DOI] [PubMed] [Google Scholar]
- Han B, Serra P, Amrani A, Yamanouchi J, Maree AF, Edelstein-Keshet L, Santamaria P. Prevention of diabetes by manipulation of anti-IGRP autoimmunity: high efficiency of a low-affinity peptide. Nat Med. 2005a;11:645–652. doi: 10.1038/nm1250. [DOI] [PubMed] [Google Scholar]
- Han B, Serra P, Yamanouchi J, Amrani A, Elliott JF, Dickie P, Dilorenzo TP, Santamaria P. Developmental control of CD8 T cell-avidity maturation in autoimmune diabetes. J Clin Invest. 2005b;115:1879–1887. doi: 10.1172/JCI24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SI, Aramata S, Yasuda K, Kataoka K. MafA stability in pancreatic beta cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol Cell Biol. 2007;27:6593–6605. doi: 10.1128/MCB.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington RH, Sharma A. Transcription factors recognizing overlapping C1-A2 binding sites positively regulate insulin gene expression. J Biol Chem. 2001;276:104–113. doi: 10.1074/jbc.M008415200. [DOI] [PubMed] [Google Scholar]
- Hay CW, Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006;55:3201–3213. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- Hutton JC, Eisenbarth GS. A pancreatic beta-cell-specific homolog of glucose-6-phosphatase emerges as a major target of cell-mediated autoimmunity in diabetes. Proc Natl Acad Sci U S A. 2003;100:8626–8628. doi: 10.1073/pnas.1633447100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller DM, McWeeney S, Arsenlis A, Drouin J, Wright CV, Wang H, Wollheim CB, White P, Kaestner KH, Goodman RH. Characterization of pancreatic transcription factor Pdx-1 binding sites using promoter microarray and serial analysis of chromatin occupancy. J Biol Chem. 2007;282:32084–32092. doi: 10.1074/jbc.M700899200. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy B, Dudek NL, McKenzie MD, Purcell AW, Brooks AG, Gellert S, Colman PG, Harrison LC, Lew AM, Thomas HE, et al. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J Clin Invest. 2006;116:3258–3265. doi: 10.1172/JCI29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz KA, Vatamaniuk MZ, Brestelli JE, Friedman JR, Matschinsky FM, Kaestner KH. Foxa2 regulates multiple pathways of insulin secretion. J Clin Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Sund NJ, Vatamaniuk MZ, Matschinsky FM, Stoffers DA, Kaestner KH. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- Lieberman SM, Evans AM, Han B, Takaki T, Vinnitskaya Y, Caldwell JA, Serreze DV, Shabanowitz J, Hunt DF, Nathenson SG, et al. Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc Natl Acad Sci U S A. 2003;100:8384–8388. doi: 10.1073/pnas.0932778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CC, Bischof LJ, Bergman B, Hornbuckle LA, Hilliker C, Frigeri C, Wahl D, Svitek CA, Wong R, Goldman JK, et al. Cloning and Characterization of the Human and Rat Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein (IGRP) Genes. J Biol Chem. 2001;276:25197–25207. doi: 10.1074/jbc.M101549200. [DOI] [PubMed] [Google Scholar]
- Martin CC, Oeser JK, O’Brien RM. Differential regulation of islet-specific glucose-6-phosphatase catalytic subunit-related protein gene transcription by Pax-6 and Pdx-1. J Biol Chem. 2004;279:34277–34289. doi: 10.1074/jbc.M404830200. [DOI] [PubMed] [Google Scholar]
- Martin CC, Oeser JK, Svitek CA, Hunter SI, Hutton JC, O’Brien RM. Identification and Characterization of a Human cDNA and Gene Encoding a Ubiquitously Expressed Glucose-6-Phosphatase Catalytic Subunit-Related Protein. J Mol Endocrinol. 2002;29:205–222. doi: 10.1677/jme.0.0290205. [DOI] [PubMed] [Google Scholar]
- Martin CC, Svitek CA, Oeser JK, Henderson E, Stein R, O’Brien RM. Upstream Stimulatory Factor (USF) and NeuroD/BETA2 Contribute toIslet-Specific Glucose-6-Phosphatase Catalytic Subunit Related Protein (IGRP)Gene Expression. Biochem J. 2003;371:675–686. doi: 10.1042/BJ20021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T, Zhao L, Stein R. The DNA binding activity of the RIPE3b1 transcription factor of insulin appears to be influenced by tyrosine phosphorylation. J Biol Chem. 2001;276:22071–22076. doi: 10.1074/jbc.M010321200. [DOI] [PubMed] [Google Scholar]
- Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci U S A. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- Mukherjee R, Wagar D, Stephens TA, Lee-Chan E, Singh B. Identification of CD4+ T cell-specific epitopes of islet-specific glucose-6-phosphatase catalytic subunit-related protein: a novel beta cell autoantigen in type 1 diabetes. J Immunol. 2005;174:5306–5315. doi: 10.4049/jimmunol.174.9.5306. [DOI] [PubMed] [Google Scholar]
- Newgard CB, Lu D, Jensen MV, Schissler J, Boucher A, Burgess S, Sherry AD. Stimulus/secretion coupling factors in glucose-stimulated insulin secretion: insights gained from a multidisciplinary approach. Diabetes. 2002;51(Suppl 3):S389–393. doi: 10.2337/diabetes.51.2007.s389. [DOI] [PubMed] [Google Scholar]
