Abstract
Folding within the crowded cellular milieu often requires assistance from molecular chaperones that prevent inappropriate interactions leading to aggregation and toxicity. The contribution of individual chaperones to folding the proteome remains elusive. We here demonstrate that the eukaryotic chaperonin TRiC/CCT (TCP1-Ring Complex or Chaperonin Containing TCP1) has broad binding specificity in vitro similar to the prokaryotic chaperonin GroEL. However, in vivo TRiC substrate selection is not based solely on intrinsic determinants; instead, specificity is dictated by factors present during protein biogenesis. The identification of cellular substrates revealed that TRiC interacts with folding intermediates of a subset of structurally and functionally diverse polypeptides. Bioinformatics analysis revealed an enrichment in multidomain proteins and regions of beta strand propensity that are predicted to be slow-folding and aggregation-prone. Thus, TRiC may have evolved to protect complex protein topologies within its central cavity during biosynthesis and folding.
Introduction
Eukaryotic cells contain several distinct chaperone families that together promote protein folding 1,2. Misregulation of this process leads to misfolding and aggregation events linked to multiple pathological disorders 3,4. It is thought that proteins differ widely in their chaperone requirements 2. It is unclear, however, what features of a folding polypeptide, if any, determine its interaction with specific chaperones. Determining whether different chaperones evolved to meet the folding requirements of specific classes of substrates is central to understanding the logic of cellular protein folding and assembly. Addressing this possibility requires a better understanding of which types of proteins require a given chaperone. However, the cellular substrates of most eukaryotic chaperones have not yet been defined.
The essential chaperonin TRiC/CCT (for TCP1 Ring Complex; or Chaperonin Containing TCP1) is distinguished from other chaperones by its unique ring-shaped architecture, which gives rise to a central cavity that serves as a folding chamber for substrate polypeptides 5,6. It is not known why some proteins require the ring-shaped TRiC to fold while others can reach their native states with the assistance of simpler chaperone systems. Indeed, the cellular function of TRiC remains ill-defined and controversial. TRiC was originally proposed to be highly specialized to recognize a few cytoskeletal proteins through specific sequence elements 7. However, the recent identification of additional TRiC substrates have called into question this original idea 8–12. Here we determine the principles of substrate selection by TRiC and define the subset of cellular proteins that interact with this chaperonin in eukaryotic cells using a combination of experimental and computational analyses.
Results
Principles of TRiC substrate selection
TRiC is part of a chaperone network linked to protein synthesis 13 and has been shown to facilitate folding of newly translated proteins in vivo 9,14. Previous studies established that TRiC interacts transiently with a subset of cellular proteins during biogenesis 14. We thus examined the flux of newly translated proteins through TRiC in mammalian cells using a previously established pulse-chase analyses, whereby newly-made proteins are specifically labeled by a brief pulse and folding and maturation occurs during the chase 14. Newly synthesized polypeptides interacting with TRiC were isolated by immunoprecipitation with antibodies against TRiC subunits β and ε (Supplementary Fig. 1; Fig. 1A). Two-dimensional PAGE analysis showed that soon after translation a large number of newly made proteins associated with TRiC. Following a period of chase, these proteins were dissociated, as expected for chaperone substrates which should be released upon completion of folding (Fig. 1A, left and center panel). Chaperonin complex assembly was also observed during the time course of the chase, whereby the β and ε subunits associated with the remaining TRiC subunits (Fig. 1A, center, see also ref.14). Mass spectrometry analysis of TRiC interacting proteins only identified highly abundant substrates, namely the WD-repeat containing translation initiation factor 3β and GAPDH, in addition to the known TRiC substrates actin and tubulin (Supplementary Table 1). Thus, identification of low abundance cellular substrates of TRiC required alternate genome-wide approaches.
Figure 1. Principles of TRiC substrate selection in the eukaryotic cytosol.
(A–B) Human fibroblast cells (TSA-201) were pulse-labeled with 35S-methionine for 5 min, followed by a 0 or 30 min chase. Total soluble protein was immunoprecipitated with anti-TRiC antibodies or a nonimmune antibody control and separated on 2D gels. (B) TRiC recognizes different proteins in vitro and in vivo. 2D gels of in vitro (green) and in vivo (fuschia) TRiC-bound proteins were compared. The two gels were merged with overlapping spots circled and illustrated in blue (B, right panel). (C) A denatured 35S-labelled cytosolic extract was diluted into buffer containing either GroEL or TRiC. Bound proteins were assessed by immunoprecipitation with anti-chaperonin (cpn) antibodies (EL or TRiC), and separated by 2D gel. 2D gel images of GroEL-bound (red) and TRiC-bound (green) proteins were merged with overlapping spots circled and illustrated in blue (C, right panel).
To better define the principles that govern TRiC substrate specificity, we next examined what determines association of cellular proteins with TRiC. In principle, chaperonin-substrate interactions may be solely determined by the presence of specific TRiC-binding motifs in the substrates, such as sequence elements, that distinguish them from the rest of the proteome. A prediction of this model is that the in vitro substrate specificity of TRiC towards denatured cytosolic proteins will mirror that observed in vivo. Accordingly, we compared the subset of cellular proteins that bind TRiC upon translation in vivo (Fig. 1B, left) with those proteins binding TRiC when the same 35S-labeled extract is denatured and presented to the chaperonin in vitro (Fig. 1B, center). The sets of eukaryotic proteins interacting with TRiC in vivo and in vitro were strikingly different. While actin and tubulin were prominently bound in either condition, examination of the merged gels revealed less than 10% overlap between the proteins selected by TRiC in vitro and in vivo (Fig. 1B, right). We conclude that TRiC does not select its substrates based solely on the presence of specific sequence motifs, as was proposed from in vitro studies 15,16. Instead, TRiC-substrate selection in the cell is strongly dependent on the context of translation, where both cotranslational folding events and cooperating chaperone systems may affect the conformation of de novo folding intermediates 11,17,18.
