Abstract
We investigated the physiological role of endogenous MAPK-activating death domain-containing protein (MADD), a splice variant of the IG20 gene, that can interact with TNFR1 in tumor necrosis factor-α (TNFα)-induced activation of NF-κB, MAPK, ERK1/2, JNK, and p38. Using exon-specific short hairpin RNAs expressing lentiviruses, we knocked down the expression of all IG20 splice variants or MADD, which is overexpressed in cancer cells. Abrogation of MADD expression rendered cells highly susceptible to TNFα-induced apoptosis in the absence of cycloheximide. It also resulted in a dramatic loss in TNFα-induced activation of MAPK without any apparent effect on NF-κB activation. This observation was substantiated by an accompanying loss in the activation of p90RSK, a key downstream target of MAPK, whereas the NF-κB-regulated interleukin 6 levels remained unaffected. Endogenous MADD knockdown, however, did not affect epidermal growth factor-induced MAPK activation thereby demonstrating the specific requirement of MADD for TNF receptor-mediated MAPK activation. Re-expression of short hairpin RNA-resistant MADD in the absence of endogenous IG20 expression rescued the cells from TNFα-induced apoptosis. The requirement for MADD was highly specific for TNFα-induced activation of MAPK but not the related JNK and p38 kinases. Loss of MADD expression resulted in reduced Grb2 and Sos1/2 recruitment to the TNFR1 complex and decreased Ras and MEKK1/2 activation. These results demonstrate the essential role of MADD in protecting cancer cells from TNFα-induced apoptosis by specifically activating MAPKs through Grb2 and Sos1/2 recruitment, and its potential as a novel cancer therapeutic target.
Genes in higher organisms generate alternate transcripts that are translated into closely related proteins with different functions. Perturbations in the tightly regulated alternate splicing of key genes in cancers can result in the accumulation of select splice variants of a particular gene or suppression of others. For instance, some cancers are known to preferentially express the more oncogenic and constitutively active RONΔ (where RON is recepteur d'origine nantais receptor tyrosine kinase) splice variant of RON receptor tyrosine kinase (1). The study of genes that undergo alternative splicing is therefore likely to unravel novel therapeutic targets against cancer (2–4). The IG20 (insulinoma-glucagonoma) is one such gene previously identified in our laboratory (4) that is implicated in cancer cell survival, proliferation, apoptosis, and other regulated functions through alternative splicing (5–20). The IG20 gene encodes at least six different splice variants (SVs)3 of which the expression of KIAA0358 and IG20-SV4 isoforms is restricted to certain neuronal tissues (17), with KIAA acting as a Rab3a-GEP (20–22). The other four, namely IG20pa, MADD, IG20-SV2, and DENN-SV, are expressed more ubiquitously (4). Of these, MADD and DENN-SV are constitutively expressed, whereas the IG20pa and IG20-SV2 may or may not be expressed.
Among the IG20 isoforms, MADD is overexpressed in cancer cells and tissues, and by acting as a negative regulator of caspase-8 activation, it contributes to cancer cell survival (18). Abrogation of MADD, but not the other IG20-SVs (IG20-SV is the insulinoma-glucagonoma clone 20 splice variant), renders cancer cells more susceptible to spontaneous as well as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis (18, 19). Moreover, expression of an shRNA-resistant MADD, and not the other isoforms of the IG20 gene, can rescue cells from undergoing apoptosis upon shRNA-mediated abrogation of expression of all isoforms of the IG20 gene (19). Endogenous MADD can prevent caspase-8 activation without directly interacting with caspase-8. The intriguing finding that MADD plays a predominant role in cancer cell survival and confers resistance to TRAIL-induced apoptosis led us to further examine the role of endogenous MADD in TNFα-induced apoptosis and the underlying signaling pathways such as NF-κB and MAPKs (ERK, JNK, and p38) in this study.
The MAPKs are serine/threonine-specific protein kinases that respond to a variety of extracellular stimuli and regulate several important and critical cellular functions such as cell cycle progression, expression of cytokines, motility, and adherence. Hence MAPKs influence cell survival, proliferation, differentiation, development, and apoptosis (23–24). The three main members of MAPK family are ERK1/2 or more commonly referred to as MAPK, JNK, and p38.
Relatively high levels of MAPK activity are noted in approximately one-third of all human cancers, thereby making MAPK an attractive target in the development of novel cancer therapies (23–25). Activated MAPKs phosphorylate several nuclear and cytoplasmic substrates involved in diverse cellular processes, including regulation of transcription and activation of kinases and phosphatases. One of the key substrates of MAPK that plays an important role in cell growth and proliferation is p90RSK (26–27). In addition, MAPK also activates the transcription factor ELK1 that triggers the transcription of c-Fos (28–29). It is important to note that p90RSK is activated in several cancers, and c-fos is a known proto-oncogene (26, 30).
Although we know a great deal about the proximal events that link activated growth factor receptors to MAPK activation, similar events underlying the TNF/TNFR1-mediated increase in MAPK activity are not fully understood. TNFα, an inflammatory cytokine, is likely to play a very important role in tumorigenesis and metastasis. Tumor stromal cells produce TNFα, which is known to promote angiogenesis and metastasis through MAPK activation (31). Because MADD is known to directly interact with TNFR1 cytoplasmic tail and overexpression of exogenous MADD promotes TNFα-induced activation of MAPK and JNK (12), we investigated the physiological role of MADD in TNFα/TNFR1-mediated signaling pathways. We provide here clear evidence that shows an essential role for endogenous MADD but not other splice variants of the IG20 gene in promoting resistance to TNFα-induced apoptosis. Our studies revealed a critical role for MADD in basal as well as TNFα-induced MAPK activation, but not JNK or p38, by facilitating the recruitment of Grb2 and Sos1/2 to the TNFα-TNFR1 complex with resultant Ras activation.
