Abstract
The high mortality rate of lung cancer is largely due to the spread of disease to other organs. However, the molecular changes driving lung cancer invasion and metastasis remain unclear. In this study, we identified fibulin-5, a vascular ligand for integrin receptors, as a suppressor of lung cancer invasion and metastasis. Fibulin-5 was silenced by promoter hypermethylation in a majority of lung cancer cell lines and primary tumors. It inhibited lung cancer cell invasion and downregulated matrix metalloproteinase-7 (MMP-7), which promoted lung cancer cell invasion. Knockdown of fibulin-5 was sufficient to stimulate cell invasion and MMP-7 expression. The expression levels of fibulin-5 and MMP-7 were inversely correlated in lung tumors. Suppression of MMP-7 expression by fibulin-5 was mediated by an integrin-binding RGD motif via the extracellular signal-regulated kinase (ERK) pathway. Furthermore, overexpression of fibulin-5 in H460 lung cancer cells inhibited metastasis in mice. Collectively, these results suggest that epigenetic silencing of fibulin-5 promotes lung cancer invasion and metastasis by activating MMP-7 expression through the ERK pathway.
Keywords: Fibulin-5, MMP-7, integrin, invasion, lung cancer
Introduction
The five-year survival rate of lung cancer is below 20%, with most patients dying from distant metastasis (1). However, the mechanism of lung cancer metastasis is poorly understood. Tumor metastasis is characterized by cell detachment from primary tumors and invasion of recipient tissues (2). A critical step in this process is degradation of the basement membrane, which contains extracellular matrix (ECM) proteins and functions as a barrier to surrounding tissues. The degradation of the basement membrane is catalyzed by proteolytic enzymes including matrix metalloproteinases (MMPs) and tissue inhibitor of metalloproteinases (TIMPs) (3). The activities of these enzymes are regulated by cell-surface receptor molecules known as integrins (4). Integrins not only mediate cell adhesion to the ECM, but also regulate intracellular signaling through kinases such as extracellular signal-regulated kinase (ERK) and focal adhesion kinase (FAK) (5). Accumulating evidence suggests that the activation of MMPs and TIMPs through integrin signaling plays an important role in cancer invasion and metastasis (5, 6). Nevertheless, the genetic and epigenetic alterations driving MMP/TIMP activation and tumor invasion remain to be identified (7).
The fibulin family, including fibulin-1-6, is a group of widely expressed ECM proteins localized to the basement membrane, stroma, and ECM fibers (8). Fibulins are characterized by repeated epidermal growth factor (EGF)-like domains and a unique carboxyl-terminal structure (9). They mediate cell-to-cell and cell-to-matrix communication, and provide organization and stabilization to ECM structures during organogenesis and vasculogenesis (9). It has been shown that fibulin family members are aberrantly expressed in tumors, and can either suppress or promote cancer cell growth depending on cell types and cellular contexts (10-13).
Fibulin-5 is a recently identified fibulin family member (14). Distinguished from other fibulins, it contains a conserved RGD motif that binds to integrins and mediates endothelial cell adhesion (15, 16). Fibulin-5 can suppress angiogenesis in an RGD-dependent manner (17). The expression of fibulin-5 is induced in response to pathological conditions such as lung injury and pulmonary hypertension (18, 19), and is regulated by transforming growth factor-β (TGF- β) (11). Fibulin-5-deficient mice exhibited emphysematous changes (16, 20), suggesting an important physiological function in pulmonary tissues. Although fibulin-5 is known to be downregulated in several types of tumors (12, 14), its functional role in lung cancer has not been characterized.
In this study, we identified fibulin-5 as a frequently silenced gene in lung cancer. Our results indicate that fibulin-5 functions as a metastasis suppressor of lung cancer, and downregulation of fibulin-5 drives ERK-mediated MMP-7 induction, and therefore lung cancer cell invasion.
Materials and Methods
Bioinformatics analysis
The expression of fibulin family members was analyzed using the National Center for Biotechnology Information (NCBI) SAGE databases (http://cgap.nci.nih.gov/SAGE). CpG islands were identified using the CpG Island Searcher (http://cpgislands.usc.edu) program.
Tissue samples
The acquisition of the tissues was approved by the Institutional Review Board at the University of Pittsburgh. Tissue microarray slides (US Biomax, Rockville, MD) are described in Table S1. Frozen specimens from the University of Pittsburgh Cancer Institute (UPCI) lung cancer program are described in Table S2.
Immunohistochemistry
Immunohistochemistry was performed with the mouse antibodies against fibulin-5 (R&D system, Minneapolis, MN) and MMP-7 (EMD BioSciences, San Diego, CA) as previously described (21).