TRiC was proposed to be a highly specific chaperone, in contrast to the broader specificity prokaryotic chaperonin GroEL 7. To further define the substrate-recognition principles of TRiC, we next determined whether TRiC recognizes a more restricted range of proteins when compared to GroEL 19. To compare the cellular proteins recognized directly by TRiC and GroEL we presented denatured 35S-labeled cytosolic proteins to purified GroEL or TRiC, followed by immunoprecipitation of chaperonin substrate complexes (Supplementary Fig. 2 and Fig. 1C). A large fraction of cellular proteins, including actin and tubulin, was recognized by both chaperonins. Comparison of the GroEL and TRiC-bound protein spectra revealed that the recognition specificities of these chaperonins were strikingly similar, with over 80% overlap between the protein sets recognized by either chaperonin (Fig. 1C, right). Given the well-established affinity of GroEL for hydrophobic substrate determinants, this result indicates that hydrophobicity is a strong component of TRiC substrate recognition, consistent with previous findings 8,9,20,21. Importantly, the similar binding specificity of both chaperonins suggests that TRiC sustains the capacity to recognize a wide breadth of proteins and is not intrinsically a highly specific chaperonin. We conclude that TRiC possesses broad recognition specificities, yet in the cell it only interacts with a defined set of substrates.
Genomic screen for TRiC substrates during biosynthesis
Our finding that TRiC substrate selection in the cell is determined in the context of protein biosynthesis raises the question of what features distinguish proteins recognized by TRiC during biogenesis from the wide spectrum of potential interactors. Accordingly, we adapted a genome-wide approach that allowed identification of physiologically relevant TRiC substrates by detecting chaperone interactions in the context of translation.
We employed a screening approach that monitored which proteins interact with TRiC during translation of cDNA expression pools in a cell-free mammalian translation system 22. This approach, termed small pool expression cloning, or SPEC, allowed detection of TRiC-substrate interactions in a physiologically relevant context (Fig. 2A). Importantly, SPEC presents several unique advantages for the identification of physiological chaperone substrates. First, these translation lysates contain the full complement of chaperones and translation components required to fold most cytosolic proteins to an active state 23. TRiC-substrate interactions are thus examined in the context of protein biosynthesis, whereby cotranslational folding can occur in the presence of physiological levels of upstream chaperones and folding cofactors. Second, proteins are translated at very low levels, in the picomolar range, and thus interactions with the chaperonin are observed without overexpression of the substrates 24. Furthermore, the lack of bias toward detection of abundant proteins in the SPEC approach permits the identification of chaperone interactions for rare, low-abundance proteins, unlike large-scale immunoprecipitation reactions.
Figure 2. Screening for the TRiC interactome in the context of translation.
(A) Small pool expression cloning screen: bacterial cDNA clones were cultured in 96-well plates and pooled in rows and columns from which plasmid DNA was purified. The orthogonal cDNA pools were expressed in rabbit reticulocyte lysate (RRL) in the presence of 35S-methionine and immunoprecipitated with anti-TRiC antibodies. (B) Total expressed cDNA pools in RRL (Total) and TRiC-bound proteins (anti-TRiC IP) were analysed by SDS-PAGE and phosphorimaging. Immunoprecipitations with nonimmune antibodies demonstrated the specificity of the TRiC immunoprecipitations (Control). Actin expression and TRiC-binding served as a positive control throughout. (B and C) Red and blue astericks indicate TRiC-interacting candidate clones present in orthogonal pools. (C) Interactions were confirmed by expression of single candidate clones B3 and D2 in RRL, followed by anti-TRiC immunoprecipitation. cDNAs were identified by sequencing from upstream promoter sequences.
A high-throughput screen was performed using single mouse cDNAs arrayed in a 96-well plate format. Orthogonal cDNA pools were made along rows and columns for each plate and analyzed by in vitro translation in rabbit reticulocyte lysate in the presence of 35S-methionine (Fig. 2A). The presence of TRiC substrates encoded by cDNAs in the pools was detected by co-immunoprecipitation with the chaperonin followed by SDS-PAGE and phosphorimaging analysis (Fig. 2B). cDNAs encoding candidate substrates expressed in two overlapping pools were isolated and their interaction with TRiC confirmed before their identification by DNA sequencing (Fig. 2C). To simplify the screen, abundant housekeeping mRNAs were subtracted from the cDNA library, thus increasing the proportion of lower abundance transcripts surveyed 25. Of note, the identification of alpha-tubulin among the TRiC interacting substrates demonstrated that housekeeping proteins were still represented in the library, thus providing a good representation of transcripts in the cell (Supplementary Table 1). In all, 2600 clones were surveyed and more than one hundred TRiC-interacting proteins were isolated. The screen was not conducted to saturation but was concluded upon the repeated identification of several clones. From these data, 6–7% of all cytosolic proteins are anticipated to interact with TRiC. Importantly, this analysis is in agreement with previous estimates from pulse-chase analysis 14 and proteomics studies 26. Because only high affinity interactions are detected by immunoprecipitation, the predicted number of TRiC-interacting proteins might be underestimated. Assuming that the eukaryotic cytosol contains approximately 4500 proteins, based on estimates from the yeast Saccharomyces cerevisiae, our analysis predicts that on the order of 300 cytosolic proteins interact with TRiC in vivo.