EXPERIMENTAL PROCEDURES
Materials—Antibodies against phospho-ERK, phospho-JNK, phospho-p38, phospho-MEK1/2, phospho-p90RSK, ERK, JNK, p38, IκB-α, caspase-3 (H-277), and the epidermal growth factor were purchased from Cell Signaling Technology Inc. (Beverly, MA). Rabbit anti-human Grbb2 antibody was obtained from Cell Signaling (Boston). Rabbit anti-human Sos1/2 antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The recombinant human TNFα, EGF, and biotinylated goat anti-human TNFR1 antibody, goat anti-human TNFR1, and goat anti-human EGF receptor antibody were fromR&D Systems (Minneapolis, MN). The anti-GFP/yellow fluorescent protein mouse monoclonal antibody (JL-8) was obtained from Clontech. Anti-actin antibody was obtained from Sigma. The anti-MADD antibody was generated at Eurogentec (Koeln, Germany). The goat anti-mouse IgG1 peroxidase-conjugated secondary antibody was obtained from Caltag Laboratories (Burlingame, CA). The donkey anti-rabbit and anti-goat peroxidase-conjugated polyclonal secondary antibody and ECL Plus were purchased from GE Healthcare. HRP-streptavidin was purchased from Pierce. The tetramethylrhodamine methyl ester was procured from Molecular Probes/Invitrogen.
Cell Culture—Cervical carcinoma cell line HeLa, ovarian carcinoma cell line PA-1, and human embryonic kidney cell line 293T were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with fetal bovine serum (10%), antibiotic/antimycotic (1%), and l-glutamine (2 mm). Cells were cultured at 37 °C with 5.0% CO2 in a humidified atmosphere by routine passage every 3 days.
Lentiviral Production—The procedures for generating recombinant lentiviruses and infecting various cells have been described previously (18–19). Specific knockdown of select combinations or all IG20 isoforms using different lentiviruses expressing different shRNAs was described previously (18–19). The cells were harvested at different time periods post-transduction as indicated. For experiments wherein MADD function was reconstituted, we used cells in which all IG20-SVs were knocked down. In these cells, devoid of endogenous IG20-SVs, we expressed a site-directed MADD mutant (SirMADD) that is not recognized by the Mid shRNA that we have described previously (18).
Reverse Transcription (RT)-PCR—Total RNA was extracted from HeLa and PA-1 cells (1 × 106 cells untransduced and transduced with SCR, Mid, 13L, and 16E shRNA expressing lentiviruses for 48 h) using TRIzol reagent (Invitrogen). RNA (1 μg) was used to perform RT-PCR using the SuperScript III One-Step RT-PCR system (Invitrogen). Briefly, the cDNAs were synthesized at 50 °C for 30 min followed by incubation at 94 °C for 2 min. The preparations were then subjected to 30 cycles of PCR (denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min). Final extension was carried out at 72 °C for 7 min. The primers used to amplify all the IG20 isoforms have been reported previously (4). Glyceraldehyde-3-phosphate dehydrogenase primers were used as an internal control for the RT-PCR, and the PCR products were visualized on a 5% PAGE gel.
Caspase Activation—Thirty six hours post-transduction, HeLa cells were treated with TNFα (50 ng/ml), and apoptosis was assessed based on caspase-3 activation. Relative levels of active caspase-3 were determined in fixed and permeabilized cells using cytofix/cytoperm solution (BD Biosciences) for 30 min at 4 °C. Cells were washed with Perm/Wash buffer (Pharmingen) and stained for 1 h in a dark at room temperature with 20 μl of active caspase-3 phycoerythrin-conjugated antibody per sample. Cells were collected, washed, and resuspended in Perm/Wash buffer, and the percentage of phycoerythrin-positive cells in GFP-positive pool for each sample was then determined using a flow cytometer.
Tetramethylrhodamine Methyl Ester Staining—HeLa cells (5 × 105) were transduced with different lentiviruses for 36 h and then treated with TNFα (50 ng/ml), without cycloheximide, for 8 h. The cells were subsequently collected, washed in cold PBS, and then stained with tetramethylrhodamine methyl ester (100 nm) for 15 min at 37 °C. Cells were then washed with ice-cold PBS and then subjected to FACS analysis by gating on the GFP+. Loss of tetramethylrhodamine methyl ester staining as a marker of apoptosis because of mitochondrial depolarization was determined.
Determination of IL-6 Levels—HeLa cells were transduced for 36 h with different lentiviral vectors and subsequently treated with TNFα (50 ng/ml) for 8 or 16 h. Culture supernatants were collected and assayed for IL-6 levels using the human IL-6 ELISA kit (BioSource International, Camarillo, CA) following the manufacturer's instructions.
Immunoprecipitation of the TNFR1 Complex—HeLa cells (4 × 105) were transduced with different lentiviruses for 72 h followed by overnight serum starvation and then treated with TNFα (50 ng/ml) for different times at 37 °C. Afterward, the cells were washed in cold PBS, pelleted, and then lysed in 0.5 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl; 1% Non-idet P-40) for 30 min at 4 °C and clarified by centrifugation. The supernatants were normalized for protein concentration, and 1 mg of lysates was pre-cleared with protein G-agarose beads and incubated with 10 μg of goat anti-human TNFR1 antibody on a Rotoshaker overnight at 4 °C. The immune complexes were immunoprecipitated using 50 μl (50% slurry) of protein G-agarose beads. The immune complexes were then subjected to 8–16% SDS-PAGE analysis and immunoblotted to detect Grb2, Sos1/2, and TNFR1. As a positive control, EGF-treated or untreated HeLa cell lysates were used for EGFR immunoprecipitation.
Immunoblotting—HeLa (3 × 106) or PA-1 (5 × 106) cells transduced with different lentiviruses for various times were serum-starved for 6 h. They were then treated with either TNFα (50 ng/ml) or other ligands as indicated at 37 °C. Cells were lysed, and equivalent amounts of lysates (50 μg of protein/sample) were separated by SDS-PAGE (12%) and transferred to polyvinylidene difluoride membranes. The membranes were then blocked and stained with primary antibodies diluted 1:5000 in TBST containing 5% bovine serum albumin at 4 °C overnight. Secondary antibodies such as HRP-conjugated goat anti-mouse IgG1 is used at 1:5000 dilution for detecting p-ERK, p-JNK, p-p38, and Ras, and HRP-conjugated donkey anti-rabbit IgG is used at 1:10,000 dilution for detecting total ERK, JNK, p38, p90RSK, IκB-α, MEK1/2, Grb2, Sos1/2, and β-actin. HRP-conjugated donkey anti-goat IgG was used at 1:5000 dilution for detecting EGFR. HRP-Streptavidin was used at 1:300 dilution for detecting biotinylated TNFR1. An enhanced chemiluminescence-plus detection system (GE Healthcare) was used to detect specific protein bands. Protein loading in all experiments was normalized by stripping the blots and then re-probing with the antibody for β-actin.