- Nishimura W, Kondo T, Salameh T, El Khattabi I, Dodge R, Bonner-Weir S, Sharma A. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci U S A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrolonis AJ, Yang Q, Tummino PJ, Fish SM, Prack AE, Jain S, Parsons TF, Li P, Dales NA, Ge L, et al. Enzymatic characterization of the pancreatic islet-specific glucose-6-phosphatase-related protein (IGRP) J Biol Chem. 2004;279:13976–13983. doi: 10.1074/jbc.M307756200. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Guo M, Huang S, Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol. 2002;22:412–420. doi: 10.1128/MCB.22.2.412-420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol Cell Biol. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza HM, Yasuda K. Roles of Maf family proteins in lens development. Dev Dyn. 2004;229:440–448. doi: 10.1002/dvdy.10467. [DOI] [PubMed] [Google Scholar]
- Rocques N, Abou Zeid N, Sii-Felice K, Lecoin L, Felder-Schmittbuhl MP, Eychene A, Pouponnot C. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell. 2007;28:584–597. doi: 10.1016/j.molcel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Samaras SE, Cissell MA, Gerrish K, Wright CV, Gannon M, Stein R. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in Pancreatic beta cells: role for hepatocyte nuclear factor 3 beta and Pax6. Mol Cell Biol. 2002;22:4702–4713. doi: 10.1128/MCB.22.13.4702-4713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis EF. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sander M, German MS. The beta cell transcription factors and development of the pancreas. J Mol Med. 1997;75:327–340. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- Sharma A, Stein R. Glucose-induced transcription of the insulin gene is mediated by factors required for beta-cell-type-specific expression. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh JJ, Pan CJ, Mansfield BC, Chou JY. In islet-specific glucose-6-phosphatase-related protein, the beta cell antigenic sequence that is targeted in diabetes is not responsible for the loss of phosphohydrolase activity. Diabetologia. 2005:48. doi: 10.1007/s00125-005-1848-6. [DOI] [PubMed] [Google Scholar]
- Sund NJ, Vatamaniuk MZ, Casey M, Ang SL, Magnuson MA, Stoffers DA, Matschinsky FM, Kaestner KH. Tissue-specific deletion of Foxa2 in pancreatic beta cells results in hyperinsulinemic hypoglycemia. Genes Dev. 2001;15:1706–1715. doi: 10.1101/gad.901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Miles RR, Halladay DL, Onyia JE. Cryptic enhancer elements in luciferase reporter vectors respond to the osteoblast-specific transcription factor Osf2/Cbfa1. Biotechniques. 2000;28:506–510. doi: 10.2144/00283st09. [DOI] [PubMed] [Google Scholar]
- Van Schaftingen E, Gerin I. The glucose-6-phosphatase system. Biochem J. 2002;362:513–532. doi: 10.1042/0264-6021:3620513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kooi BT, Onuma H, Oeser JK, Svitek CA, Allen SR, Vander Kooi CW, Chazin WJ, O’Brien RM. The Glucose-6-Phosphatase Catalytic Subunit Gene Promoter Contains Both Positive and Negative Glucocorticoid Response Elements. Mol Endocrinol. 2005;19:3001–3022. doi: 10.1210/me.2004-0497. [DOI] [PubMed] [Google Scholar]
- Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007a;50:348–358. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gauthier BR, Hagenfeldt-Johansson KA, Iezzi M, Wollheim CB. Foxa2 (HNF3beta ) controls multiple genes implicated in metabolism-secretion coupling of glucose-induced insulin release. J Biol Chem. 2002;277:17564–17570. doi: 10.1074/jbc.M111037200. [DOI] [PubMed] [Google Scholar]
- Wang YCMC, Oeser JK, Sarkar S, McGuinness OP, Hutton JC, O’Brien RM. Deletion of the Gene Encoding the Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein Autoantigen Results in a Mild Metabolic Phenotype. Diabetologia. 2007b;50:774–778. doi: 10.1007/s00125-006-0564-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Flemming BP, Martin CC, Allen SR, Walters J, Oeser JK, Hutton JC, O’Brien RM. Long-range enhancers are required to maintain expression of the autoantigen islet-specific glucose-6-phosphatase catalytic subunit-related protein in adult mouse islets in vivo. Diabetes. 2008;57:133–141. doi: 10.2337/db07-0092. [DOI] [PubMed] [Google Scholar]
- Wang Y, Oeser JK, Yang C, Sarkar S, Hackl SI, Hasty AH, McGuinness OP, Paradee W, Hutton JC, Powell DR, et al. Deletion of the gene encoding the ubiquitously expressed glucose-6-phosphatase catalytic subunit-related protein (UGRP)/glucose-6-phosphatase catalytic subunit-beta results in lowered plasma cholesterol and elevated glucagon. J Biol Chem. 2006;281:39982–39989. doi: 10.1074/jbc.M605858200. [DOI] [PubMed] [Google Scholar]
- Wong CP, Li L, Frelinger JA, Tisch R. Early autoimmune destruction of islet grafts is associated with a restricted repertoire of IGRP-specific CD8+ T cells in diabetic nonobese diabetic mice. J Immunol. 2006;176:1637–1644. doi: 10.4049/jimmunol.176.3.1637. [DOI] [PubMed] [Google Scholar]
- Wu KL, Gannon M, Peshavaria M, Offield MF, Henderson E, Ray M, Marks A, Gamer LW, Wright CV, Stein R. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell- specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Danke NA, Berger D, Reichstetter S, Reijonen H, Greenbaum C, Pihoker C, James EA, Kwok WW. Islet-specific glucose-6-phosphatase catalytic subunit-related protein-reactive CD4+ T cells in human subjects. J Immunol. 2006;176:2781–2789. doi: 10.4049/jimmunol.176.5.2781. [DOI] [PubMed] [Google Scholar]
- Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.