Distinct in vivo kinetics of substrate flux through TRiC
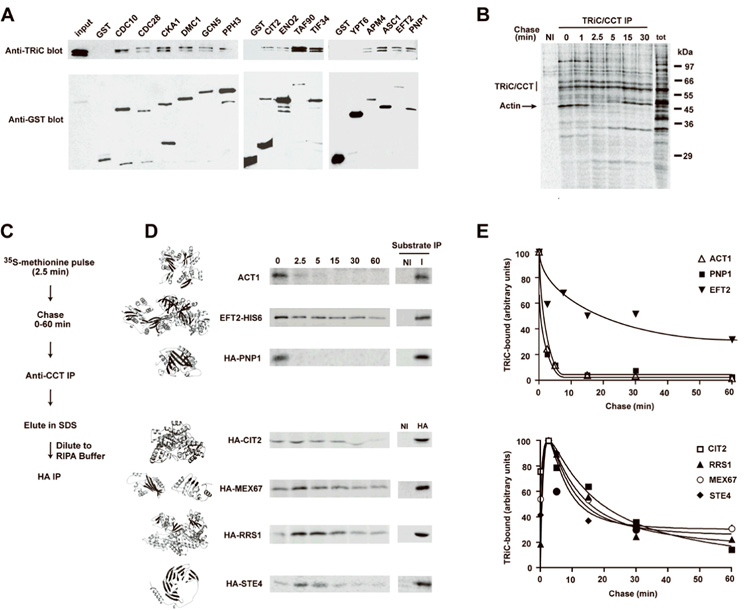
Our screen revealed that a wide range of proteins interact with TRiC upon translation, most of which are conserved across eukaryotes (Supplementary Table 1). Accordingly, we used S. cerevisiae to validate the TRiC-interactions in vivo taking advantage of the extensive annotation of the yeast genome. Initially, candidates carrying an N-terminal GST-tag 27 were affinity isolated, and association with TRiC assessed by immunoblot analysis. Remarkably, all of the yeast homologues of SPEC-derived candidates surveyed bound to TRiC in vivo but neither GST alone nor GST-tagged to Ypt6, a control GTP-binding protein, associated with the chaperonin (Fig. 3A). This supports the idea that the screen identified bona fide TRiC substrates and that the interaction with TRiC is evolutionarily conserved across eukaryotic organisms.
Figure 3. In vivo analysis of the TRiC interactome reveals distinct kinetics of substrate flux through the chaperonin.
(A) GST-fusion proteins were expressed in S. cerevisiae behind a galactose-inducible promoter and affinity-purified with glutathione sepharose. Purified proteins were separated by SDS-PAGE and subject to western blot analysis with anti-TRiC or anti-GST antibodies. (B–C) The flux of newly synthesized proteins binding to TRiC was assessed by pulse-chase analysis in wild-type yeast. 35S-labelled cells were harvested at the indicated timepoints, lysed and immunoprecipitated with anti-TRiC antibodies. (C–E) The flux of individual proteins through TRiC was assessed by pulse-chase analysis of wild type yeast expressing tagged-substrates. TRiC-bound proteins from the pulse-chase analysis were eluted with SDS and subjected to a second immunoprecipitation with either nonimmune (NI) antibodies or antibodies recognizing tagged substrates (I). Structural models for each of the substrates were generated by multiple sequence alignment with homologous proteins using SWISS-MODEL 49. (E) TRiC-bound substrates isolated in the pulse-chase were quantitated with ImageQuant software (v. 5.2, Amersham Biosciences) and the kinetics of their association plotted over time.
One hallmark of chaperones that facilitate de novo folding is that they associate transiently with newly made proteins and dissociate during the course of polypeptide folding and maturation. As such, we examined the in vivo flux of SPEC-derived TRiC substrates through the chaperonin in yeast cells. Pulse-chase analysis followed by anti-TRiC immunoprecipitation indicated that, as in mammalian cells, a range of yeast proteins transiently associate with TRiC early in their biogenesis, and dissociate during the chase, consistent with release upon folding (Fig. 3B, 15 min). To observe the flux of single substrates through the chaperonin, TRiC immunoprecipitations were subjected to a second round of immunoprecipitation against a unique tag in the substrates (Fig. 3C, 3D). Transient association kinetics were observed for SPEC-identified substrates, indicating these proteins transit through the chaperonin during their biogenesis (Fig. 3D). Notably, two types of association kinetics were observed among TRiC substrates. One class of proteins, including actin and the small purine nucleoside phosphorylase (PNP1), bound early to TRiC and rapidly dissociated (Fig. 3D, 3E). Elongation factor 2 (EFT2) also bound rapidly to TRiC, but dissociated very slowly from the chaperonin (Fig. 3D, 3E). Another set of substrates exhibited much slower kinetics of association with TRiC. For instance, the mRNA export factor MEX67, the homotetrameric citrate synthase CIT2, the multidomain protein arginyl tRNA synthetase RRS1 (or ArgRS), as well as the WD40 Gβ-protein STE4 did not bind to TRiC at the earliest chase times but instead their binding seemed to peak later in the time-course (Fig 3D, 3E). Notably, the TRiC association kinetics was independent of substrate size, since the larger EFT2 associated rapidly with TRiC while the smaller STE4 did not. It is tempting to speculate that the slower TRiC-binding kinetics arises from the action of cooperating upstream chaperones delaying binding to TRiC during translation 13,18. Intriguingly, all the slower binding proteins share the common property of belonging to larger protein complexes, raising the possibility that these substrates accumulate on TRiC, before their release into oligomeric assemblies. In support of this idea, several WD40-containing F-box proteins translated in a cell-free system accumulated on TRiC in the absence but not in the presence of their oligomeric partner protein Skp1 (Supplementary Fig 3). We conclude that a genome-wide screen for the TRiC interactome identified bona fide TRiC substrates that flux through the chaperonin during biogenesis in vivo.