RAS Activation Assay—We used a commercially available kit for determining Ras activity as per manufacturer's protocol (Millipore, Temecula, CA). HeLa cells (4 × 106) transduced with respective lentiviruses for 36 h and serum-starved for 18 h were treated with TNFα (100 ng/ml) for 5 min at 37 °C. Cells were then lysed, and the lysates were incubated for 45 min at 4 °C with 5 μl of Ras assay reagent (glutathione S-transferase fusion protein, corresponding to the human Ras binding domain (RBD, residues 1–149) of Raf-1 bound to glutathione-agarose). The sample beads were then washed and resuspended in 40 μl of 2× Laemmli reducing sample buffer and boiled for 5 min. The presence of activated RAS (RAS-GTP) was determined by anti-RAS immunoblotting.
RESULTS
Down Modulation of MADD Renders HeLa Cells Susceptible to TNFα-induced Apoptosis—TNFα is an inflammatory and pro-survival cytokine. Unlike some of the other members of the TNF family such as TRAIL and FasL, TNFα is a potent activator of NF-κB, and for TNFα-induced apoptosis the cells need to be pretreated with cycloheximide, a protein translation inhibitor (32). Because previous studies from our laboratory demonstrated a role for endogenous MADD in promoting cell survival (18, 19), we determined the effect of MADD knockdown on TNFα-induced cell survival. We treated HeLa cells with recombinant lentiviruses that express either scrambled or exon specific shRNAs and monitored the effects of this treatment on IG20 protein by Western blot using the 13L antibody that specifically reacts with amino acid sequence encoded by exon 13L. Cells treated with either 16E or 13L shRNA showed a moderate reduction presumably because of reduced expression of IG20pa/IG20-SV2 and IG20pa/MADD, respectively. However, cells treated with Mid shRNA showed a marked reduction in the levels of expression of “IG20 protein” because of knock-down of all isoforms. Because antibodies that can specifically react with each IG20 isoform are not available and to determine whether the shRNA treatments had the desired effect, we tested for the expression of specific transcripts by RT-PCR (4). As evident from the results shown in Fig. 1B, IG20 transcripts were significantly down-modulated at 48 h of transduction. Lentivirus expressing Mid shRNA knocked down the expression of all four IG20-SVs, whereas 13L knocked down MADD and IG20pa and 16E knocked down IG20pa and IG20-SV2. We then treated the cells with TNFα and determined the percentage apoptosis by monitoring the expression of the active form of caspase 3 (Fig. 1, C and D). Down-modulation of all IG20-SVs using Mid shRNA or MADD and DENN-SV using 13L shRNA resulted in significant increases in active caspase-3 levels upon TNFα treatment (Fig. 1, C and D). Knocking down IG20pa and IG20-SV2 splice variants and sparing MADD and DENN-SV (using 16E shRNA) isoforms yielded results similar to that seen with control cells. Also, down modulation of MADD using 13L shRNA resulted in consistently higher spontaneous as well as TNFα-induced apoptosis. It should be noted that we did not pretreat the cells with cycloheximide in the above experiments. Using the above three shRNAs, we deduced that the effects seen in 13L-treated cells are essentially because of knockdown of MADD (18, 19). To understand the survival role of MADD in TNFα-induced signaling, we systematically examined the effect of MADD abrogation on key signaling pathways.
FIGURE 1.
Down-modulation of MADD or all IG20-SVs leads to TNFα-induced apoptosis. A, HeLa cells were transduced with different lentiviral vectors expressing SUP (empty vector, control (CTL)), SCR (scrambled) control, Mid, 13L, and 16E shRNA for 36 h. Whole cell lysates were analyzed by immunoblotting using 13L antibody that specifically detects IG20pa and MADD proteins that contain exon 13L, and here we refer to them as MADD protein. B, 1 μg of total RNA obtained from HeLa cells 48 h post-transduction with the indicated shRNA-encoding lentiviruses was used for reverse transcription-PCR. The products were separated by PAGE using a 5% gel. Amplification of all four IG20-SVs was carried out using the F1-B1 primers (15, 16). MID virus transduction resulted in the knockdown of all IG20 isoforms. Expression of 13L shRNA resulted in the specific knockdown IG20pa and MADD, whereas expression of 16E abrogated IG20pa and IG20-SV2 isoforms. C, subsequently, the transduced cells were treated with 50 ng/ml TNFα for 8 h, and active caspase-3 was detected by FACS. Representative histogram (C) and summarized data (D) on active caspase-3 detection are shown.
Down-modulation of IG20-SVs or MADD Does Not Affect TNFα-induced NF-κB Activation—TNFα is a known potent activator of the NF-κB-mediated pro-survival signaling pathway (33). Using the above described knockdown and control HeLa cells, we determined the status of TNFα-induced changes in NF-κB activity as reflected by the degree of IKα degradation (Fig. 2A). As shown, controls, Mid, and 13L shRNA-expressing cells (36 h post-transduction) were treated with TNFα for various times. Cell lysates at the indicated times were immunoblotted with anti-IκB-α antibody. As observed in Fig. 2A, the pattern of IκB-α degradation upon MADD down-modulation (in Mid and 13L cells) was similar to that of control cells (SCR), suggesting that loss of MADD or any of the other IG20-SVs had no discernible effect on TNFα-induced NF-κB activation. We then substantiated the above finding by determining the effect of MADD abrogation on one of the NF-κB target genes, namely IL-6 (Fig. 2B). In a similar experiment as described above, we determined the relative levels of IL-6 secreted into the cell culture supernatant using IL-6-specific ELISA. In accordance with the results shown in Fig. 2A, loss of MADD expression did not have a significant effect on TNFα/NF-κB-mediated increase in IL-6 secretion (Fig. 2B). We next examined the effect of MADD knock-down on TNFα-induced activation of various MAPKs such as MAPK (ERK1/2), JNK, and p38 (33).
FIGURE 2.