Physical and structural properties of the TRiC Interactome
We next sought to define what distinguishes the relatively small fraction of TRiC-interacting proteins from the cytosolic proteome at large. Having identified a large set of proteins that interact with TRiC in a physiologically relevant context (herein the TRiC Interactome, Suppl. Table 1) we applied a bioinformatics approach to identify common features among TRiC substrates that may underlie their physiological association with TRiC. Proteins from the Interactome spanned a breadth of cellular processes, most of which are conserved across eukaryotic organisms. Interestingly, 40% of the TRiC Interactome comprises essential genes, more than twice the proportion of essential genes in the yeast genome 28. Of note, the frequency of TRiC substrates belonging to oligomeric protein complexes was also enriched in the TRiC Interactome to 90% compared to less than 50% of all yeast proteins in the MIPS complex database29. These two observations may be linked since proteins that belong to oligomeric complexes are more highly connected in the protein network and, thus, disproportionately essential 30. We hypothesize that the eukaryotic chaperonin assists in folding proteins belonging to oligomeric assemblies and may serve as a reservoir to stabilize them against aggregation or degradation prior to complex formation.
Previous biochemical experiments using actin and tubulin, led to the proposal that TRiC selects its substrates by specific recognition of a set of polar sequence elements 15,16. Accordingly, we analyzed the TRiC Interactome to search for the presence of specific sequence motifs that may confer TRiC binding. Sequence analysis of the TRiC Interactome failed to reveal statistically significant consensus sequences, or an enrichment in previously proposed TRiC-binding sequence elements 15,16. This is consistent with the finding that TRiC is not a highly sequence-specific chaperonin (Fig. 1C).
In principle, physical and/or structural properties could influence the folding characteristics and contribute to chaperonin binding. A comparison of the size distribution of TRiC-interacting proteins to the naturally occurring distribution of cytosolic proteins in the S. cerevisiae proteome (Fig. 4A) or the murine proteome (Supplementary Fig. 4) demonstrated a statistically significant enrichment for proteins ranging between 40–75 kDa in size, in good agreement with results obtained using pulse-chase approaches, 14 (Fig. 1 & 4A). Of note, the chaperonin chamber is predicted to accommodate polypeptides in this size range 31. Since a single folding domain is approximately 25–30 kDa 1, this finding suggests that the TRiC-interactome consists mostly of multidomain proteins. Interestingly, a number of larger substrates were also identified in our screen, raising the possibility that some substrates are not completely encapsulated during folding. In support of this idea, the 100 kDa protein myosin is an known to be an obligate substrate of TRiC 32. Furthermore, the bacterial chaperonin GroEL similarly supports the folding of several large proteins 33.
Figure 4. Physical and structural properties of the TRiC/CCT Interactome.
Prevalence of physical and structural properties of TRiC/CCT-interacting proteins was compared to their natural occurrence in the yeast cytosolic proteome. Protein size (number of residues) (A), maximal number of hydrophobic residues in a 60 amino acid window (B), percent helical content (C), percent beta sheet content (D), and maximal number of beta sheet residues in a 60 amino acid window (E) were considered. All differences between the TRiC/CCT Interactome and the yeast cytosolic proteome were statistically significant with p-value ≤ 10−4, except for protein size which had a p-value of < 0.05.
The proteins in the TRiC Interactome exhibited an enrichment in hydrophobic sequences (Fig. 4B). This finding could be explained by substrate size, as larger proteins retain correspondingly larger hydrophobic cores. Alternatively, hydrophobic inter-domain contacts within substrate proteins could contribute to the higher proportion of hydrophobic sequences. Inappropriate interactions among these hydrophobic surfaces, for instance in domain swapping, could complicate the folding of multidomain proteins 34. Previous observations that TRiC can recognize hydrophobic determinants 8,9,20,21 (see also Fig. 1C) suggests the chaperonin may prevent these inappropriate interactions.
Analysis of the structural propensities of the TRiC Interactome revealed the most striking shared property among TRiC substrates. While no specific protein fold was enriched in the TRiC interactome, we observed that proteins of high beta sheet propensity and/or low alpha helical content were highly enriched among TRiC substrates (Fig. 4C & 4D). Additional scanning window analysis demonstrated that a high proportion of TRiC-interacting proteins contain long continuous stretches of beta sheet propensity, with a particular bias toward stretches of 35–45 amino acids (Fig. 4E). Thus, analysis of secondary structure propensity identifies a clear commonality among proteins in the TRiC interactome. Importantly, the enrichment in beta-rich proteins suggests that TRiC substrates have complex topologies that are predicted to be slow-folding and aggregation-prone.
Discussion
Here we combine genomic and proteomic approaches to identify the interactome of the eukaryotic chaperonin TRiC/CCT. Contrary to initial proposals that TRiC is specialized for the folding of only a small subset of eukaryotic proteins, we find that the chaperonin interacts with a broad range of polypeptides that function in many cellular processes (Table 1). TRiC specificity is strongly influenced by the cellular context of translation and a wide range of structural conformations are recognized by the chaperonin, indicating that TRiC functions to facilitate the folding of many structural protein families. A bioinformatics analysis of the TRiC substrates demonstrated that the TRiC-interactome is enriched in large, hydrophobic polypeptides with complex topologies predicted to be slow-folding and aggregation-prone. Strikingly, the analysis also suggests that TRiC assists in folding subunits belonging to oligomeric protein complexes. These findings allow us to propose a model for TRiC function in the stabilization of slow-folding proteins susceptible to the formation of kinetically-trapped intermediates and in the subsequent coordination of substrates into protein assemblies.