Down-modulation of MADD or IG20-SVs does not affect TNFα-induced NF-κB activation or the resultant IL-6 production. A, 36 h posttransduction, HeLa cells were treated with TNFα (50 ng/ml), and at indicated time points the cells were collected, lysed, subjected to Western blotting, and probed for IκB-α and actin. B, HeLa cells transduced with respective lentiviral vectors for 36 h were either left unstimulated or stimulated with TNFα (50 ng/ml) for 8 and 16 h, respectively. The supernatants were collected and IL-6 levels measured using an ELISA.
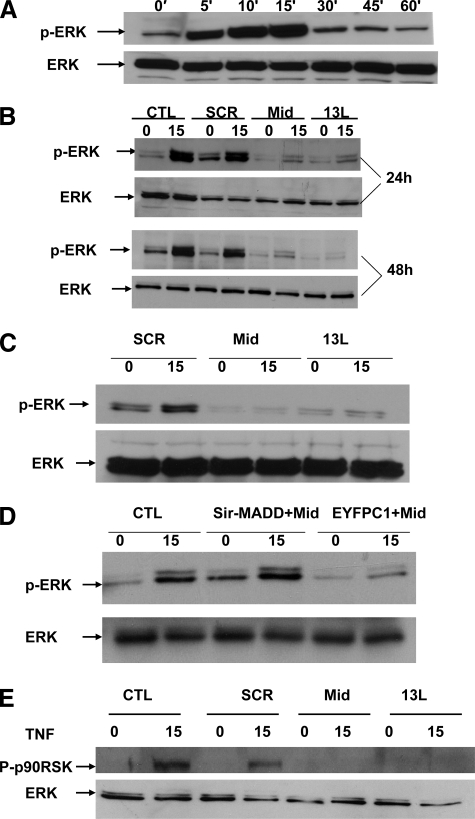
Endogenous MADD Is Essential and Sufficient for TNFα-induced ERK Activation—Once again, using the experimental approach described above, we treated the control, Mid, and 13L shRNA-expressing HeLa cells harvested at three different times after transduction with TNFα. The lysates at the indicated time points were immunoblotted with anti-phospho-ERK1/2 and anti-ERK1/2 antibodies. As shown in Fig. 3A, TNFα induced rapid phosphorylation of ERK1/2 at 15 min in untransduced HeLa cells. However, we noted a reduction in both basal as well as TNFα-induced MAPK activation as shown by reduced phosphorylation of ERK1/2 in Mid- and 13L-expressing cells relative to control cells (Fig. 3B). These data indicated that MADD is essential for MAPK activation.
FIGURE 3.
IG20-SVs regulate TNFα-induced ERK activation. A, kinetics of TNFα-induced ERK activation. HeLa cells (3 × 106 per 100-mm dish) were serum-starved for 6 h and were either left untreated or treated with TNFα (50 ng/ml) for various times, as indicated. Cell lysates were analyzed for the detection of phosphorylated and total ERK by immunoblotting. B, down-modulation of IG20-SVs or MADD significantly decreased TNFα-induced ERK phosphorylation. HeLa cells (3 × 106 per 100-mm dish) were either untransduced or transduced with different lentiviruses expressing SCR, Mid, and 13L shRNA for different time periods as indicated. At the end of each time period, the cells were serum-starved for 6 h and were left either untreated or treated with TNFα (50 ng/ml) for 15 min. Cell lysates were analyzed for phosphorylated ERK or total ERK by immunoblotting. The data shown are representative of at least three independent experiments. C, PA-1 cells (5 × 106 per 100-mm dish) that express only the MADD and the DENN-SV isoforms were transduced with different lentiviruses expressing SCR, Mid (to knockdown both isoforms), and 13L (to knock down only MADD) shRNA for 48 h. The cells were then serum-starved for 6 h and either left unstimulated or stimulated with TNFα (50 ng/ml) for 15 min. Cell lysates were analyzed for phosphorylated ERK or total ERK by immunoblotting. D, HeLa cells (3 × 105 per 60-mm dish) were either left untransfected or transfected with either Mid shRNA-resistant MADD (Sir-MADD) or EYFPC1 plasmids. Twenty four hours post-transfection, the cells were infected with Mid shRNA, and 48 h later the cells were either left unstimulated or stimulated with TNFα (50 ng/ml) for 15 min. Cell lysates were analyzed for phosphorylated ERK or total ERK by immunoblotting. The data presented are representative of at least three independent experiments. E, HeLa cells (3 × 106 per 100-mm dish) were transduced with different lentiviruses expressing SCR and Mid shRNA for 48 h. The cells were then serum-starved for 6 h and were either left untreated or treated with TNFα (50 ng/ml) for 15 min. Cell lysates were analyzed for phosphorylated p90RSK or total ERK by immunoblotting. CTL, control.
To further substantiate this, we also carried out similar experiments in PA-1 ovarian cancer cells that express only the MADD and the DENN-SV isoforms of the IG20 gene (14, 15, 19). Similar to the observation made in HeLa cells, TNFα induced significant increases in MAPK activity in control PA-1 cells; however, this was dramatically decreased in cells expressing either Mid (which knocks down both MADD and DENN-SV) or 13L (which knocks down only MADD) shRNAs (Fig. 3C), thereby supporting our previous findings that endogenous MADD is indeed required for TNFα-induced ERK1/2 activation. To further confirm the central role of MADD, we generated a mutated MADD cDNA construct that is resistant to Mid shRNA (pEYFPC1-Sir-MADD) (19). We expressed Sir-MADD in Mid shRNA-expressing cells in which endogenous IG20 expression was down-modulated. As shown in Fig. 3D, transfection of pEYFPC1-Sir-MADD but not the empty vector restored TNFα-induced ERK1/2 activation to levels shown by control cells indicating that expression of MADD isoform alone is sufficient to mediate TNFα-induced ERK1/2 activation.
Next, we determined the effect on an important intracellular substrate of MAPK, namely p90RSK. As shown in Fig. 3E, down-modulation of all IG20-SVs (Mid ShRNA) or MADD (13L shRNA) led to a dramatic decrease in phosphorylation of p90RSK. Even considering the slight variation in the amounts of total p90Rsk in the whole cell lysates, loss of MADD expression had profound inhibitory effect on TNFα-induced p90RSK activation.