Cellular Principles of TRiC substrate selection
Our studies resolve a long-standing controversy over TRiC substrate recognition and specificity. The data presented do not support the idea that TRiC selects its substrates in the cell based solely on intrinsic determinants, such as specific sequence elements. Instead, TRiC recognition is strongly influenced by the context of protein biogenesis and may involve structural and physical features of the biosynthetic intermediates as well as cooperating chaperone systems (Fig. 5).
Figure 5. Function of TRiC during de novo protein folding.
TRiC (type II) has a broad substrate binding specificity in vitro similar to the type I chaperonin GroEL. However, in vivo TRiC substrate selection is not based solely on intrinsic determinants, but rather specificity is dictated by factors present during protein biogenesis, such as cotranslational folding or upstream chaperone systems. TRiC binds to a subset of newly synthesized polypeptides to stabilize exposed sequences during folding and prevent aggregation. The TRiC substrate set is preferentially enriched in proteins with slow-folding kinetics and aggregation propensity, such as beta-domain containing proteins. Folded substrates are finally released either in their monomeric state or in the context of oligomeric protein assemblies.
In vitro experiments comparing TRiC with the broad spectrum chaperonin GroEL indicate that TRiC is not a highly specific chaperone. Since these in vitro substrates differ substantially from the substrates binding to TRiC in vivo, this result implies that TRiC substrate interaction is dictated by the folding properties of the polypeptide in the context of translation. The substantial differences between the spectrum of cellular proteins that bind to TRiC during biosynthesis and those that TRiC selects when the same labeled proteins are presented in a denatured form highlights the importance of examining chaperone substrates in a physiologically relevant context. Furthermore, the similarities between the proteins bound by TRiC and GroEL in vitro suggest that the specificity of the eukaryotic chaperonin stems from the particular combination of folding kinetics and exposed recognition motifs in the co- or post-translationally-generated intermediates of its cellular substrates. The lack of sequence specificity in the TRiC substrate interaction is reinforced by the absence of sequence motifs in the in vivo subset that might confer TRiC binding propensity. Future studies should uncover the molecular determinants and the possible role of upstream cofactors in TRiC recognition.
Properties of TRiC substrates
We find that approximately 7% of the proteins we screened by genomic approaches associated with TRiC upon translation. This is consistent with our pulse-chase analysis, which indicates that approximately 5–10% of cytoplasmic proteins flux through the chaperonin. Analysis of the shared features of the Interactome identified some common properties in these substrates that may illuminate the principles of substrate selection by TRiC. In particular, the enrichment of beta-rich proteins among TRiC substrates provides a link between chaperonin interaction and one of the major challenges faced by the biosynthetic machinery that must facilitate cellular folding. Indeed, regions with high beta-strand propensity are inherently aggregation-prone and difficult to fold, such that evolutionary pressures have introduced elements of natural design to protect exposed beta-edges in native protein structures 35. Importantly, these design elements would not be in place until completion of folding. Thus, folding intermediates with regions of high beta-sheet propensity are especially vulnerable to misfolding and aggregation. It is tempting to speculate that the chaperonin TRiC functions to stabilize these exposed beta edges against aggregation until loops or helices in the folded protein architecture emerge to protect the beta strand in the native state.
It is also possible that the enrichment in beta-sheet propensity emerges from a more overarching characteristic of TRiC substrates. The observation that beta-sheet propensity correlates with slow-folding kinetics provides another intriguing link between our analysis of the TRiC interactome and folding in the cell. Emerging studies find that complex protein topology, such as that found in beta-rich proteins, is a major contributor to slow folding rates in vitro 36–38. Of note, the TRiC Interactome also contains all-helical proteins such as citrate synthase, raising the possibility that the enrichment in beta sheet content is diagnostic of a more general property recognized by TRiC, such as complex topology or slow-folding kinetics. These features can lead to accumulation of toxic aggregates which have been linked to a number of amyloid diseases 4. The idea that TRiC may play a role in preventing aggregation of amyloidogenic folding intermediates resonates with the recent identification of this chaperonin as a potent suppressor of huntingtin amyloid formation, a polyglutamine protein that is highly aggregation-prone 39–41.
Contribution of TRiC to the Cellular Folding Network
Our data provides a clue on how the cell solves the problem of folding proteins with complex topologies, such as those with beta-rich domains. During synthesis, these proteins are in a precarious position as they need to bring together many discontinuous regions to reach the native state, and thus are susceptible to aggregation with neighboring proteins. Binding to the ring-shaped chaperonin TRiC would solve several problems posed by this process: first, the polyvalent binding of multiple substrate regions (e.g. hydrophobic beta-strands) to different subunits in a ring would sequester several aggregation-prone sequences during translation; secondly, the allosteric communication between subunits in one ring 42 would provide a mechanism for concerted release of all folding elements to the central chaperonin chamber; and finally, this chamber would provide a sequestered environment to protect slow-folding proteins until they reach the native state. Taken together, the combination of proteomic and bioinformatics analyses reveal TRiC substrates have complex topologies that are slow-folding and aggregation-prone, and provide a compelling rationale for the function of this oligomeric ring shaped chaperonin in assisting cellular folding.
The additional finding that TRiC substrates are enriched in proteins belonging to oligomeric assemblies suggests that TRiC also functions to facilitate complex assembly. We envision two possible mechanisms for this role. In principle, the processes of TRiC-mediated folding and assembly could be directly coupled. Alternatively, TRiC could fold monomeric subunits and maintain them in an assembly-competent state until association with oligomeric partners. Thus, TRiC may contribute to the regulation of complex cellular processes such as signaling or cell cycle regulation by maintaining a pool of assembly-competent but inactive protein molecules. Indeed, other chaperones such as Hsp90 have also been implicated in the regulation of signaling cascades by a similar mechanism 43.