Requirement of Endogenous MADD for Specific Activation of ERK upon TNFα Treatment—In addition to MAPK, TNFα is also known to activate related kinases such as JNK and p38. To see whether the requirement for MADD is specific for TNFα-induced activation of ERK1/2, we determined the effect of knockdown of MADD and IG20-SVs on JNK and p38 kinases. Down-modulation of all IG20-SVs or a combination of MADD along with IG20pa did not affect TNFα-induced JNK1/2 activation (Fig. 4A) or p38 phosphorylation (Fig. 4B), thereby demonstrating the specific requirement of endogenous MADD for TNFα-induced ERK1/2 activation.
FIGURE 4.
IG20-SVs do not affect TNFα-induced activation of JNK and p38 and are not involved in EGF-induced ERK phosphorylation. A and B, TNFα-induced JNK and p38 phosphorylation in HeLa cells with knockdown of IG20-SVs. HeLa cells (3 × 106 per 100-mm dish) were transduced with different lentiviruses expressing SCR, Mid, and 13L shRNA for 48 h. The cells were then serum-starved for 6 h and were either left un-stimulated or stimulated with TNFα (50 ng/ml) for 15 min. Cell lysates were analyzed for phosphorylated JNK and total JNK (A) or phosphorylated p38 and p38 (B) by immunoblotting. The data presented are representative of at least three independent experiments. C, HeLa cells (3 × 106 per 100-mm dish) were transduced with different lentiviruses expressing SCR and Mid ShRNA for 48 h. The cells were then serum-starved for 6 h and were either left unstimulated or stimulated with 100 ng/ml EGF or 50 ng/ml TNFα for 15 min (C). Cell lysates were analyzed for phosphorylated ERK and total ERK by immunoblot. The data are representative of at least three independent experiments. CTL, control.
Apart from TNFα, growth factors such as EGF (through EGFR) are also known to activate MAPK (23, 24). To determine whether endogenous MADD is required for MAPK activation by EGF, we treated control and Mid shRNA-expressing cells with EGF and compared them with results obtained using TNFα treatment. As expected, EGF treatment resulted in a significant MAPK activation. However, unlike TNFα-induced activation of MAPK that was dramatically suppressed in Mid shRNA-expressing cells, there was no significant change in MAPK activity in EGF-treated cells (Fig. 4C). Taken together, these data clearly demonstrated the specific requirement of MADD isoform in TNFα-induced activation of MAPK.
Endogenous MADD Is Required for Ras Activation and Grb2 and Sos1/2 Recruitment to TNFR1—To determine the most proximal event targeted by MADD in the TNFα-induced MAPK activation pathway, we systematically traced the effect of MADD knockdown on key upstream events in the MAPK activation pathway. We determined the effect of MADD on MEKK1/2 activation. As shown in Fig. 5A, 15 min after TNFα treatment there was a significant activation of MEKK1/2 in control cells but not in cells treated with either Mid or 13L shRNA with resultant MADD knockdown. Next, we determined Ras activation in control and MADD knockdown cells treated with TNFα, using Raf-RBD pulldown followed by immunoblotting with an anti-Ras antibody. As shown in Fig. 5B, although the total Ras levels were comparable in all lysates, the TNFα-stimulated Ras-GTP levels were highly suppressed in cells with MADD knockdown. Based on the above findings, we conclude that MADD is necessary for Ras and MEKK1/2 activation.
FIGURE 5.
MADD facilitates TNFα-induced signaling by recruiting Grb2 to TNFR1. A, HeLa cells (3 × 106 per 100-mm dish) were either left untransduced or were transduced with different lentiviruses expressing SCR, Mid, and 13L shRNA for 48 h. The cells were then serum-starved for 6 h and were either left untreated or treated with TNFα (50 ng/ml) for 15 min. Cell lysates were probed for phosphorylated MEKK1/2 or total MEKK by immunoblotting. B, HeLa cells (4 × 106) transduced with SCR and Mid lentiviruses for 36 h were serum-starved for 18 h and treated with TNFα (100 ng/ml) for 5 min at 37 °C. Activated Ras in the cell lysates was precipitated by using RBD-coupled agarose. The presence of activated Ras (Ras-GTP) was determined by anti-Ras immunoblotting. The total RAS in the whole cell lysate served as the loading control. C, HeLa cells (4 × 106) transduced with SCR and Mid lentiviruses for 72 h were serum-starved overnight and treated with TNFα (50 ng/ml) for different times at 37 °C. Cell lysates were prepared and immunoprecipitated using an anti-TNFR1 antibody and then subjected to SDS-PAGE followed by Western blotting using Grb2, Sos1/2, or a biotinylated TNFR1-specific antibody. The total Grb2, Sos1/2, and TNFR1 in the whole cell lysate served as loading controls. D, EGFR was immunoprecipitated from EGF treated or untreated HeLa cell lysates, and proteins were separated and immunoblotted (IB) using Grb2- or Sos1/2-specific antibodies. CTL, control.
We then determined the effect of loss of MADD expression on the event immediately upstream of Ras and downstream of TNFR1, namely the recruitment of Grb2 that occurs immediately after the ligation of TNFα with TNFR1. We treated both SCR and Mid shRNA-expressing cells with 50 ng/ml of TNFα for the indicated times, lysed the cells, and immunoprecipitated TNFR1 complexes from clarified cell lysates. We then determined the relative levels of TNFR1 in the immunoprecipitate and also the levels of co-precipitated Grb2 and Sos1/2 using immunoblotting. As shown in Fig. 5C, the amounts of TNFR1 precipitated in the SCR and MADD knockdown cells at various times after TNFα treatment were comparable indicating that abrogation of MADD expression had no adverse effect on the cell surface levels of TNFR1 at the time points examined. Also the levels of TNFR1 were very similar in all whole cell lysates clearly demonstrating that we used equivalent amounts of cell lysates for immunoprecipitation. In control cells treated with TNFα, we observed a rapid increase in the amounts of co-precipitated Grb2 and Sos1/2, which peaked at 2 min. In comparison, in cells with MADD knockdown and treated with TNFα, Grb2 and Sos1/2 failed to co-precipitate. The relative expression levels of Grb2 and Sos1/2 were similar in the total whole cell lysates (Fig. 5C). As a control, we also performed EGFR immunoprecipitation using EGF-treated HeLa cell lysate. Consistent with earlier reports (34), upon treatment with EGF for 2.5 min, a dramatic increase in the recruitment of Grb2 and Sos1/2 to EGFR was noted (Fig. 5D). The above results clearly demonstrated the requirement of MADD for a more efficient recruitment of Grb2 and Sos1/2 to the TNFR1 complex upon TNFα treatment; failing which Ras activation is compromised.
DISCUSSION
TNFα is the prototypic member of the TNF family of ligands and mainly transduces signals through TNFR1 and -2. However, in non-lymphoid cells it signals through TNFR1, which is the dominant receptor (35). Our recent study using thyroid cancer cells (36) and this study clearly show that MADD can protect cells from TNFα-induced apoptosis. The pro-survival nature of MADD became apparent when TNFα-induced apoptosis was enhanced upon abrogation of MADD expression. Generally, TNFα-induced apoptosis is only seen in cells treated with cycloheximide, which is necessary to block new protein synthesis that includes several anti-apoptotic proteins whose transcription is triggered because of TNFα-induced activation of NF-κB (32). However, we observed significant apoptosis upon TNFα treatment in cells expressing reduced levels of MADD without pre-treatment with cycloheximide (36 and Fig. 1). Furthermore, our study clearly demonstrated that MADD is essential for basal as well as TNFα-induced activation of ERK1/2 but not for the activation of NF-κB (Fig. 6). Moreover, our results showed that MADD is required for the recruitment of Grb2 and Sos1/2 to the TNFR1 signaling complex and for Ras activation, two initial events that are absolutely essential for downstream activation of MEKK followed by ERK1/2 and Rsk activation.
FIGURE 6.
Model depicting the essential role of MADD in TNFR1-mediated MAPK activation. A, MADD can constitutively bind to TNFR1, and upon TNFα binding to the TNFR, MADD facilitates rapid recruitment of Grb2 to TNFR1 that leads to the activation of Ras and other downstream MAPK signaling molecules. B, TNFα treatment of cells wherein MADD is abrogated results in the prevention or delay in Grb2 recruitment to the TNFR1 signaling complex and consequent activation of ERK1/2 with no apparent effect on NF-κB activation.
Most of the cancer cells are known to be highly susceptible to death receptor-induced apoptosis when treated with death ligands such as FasL and TRAIL. In contrast, TNFα per se is not known to induce such apoptosis. Our results unambiguously show that it is MADD that offers natural protection against TNFα/TNFR1-induced cell death and indicates a physiological role for TNFα/TNFR1-induced apoptosis. The Fas, unlike TNFR1, is known to mediate apoptosis not only in transformed cells but also in normal cells (37). Mutations that generate nonfunctional Fas are known to result in autoimmune lymphoproliferative disorders, both in humans and mice (38). Although TNFR1 knock-out mice do not develop such disease, upon induction of peptide-induced chronic demyelinating autoimmune encephalomyelitis, there is a 50% reduction in T cell apoptosis compared with their wild type littermates (39). In addition, compared with Fas knock-out (lpr) mice, Fas/TNFR1 double knock-out mice suffer from earlier onset of lymphoproliferative syndrome and increased mortality (40). These observations provide further evidence in support of a physiological role for TNFα/TNFR1-mediated apoptosis.
One of the key pro-survival pathways activated by TNFα/TNFR1 is the NF-κB pathway, and NF-κB-regulated genes, such as BCL-XL and c-FLIP (c-FLIP is cellular-FLICE inhibitory protein), play a key role in inhibiting both extrinsic and intrinsic cell death (41–42). Our studies, however, clearly show that abrogation of MADD expression had no discernible effect on TNFα-enhanced NF-κB activity (Fig. 2). IL-6, a pro-inflammatory cytokine, is one of the targets of activated NF-κB (43). IL-6 levels in TNFα-treated MADD knockdown cells were comparable with those observed in control cells thereby strengthening our observation that loss of MADD expression has no effect on TNFα-induced NF-κB activity (Fig. 2). In lymphocytes wherein TNFR2 is also expressed, TNFα is known to interact with both TNFR1 and -2. Both TNFRs then mediate signaling cascades through MAPKs and NF-κB (33). We are currently determining the role of MADD, if any, in TNFR2 signaling.
We and others have shown that MADD interacts with some of the TNFR family members that have the death domain in their cytoplasmic tails, namely TNFR1, death receptor 4, and death receptor 5 (12, 18, 19). It appears to serve a very specific function with respect to TNFR1. As shown in Fig. 4C, MADD can be dispensed with in case of the growth factor EGF-induced MAPK activation. Ligation of TNFα to TNFR1 also results in the activation of other stress-induced kinases such as JNK and p38; however, MADD appears to have no role in these pathways (Fig. 4, A and B). There are other reports that show that activation of JNK and/or p38 kinases, although MAPK activity is suppressed, can facilitate apoptosis (44). Our results demonstrated a direct relationship between loss of MADD expression and inhibition of TNFα-induced activation of MAPK without adversely affecting JNK and p38 kinase activation; conditions that resulted in a significantly increased TNFα-induced cell death. Whether the TNFα-induced apoptosis in MADD knock-down cells is solely because of loss in MAPK activity or is because of the combined effects of increased JNK and p38 kinase activities in the absence of MAPK activation is currently being investigated.
The signaling cascade that links growth factor receptors such as EGFR to MAPK has been clearly elucidated (23, 24). In the case of EGFR signaling, ligation of the receptor results in increased autophosphorylation of the cytoplasmic tail resulting in the generation of tyrosine-phosphorylated motifs that can recruit the SH2 domain of Grb2. Subsequently, the Sos1/2 molecules are recruited to the EGFR complex through a prolinerich region that specifically interacts with the SH3 domain of Grb2 (45). The cytoplasmic tail of TNFR1 neither has the intrinsic kinase activity nor a suitable tyrosine motif that can be phosphorylated by intracellular tyrosine kinases. Therefore, it was not clear as to how Grb2 and Sos1/2 proteins are recruited to the TNFR1 complex upon ligation with TNFα. However, it has a death domain that interacts with the death domain homology region in MADD (12). The precise molecular events that lead to the formation of the TNFR1 signaling complex and activation of MAPK have not been fully elucidated. Our results do clearly demonstrate that MADD, which is constitutively bound to TNFR1, is necessary for the recruitment of Grb2 and Sos1/2 (Fig. 5). Additional key support comes from the fact that in MADD knockdown cells, TNFα treatment failed to activate immediately the downstream Ras as well as MEKK, which are indispensable for the ultimate activation of MAPK. Hildt and Oess (46) previously showed that Grb2 and TNFR1 are direct binding partners and that the SH3 domain in Grb2 interacted with the PALP motif in the cytoplasmic tail of TNFR1. Our results demonstrate that MADD, which at that time had not been identified, is necessary for Grb2 recruitment to TNFR1 complex. Whether MADD facilitates the direct interaction of Grb2 with the proline-rich motif in the cytoplasmic tail of TNFR1 remains to be seen.