While our data highlights the contribution of TRiC to cellular folding, it will be interesting to examine how TRiC cooperates with other components of the cellular folding machinery. While the few TRiC substrates characterized to date, such as actin, tubulin, Cdc20 and Cdh1 9,44,45, exhibit an obligate requirement for the chaperonin, it is possible that other TRiC-interacting proteins can be folded by alternative chaperone pathways. Indeed, an in vitro analysis of GroEL interacting proteins revealed that only a small proportion have an absolute requirement for GroEL to fold, while most GroEL substrates can fold spontaneously or use alternate chaperones 46. It is likely that TRiC substrates fall into similar categories. Future studies should determine whether some TRiC substrates can fold via alternate chaperone systems. However, even for the well-studied chaperonin GroEL, comparison of in vitro and in vivo analyses illustrates the complexity of chaperone pathways in the cell. Thus, several GroEL-interacting proteins that can fold spontaneously in vitro nonetheless aggregate in vivo upon GroEL impairment 33,46. Clearly, much remains to be learned about the function of chaperonins and how they cooperate with other chaperone machinery to facilitate folding in the cell.
Methods
Tissue Culture and Pulse-Chase Analysis of Mammalian Proteins
Human fibroblast TSA-201 cells used for pulse-chase analysis were cultured in complete medium at 37°C (DMEM with 10% (v/v) FBS, 100 U ml−1 penicillin, 100 µg ml−1 streptomycin, and 2 mM L-glutamine). Cells were starved for 15 minutes in starvation media (DMEM - methionine and cysteine, supplemented with 5% (v/v) dialyzed FBS to remove amino acids: Sigma, 10 kDa MWCO), before they were labeled with 0.8 mCi ml−1 35S-methionine for 5 minute, and then chased for 30 minutes in chase media (complete medium supplemented with 0.4% (w/v) cysteine and methionine). Then cells were harvested in ATP depletion buffer (ice-cold PBS with 1 mM azide, 5 mM EDTA, and 2 mM deoxyglucose), lysed by dounce homogenization in buffer A (20 mM HEPES pH 7.4, 100 mM NaCl, 5 mM EDTA, 5% (v/v) glycerol), and clarified by centrifugation. Equivalent amounts of protein were then immunoprecipitated with 2 µl anti-TCP1 β antibodies as described 14. Briefly, lysates were incubated with antibodies for 40 minutes, rotated with 10 µl Protein G Sepharose for another 40 minute, and immunoprecipitates were washed in TBS + Tween20 buffer as described 14.
2D Gel Analysis and Mass Spectrometry
TSA-201 cells were briefly labeled with 35S-methionine for 5 minutes, followed by a 0 or 30 minute chase in chase media as described above. Cells lysates were immunoprecipitated with anti-TCP1 β antibodies, and immunoprecipitates were resuspended in 250 µl IEF rehydration buffer (8M urea, 2% (w/v) CHAPS, 2% (v/v) IPG buffer pH 4–7, 2.8 mg ml−1 DTT, trace bromophenol blue) with 400 µg unlabelled lysate (~15 mg ml−1), before being separated on 13 cm Immobiline DryStrips pH 4–7 (Pharmacia Biotech), according to manufacturer’s instructions. The proteins were separated in the second dimension by 10% SDS-PAGE, and gels were silver-stained and analyzed by phosphorimaging. Superposition of silver-stained and radiolabelled gel images identified proteins to be further analyzed by mass spectrometry. Protein spots were excised from a keratin-free gel run in a parallel sample and digested with 12.5 ng µl−1 trypsin at 37°C for 4 hr - overnight, and identified by LC/MS as described 47. Data analysis was performed with the algorithm Protein Prospector.
Specificity of in vivo vs. in vitro binding
TSA-201 cells were labeled overnight with 10 µCi ml−1 35S-methionine in starvation medium supplemented with 1% (v/v) complete medium. To generate a cytosolic protein extract, the cells were harvested, lysed, and then denatured with 6 M guanidinium chloride. In vitro binding to the chaperonin was assessed by diluting the denatured extract (~5 mg ml−1) by 100 times into 200 µl buffer B (25 mM HEPES pH 7.4, 100 mM KCl, 5 mM EDTA, 1 mM 2-deoxyglucose, 1mM NaN3) containing either the chaperonin GroEL (0.1 µM) or TRiC (0.25 µM). Samples were incubated for 20 minutes at 30°C, followed by 20 minutes on ice, and then clarified by ultracentrifugation for 25 minutes at 20,000xg. Bound proteins were then coimmunoprecipitated with anti-chaperonin antibodies (5 µl rabbit anti-GroEL or 2 µl rabbit anti-TCP1 β antibodies), separated simultaneously by 2D gel, and then visualized by autoradiography.
To compare proteins bound to GroEL and TRiC, gel images were differentially colorized in Adobe Photoshop and overlayed. Minor rescaling of images was performed using actin and tubulin spots as points of reference.