A seminal discovery in TNFR1 signaling demonstrated a clear demarcation between signals that end in NF-κB activation and apoptosis through the formation of signaling Complex 1 and 2, respectively (33). TNFα binding to TNFR1 results initially in the formation of Complex 1 that consists of TNF-R1, TNF-R1-associated death domain, RIPK1 (receptor interacting protein kinase 1), and cIAP1 and cIAP2 (cellular inhibitor of apoptosis) (Fig. 6B). This complex by default activates NF-κB that results in promoting cell survival (activation of BCL-xL, and c-FLIP) and secretion of pro-inflammatory cytokines (TNFα and IL-6). However, when the above survival pathway is inhibited by cycloheximide, TNFα-induced signaling leads to the formation of Complex II that is made up of TNF-R1-associated death domain, Fas-associated death domain-containing protein, RIPK1, and procaspase-8 that can lead to the activation of caspase-8 and cell death (Fig. 6C) (33). Although the above model indicates the apoptotic potential of TNFR1 signaling, the use of cycloheximide is pharmacological rather than physiological.
Although knockdown studies presented here show an important pro-survival role for MADD in TNFα/TNFR1 signaling (Fig. 6), they do not indicate how this function is physiologically regulated. In an earlier study we showed that overexpression of the IG20pa splice variant of the IG20 gene, which can act as a dominant negative MADD, can render cells more susceptible to TNFα-induced apoptosis (4). This raises the possibility that under physiological conditions, through regulation of specific splice factors, the ratio of MADD to IG20pa expression could be reduced, thus rendering cells susceptible to TNFα-induced apoptosis. Because knockdown of MADD had no effect on TNFα-induced activation of NF-κB, it is unlikely to influence the formation of Complex-1, but it may facilitate more rapid formation of Complex-2 in cells treated with TNFα (Fig. 6). This would also suggest that expression of NF-κB activated anti-apoptotic genes such as BCL-XL and c-FLIP are also likely to remain unaffected in MADD knockdown cells. However, an important downstream target of MAPK is the ELK1 transcription factor that triggers the expression of c-Fos (29). JNK phosphorylates c-Jun that combines with c-Fos and forms a key transcription factor AP-1 (28). In MADD knockdown cells the above connection is likely to be severed, and whether a resultant deficiency in AP-1 is the underlying cause of TNFα-induced apoptosis in the MADD knockdown cells needs to be verified.
Tumors are enclosed by stromal cells composed of macrophages and infiltrating lymphocytes that are known to be responsible for some of the chronic inflammation seen in cancer patients. TNFα is one of the inflammatory cytokines secreted by the activated stromal cells (31), and it is likely to be responsible for the elevated levels of MAPK usually encountered in tumor cells (25). MAPK activity plays an important role in promoting tumor angiogenesis and metastasis (23, 24). Therefore, our results raise the possibility that MADD could be a potential target in cancer therapy. Because loss of MADD expression does not appear to affect the survival of primary cells (13, 17), it may be a suitable target to treat cancers either by expressing MADD-specific small interfering RNA or by developing small molecule MADD antagonists.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA107506 (to B. S. P.).
Footnotes
The abbreviations used are: SV, splice variant; MADD, MAPK-activating death domain-containing protein; DENN, differentially expressed in normal and neoplastic cells; Sir-MADD, Mid sh-RNA-resistant MADD; SCR, scrambled; shRNA, short hairpin RNA; TNF-α, tumor necrosis factor-α; TRAIL, TNF-related apoptosis-inducing ligand; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; TNFR1, tumor necrosis factor receptor 1; MAPK, mitogen-activated protein kinase; MEK1/2, mitogen-activated protein kinase kinases; Raf-RBD, Raf-Ras binding domain; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; NF-κB, nuclear factor κB; PBS, phosphate-buffered saline; HRP, horseradish peroxidase; IL-6, interleukin 6; GFP, green fluorescent protein; FACS, fluorescence-activated cell sorter; ELISA, enzyme-linked immunosorbent assay; SH, Src homology.