Small Pool Expression Cloning
TRiC-interacting proteins were identified by screening for TRiC coimmunoprecipitation of expressed cDNAs from a substracted murine library. The cDNA library was generated from mRNAs differentially expressed in G1/S and G2/M C2C12 cells as described 25. cDNAs were inserted into the pCS+ vector behind the SP6 promoter and transformed into bacteria. Bacterial cDNA clones were grown in a 16×12 array and then pooled by rows and columns from which plasmid DNA were purified (Qiagen miniprep). cDNA pools (~250 ng) were expressed in vitro in a rabbit reticulocyte lysate coupled transcription-translation system (8 µl, Promega TNT Quick Coupled Transcription/Translation Systems) in the presence of 35S-methionine (0.75 µCi µl−1) at 30°C for 45 minutes. Translations were stopped by incubation with 10 mM EDTA, 50 µg ml−1 RNase A, and 5 mg ml−1 methionine for 10 minutes on ice before they were immmunoprecipitated with anti-TCP1 β antibodies. TRiC-substrate complexes were separated by 12% SDS-PAGE and analyzed by phosphorimaging. Proteins of similar size present in two orthogonal pools were selected as candidate clones. Then the interactions of individual expressed cDNAs were confirmed by in vitro expression, followed by anti-TRiC immunoprecipitation. Confirmed clones were finally identified by SP6-primed sequencing and BLAST analysis. In total, 2600 clones were screened, from which approximately 1500 clones were estimated to have an insert generating a detectable translation product.
In vivo Validation of Substrates
Interaction of GST-fusion proteins
Amino-terminal GST-fusion proteins were expressed in yeast (strain Y258) behind a galactose-inducible promoter from the pEGH expression vector as described 27. Cells were grown to log phase in –URA raffinose synthetic media before protein expression was induced with 2% (w/v) galactose for 3 hours. Cells were harvested and lysed by bead beating for 10 minutes at 4°C in buffer C (25 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 10% (v/v) glycerol, 0.1% (v/v) TritonX-100) with 20 mM EDTA. Extracts were clarified and equivalent protein amounts were incubated with glutathione sepharose for one hour before washing four times with TBS 0.1% (v/v) Tween20. Purified complexes were separated by 12% SDS-PAGE and immunoblotted with anti-GST mAb (Covance, clone 4C10) or rabbit affinity-purified CCT antibodies raised against the apical domains of yeast CCT3, 5, 6, and 8.
Yeast pulse-chase analysis
Wild type cells (strain BY4743) expressing plasmid-borne copies of HA-tagged substrates were grown to log phase, induced via a copper-inducible promoter with 200 µM CuSO4 for 30 minutes, and starved for 30 minutes in CSM without methionine. The cells were then labeled with 100 µCi ml−1 35S-methionine for 2.5 minutes and chased with 20 mM cold methionine. At the indicated time points, aliquots were quickly chilled and harvested in 250 mM cold azide to deplete of ATP and 0.5 mg ml−1 cycloheximide to stop protein synthesis. Lysates prepared in buffer C were clarified and TRiC-substrate complexes isolated by immunoprecipitation using 4 µl CCT-specific antibodies. CCT-interacting proteins were eluted with 1% (w/v) SDS and 5 mM DTT for 15 minutes at 30°C, and then diluted to the final concentration of RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% (v/v) NP40, 0.1% (w/v) DOCA, 0.1% (w/v) SDS). HA-tagged substrates were re-immunoprecipitated from the eluate with 2 µl anti- HA antibodies (clone HA.11, Babco), separated by 12% SDS-PAGE, and analyzed by phosphorimaging. Isolated protein substrates were quantitated with ImageQuant software (v. 5.2, Amersham Biosciences) and plotted over the time course of the chase. All substrates were amino-terminally HA-tagged and cloned into the pCu426 vector under the control of a copper-inducible promoter 48, with the exception of EFT2, which was carboxy-terminally HIS-tagged. Protein structures were retrieved from the Protein Data Bank, or modeled against homologous structures by multiple sequence alignment via SWISS-MODEL 49.
Bioinformatic Characterization of the TRiC Interactome
A comprehensive list of TRiC-interacting proteins was compiled, including proteins identified in this study as well as other reports 9,11,12,19,32,44,45,50–54. TRiC-interacting proteins identified in largescale proteomics studies were also included if they bound to three or more TRiC subunits 26. Trends found among the TRiC-Interacting proteins were compared with the distribution of cytosolic proteins occurring in the yeast proteome (Figure 4) or the murine proteome (Suppl. Fig. 3). The cytosolic protein class was estimated by removing proteins with one or more transmembrane helices from the proteome, using the prediction server TMHMM 55, resulting in a reduction of the proteome by approximately 25%, in agreement with predictions by Wolf and colleagues 56.
Physical and structural properties of proteins were predicted by computer algorithms: the Kyte-Doolittle scale was used to measure hydrophobicity 57 and PSIPRED was used to determine secondary structure 58. Scanning window analysis determined the maximal number of hydrophobic (or beta strand) residues within a 60 residue window for each protein. P-values were calculated by chi-squared analysis. P-values calculated using the unequal variance t-test 59 also confirmed significant differences in the means of the distributions. Functional properties of proteins such as essentiality and complex formation were also determined. Data on protein essentiality was downloaded from the Saccharomyces Genome Database (SGD) 60. A list of yeast complexes containing at least two non-TRiC protein members was obtained from the MIPS database 29.
Supplementary Material
Acknowledgements
The authors thank members of the Frydman lab and Raul Andino for comments and discussion. The cDNA library was a generous gift from Dr. Wei-meng Zhao and Dr. Guowei Fang (Department of Biology, Stanford University, CA). We thank Drs. Wade Harper (Department of Pathology, Harvard Medical School, MA), Terri Kinzy (Department of Molecular Genetics & Microbiology, Rutgers, NJ), K. Abel (Molecular Biology Department, Behringwerke AG, Marburg, F.R.G), and Michael Rexach (Department of Biological Sciences, Stanford University, CA) for gifts of plasmids and antibodies. This work was supported by grants from NIH and the W.M. Keck Foundation.