References
- 1.Collesi, C., Santoro, M. M., Gaudino, G., and Comoglio, P. M. (1996) Mol. Cell. Biol. 16 5518-5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalnina, Z., Zayakin, P., Silina, K., and Line, A. (2005) Genes Chromosomes Cancer 42 342-357 [DOI] [PubMed] [Google Scholar]
- 3.Venables, J. P. (2006) Bioessays 28 378-386 [DOI] [PubMed] [Google Scholar]
- 4.Al-Zoubi, A. M., Efimova, E. V., Kaithamana, S., Martinez, O., El-Idrissi Mel, A., Dogan, R. E., and Prabhakar, B. S. (2001) J. Biol. Chem. 276 47202-47211 [DOI] [PubMed] [Google Scholar]
- 5.Del Villar, K., and Miller, C. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4210-4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, Y., Zhou, L., and Miller, C. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2586-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi, K., Tanaka, M., Mizoguchi, A., Hirata, Y., Ishizaki, H., Kaneko, K., Miyoshi, J., and Takai, Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14536-14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka, M., Miyoshi, J., Ishizaki, H., Togawa, A., Ohnishi, K., Endo, K., Matsubara, K., Mizoguchi, A., Nagano, T., Sato, M., Sasaki, T., and Takai, Y. (2001) Mol. Biol. Cell 12 1421-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, T. L., and Howe, P. H. (1998) Curr. Biol. 8 R191. [PubMed] [Google Scholar]
- 10.Chow, V. T., and Lee, S. S. (1996) DNA Seq. 6 263-273 [DOI] [PubMed] [Google Scholar]
- 11.Chow, V. T., Lim, K. M., and Lim, D. (1998) Genome 41 543-552 [DOI] [PubMed] [Google Scholar]
- 12.Schievella, A. R., Chen, J. H., Graham, J. R., and Lin, L. L. (1997) J. Biol. Chem. 272 12069-12075 [DOI] [PubMed] [Google Scholar]
- 13.Lim, K. M., and Chow, V. T. (2002) Mol. Carcinog. 35 110-126 [DOI] [PubMed] [Google Scholar]
- 14.Efimova, E. V., Al-Zoubi, A. M., Martinez, O., Kaithamana, S., Lu, S., Arima, T., and Prabhakar, B. S. (2004) Oncogene 23 1076-1087 [DOI] [PubMed] [Google Scholar]
- 15.Efimova, E., Martinez, O., Lokshin, A., Arima, T., and Prabhakar, B. S. (2003) Cancer Res. 63 8768-8776 [PubMed] [Google Scholar]
- 16.Ramaswamy, M., Efimova, E. V., Martinez, O., Mulherkar, N. U., Singh, S. P., and Prabhakar, B. S. (2004) Oncogene 23 6083-6094 [DOI] [PubMed] [Google Scholar]
- 17.Li, L. C., Sheng, J. R., Mulherkar, N., Prabhakar, B. S., and Meriggioli, M. N. (2008) Cancer Res. 68 7352-7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulherkar, N., Prasad, K. V., and Prabhakar, B. S. (2007) J. Biol. Chem. 282 11715-11721 [DOI] [PubMed] [Google Scholar]
- 19.Mulherkar, N., Ramaswamy, M., Mordi, D. C., and Prabhakar, B. S. (2006) Oncogene 25 6252-6261 [DOI] [PubMed] [Google Scholar]
- 20.Coppola, T., Perret-Menoud, V., Gattesco, S., Magnin, S., Pombo, I., Blank, U., and Regazzi, R. (2002) Biochem. J. 362 273-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wada, M., Nakanishi, H., Satoh, A., Hirano, H., Obaishi, H., Matsuura, Y., and Takai, Y. (1997) J. Biol. Chem. 272 3875-3878 [DOI] [PubMed] [Google Scholar]
- 22.Niwa, S., Tanaka, Y., and Hirokawa, N. (2008) Nat. Cell Biol. 10 1269-1279 [DOI] [PubMed] [Google Scholar]
- 23.Dhillon, A. S., Hagan, S., Rath, O., and Kolch, W. (2007) Oncogene 26 3279-3290 [DOI] [PubMed] [Google Scholar]
- 24.Boutros, T., Chevet, E., and Metrakos, P. (2008) Pharmacol. Rev. 60 261-310 [DOI] [PubMed] [Google Scholar]
- 25.Hoshino, R., Chatani, Y., Yamori, T., Tsuruo, T., Oka, H., Yoshida, O., Shimada, Y., Ari-i, S., Wada, H., Fujimoto, J., and Kohno, M. (1999) Oncogene 18 813-822 [DOI] [PubMed] [Google Scholar]
- 26.Ostrowski, J., Woszczynski, M., Kowalczyk, P., Wocial, T., Hennig, E., Trzeciak, L., Janik, P., and Bomsztyk, K. (2000) Br. J. Cancer 82 1041-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frodin, M., and Gammeltoft, S. (1999) Mol. Cell. Endocrinol. 151 65-77 [DOI] [PubMed] [Google Scholar]
- 28.Eferl, R., and Wagner, E. F. (2003) Nat. Rev. Cancer 3 859-868 [DOI] [PubMed] [Google Scholar]
- 29.Janknecht, R., Ernst, W. H., Pingoud, V., and Nordheim, A. (1993) EMBO J. 12 5097-5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker, P. E., Rabin, M., Watson, M., Breg, W. R., Ruddle, F. H., and Verma, I. M. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 5826-5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Baruch, A. (2003) Breast Cancer Res. 5 31-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao, C. W., Asselin, E., and Tsang, B. K. (2002) Biol. Reprod. 67 436-441 [DOI] [PubMed] [Google Scholar]
- 33.Micheau, O., and Tschopp, J. (2003) Cell 114 181-190 [DOI] [PubMed] [Google Scholar]
- 34.Sasaoka, T., Langlois, W. J., Leitner, J. W., Draznin, B., and Olefsky, J. M. (1994) J. Biol. Chem. 269 32621-32625 [PubMed] [Google Scholar]
- 35.Goeddel, D. V. (1999) Chest 116 S69-S73 [Google Scholar]
- 36.Subramanian, M., Pilli, T., Bhattacharya, P., Pacini, F., Nikiforov, Y. E., Kanteti, P. V., and Prabhakar, B. S. (2009) J. Clin. Endocrinol. Metab., in press [DOI] [PMC free article] [PubMed]
- 37.Nagata, S. (1999) Annu. Rev. Genet. 33 29-55 [DOI] [PubMed] [Google Scholar]
- 38.Buday, L., and Downward, J. (1993) Cell 73 611-620 [DOI] [PubMed] [Google Scholar]
- 39.Bachmann, R., Eugster, H. P., Frei, K., Fontana, A., and Lassmann, H. (1999) Am. J. Pathol. 154 1417-1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, T., Edwards, C. K., III, Yang, P., Wang, Z., Bluethmann, H., and Mountz, J. D. (1996) J. Immunol. 156 2661-2665 [PubMed] [Google Scholar]
- 41.Micheau, O., Lens, S., Gaide, O., Alevizopoulos, K., and Tschopp, J. (2001) Mol. Cell. Biol. 21 5299-5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen, C., Edelstein, L. C., and Gelinas, C. (2000) Mol. Cell. Biol. 20 2687-2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Y., Broser, M., and Rom, W. N. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 2225-2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Y., Zhu, X., Chen, Y., Wang, X., and Chen, R. (2007) Eur. J. Pharmacol. 576 26-33 [DOI] [PubMed] [Google Scholar]
- 45.McCormick, F. (1994) Curr. Opin. Genet. Dev. 4 71-76 [DOI] [PubMed] [Google Scholar]
- 46.Hildt, E., and Oess, S. (1999) J. Exp. Med. 189 1707-1714 [DOI] [PMC free article] [PubMed] [Google Scholar]