References
- 1.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 3.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 4.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 5.Spiess C, Meyer ASQ, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Puertas P, Martin-Benito J, Carrascosa JL, Willison KR, Valpuesta JM. The substrate recognition mechanisms in chaperonins. J Mol Recognit. 2004;17:85–94. doi: 10.1002/jmr.654. [DOI] [PubMed] [Google Scholar]
- 8.Kubota S, Kubota H, Nagata K. Cytosolic chaperonin protects folding intermediates of Gbeta from aggregation by recognizing hydrophobic beta-strands. Proc Natl Acad Sci U S A. 2006;103:8360–8365. doi: 10.1073/pnas.0600195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 10.Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16:3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 14.Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. Embo J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pappenberger G, et al. Crystal structure of the CCTgamma apical domain: implications for substrate binding to the eukaryotic cytosolic chaperonin. J Mol Biol. 2002;318:1367–1379. doi: 10.1016/s0022-2836(02)00190-0. [DOI] [PubMed] [Google Scholar]
- 16.Hynes GM, Willison KR. Individual subunits of the eukaryotic cytosolic chaperonin mediate interactions with binding sites located on subdomains of beta-actin. J Biol Chem. 2000;275:18985–18994. doi: 10.1074/jbc.M910297199. [DOI] [PubMed] [Google Scholar]
- 17.Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 18.Melville MW, McClellan AJ, Meyer AS, Darveau A, Frydman J. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol Cell Biol. 2003;23:3141–3151. doi: 10.1128/MCB.23.9.3141-3151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melki R, Batelier G, Soulie S, Williams RC., Jr Cytoplasmic chaperonin containing TCP-1: structural and functional characterization. Biochemistry. 1997;36:5817–5826. doi: 10.1021/bi962830o. [DOI] [PubMed] [Google Scholar]
- 20.Feldman DE, Spiess C, Howard DE, Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 21.Rommelaere H, De Neve M, Melki R, Vandekerckhove J, Ampe C. The cytosolic class II chaperonin CCT recognizes delineated hydrophobic sequences in its target proteins. Biochemistry. 1999;38:3246–3257. doi: 10.1021/bi9815905. [DOI] [PubMed] [Google Scholar]
- 22.King RW, Lustig KD, Stukenberg PT, McGarry TJ, Kirschner MW. Expression cloning in the test tube. Science. 1997;277:973–974. doi: 10.1126/science.277.5328.973. [DOI] [PubMed] [Google Scholar]
- 23.Nimmesgern E, Hartl FU. ATP-dependent protein refolding activity in reticulocyte lysate. Evidence for the participation of different chaperone components. FEBS Lett. 1993;331:25–30. doi: 10.1016/0014-5793(93)80290-b. [DOI] [PubMed] [Google Scholar]
- 24.Jermutus L, Ryabova LA, Pluckthun A. Recent advances in producing and selecting functional proteins by using cell-free translation. Curr Opin Biotechnol. 1998;9:534–548. doi: 10.1016/s0958-1669(98)80042-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 26.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 28.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 29.Mewes HW, et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Nature. 2001:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 31.Ditzel L, et al. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 32.Srikakulam R, Winkelmann DA. Myosin II folding is mediated by a molecular chaperonin. J Biol Chem. 1999;274:27265–27273. doi: 10.1074/jbc.274.38.27265. [DOI] [PubMed] [Google Scholar]
- 33.Chapman E, et al. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci U S A. 2006;103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 35.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong H, Isom DG, Srinivasan R, Rose GD. Local secondary structure content predicts folding rates for simple, two-state proteins. J Mol Biol. 2003;327:1149–1154. doi: 10.1016/s0022-2836(03)00211-0. [DOI] [PubMed] [Google Scholar]
- 37.Miller EJ, Fischer KF, Marqusee S. Experimental evaluation of topological parameters determining protein-folding rates. Proc Natl Acad Sci U S A. 2002;99:10359–10363. doi: 10.1073/pnas.162219099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 39.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura A, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 41.Behrends C, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 42.Reissmann S, Parnot C, Booth CR, Chiu W, Frydman J. Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Mol Biol. 2007;14:432–440. doi: 10.1038/nsmb1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 44.Vinh DB, Drubin DG. A yeast TCP-1-like protein is required for actin function in vivo. Proc Natl Acad Sci U S A. 1994;91:9116–9120. doi: 10.1073/pnas.91.19.9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yaffe MB, et al. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- 46.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 47.Hellman U, Wernstedt C, Gonez J, Heldin CH. Improvement of an "In-Gel" digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 48.Labbe S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 49.Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farr GW, Scharl EC, Schumacher RJ, Sondek S, Horwich AL. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 51.Frydman J, et al. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCallum CD, Do H, Johnson AE, Frydman J. The interaction of the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) with ribosome-bound nascent chains examined using photo-cross-linking. J Cell Biol. 2000;149:591–602. doi: 10.1083/jcb.149.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong S, et al. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J Virol. 2001;75:2526–2534. doi: 10.1128/JVI.75.6.2526-2534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pijnappel WW, et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 56.Wolf YI, Brenner SE, Bash PA, Koonin EV. Distribution of protein folds in the three superkingdoms of life. Genome Res. 1999;9:17–26. [PubMed] [Google Scholar]
- 57.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 58.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 59.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in C: The Art of Scientific Computing. Cambridge University Press; 1992. [Google Scholar]
- 60.Dwight SS, et al. Saccharomyces genome database: underlying principles and organisation. Brief Bioinform. 2004;5:9–22. doi: 10.1093/bib/5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.