Bisulfite sequencing and methylation-specific PCR (MSP)
Isolation of genomic DNA and bisulfite modification were performed as previously described (21). The primers for fibulin-5 bisulfite sequencing were 5′-AGGGAGAATTGGGGAATGAGG-3′ and 5′-CCCACCTTTTTATTCCTAACA-3′. For MSP, methylated fibulin-5 promoter was amplified using the primer pair 5′-TGTAGTGGTTGGGAGGATTTTCGGC-3′/5′-TTCCTAACATATCCAAAACGCGCG-3′, while unmethylated fibulin-5 promoter was amplified using the primer pair 5′-TGTAGTGGTTGGGAGGATTTTGGTG-3′/5′-TTCCTAACATATCCAAAACACACAA-3′. MSP products were analyzed by electrophoresis on 2% agarose gels.
Stable cell clones
A549, H1299 and H460 cells were transfected with fibulin-5 or control empty vector, and were selected by G418 (400 ng/ml for A549 and H1299; 600 ng/ml for H460). Stable clones expressing fibulin-5 were identified by Western blotting.
Antibodies and Western blotting
Western blotting was performed as previously described (22). The antibodies included monoclonal antibodies against V5 (Invitrogen, Carlsbad, CA), fibulin-5 (R&D system), MMP-7, α-tubulin (EMD BioSciences), and FAK (BD Biosciences, San Diego, CA), as well as rabbit antibodies against ERK, phospho-ERK (Thr202/Tyr204), JNK, phospho-JNK (Thr183/Tyr185), p38, phospho-p38 (Thr180/Tyr204) (Cell Signaling Technology, Beverly, MA), and phospho-FAK (Tyr-397, Biosource International, Camarillo, CA). Monoclonal anti-ILK and rabbit anti-PINCH antibodies were previously described (23).
Matrigel invasion assay
Invasion assays were performed in triplicate in 6-well trans-well units with 8-μm filters coated with Matrigel at 1:6 dilution (BD Biosciences). Each well was loaded with ∼2×106 cells. After incubation for 36 hr, cells passing through the filters into bottom wells were fixed in formalin and stained with Crystal Violet (Sigma-Aldrich, St. Louis, MO). Cell numbers in 10 randomly selected fields (×200) from each well were counted.
Analysis of secreted MMP-7
Concentrations of secreted MMP-7 were determined in triplicate by ELISA using Human Total MMP-7 Quantikine ELISA Kit (R&D Systems) according to the manufacturer's protocol.
RNA interference
Small interfering RNA (siRNA) duplexes were from Dharmacon (Chicago, IL). Fibulin-5 was knocked down using ON-TARGETplus siRNA J-017621-05 and -06. MMP-7 was knocked down by MMP-7 757 (GGCAUUCAGAAACUAUA UG) and MMP-7 877 (GCACUGUUCCUCCACUCCA).
Metastasis assay
All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. Parental and stable fibulin-5-expressing NCI-H460 cells (H460/Fibulin-5) were injected intravenously (i.v.) by tail vein into BALB/c nude mice (Harlan, Indianapolis, IN). For each injection, 1×10 6 cells suspended in 200 μl PBS were used. Following sacrifice of mice at 3, 5 and 7 weeks after injection, lung metastasis nodules were counted. The lungs were then inflated with 1-2 ml 10% buffered formalin and fixed for 24 hr before paraffin embedding. Serial midsagittal 5-μm sections were used for histologic analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism IV software. P values <0.05 were considered to be statistically significant. The means +/- one standard deviation were displayed in the figures.
Results
Downregulation of fibulin-5 in lung cancer
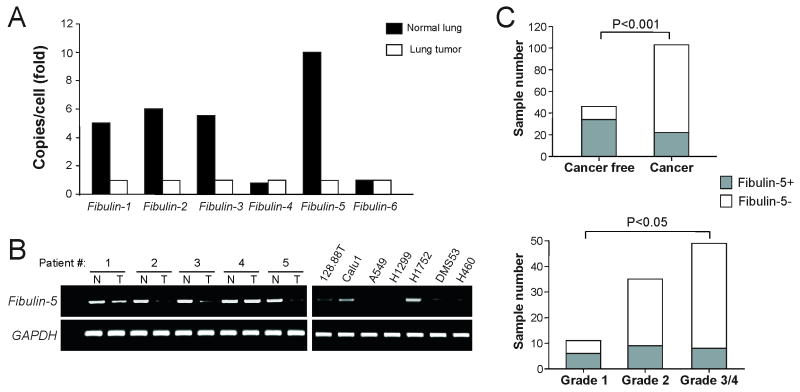
In light of our previous finding of fibulin-3 (EFEMP-1) inactivation in lung cancer (21), we analyzed the expression of all 6 fibulin family genes using the NCBI SAGE databases containing 159,059 transcripts from lung cancer, and 159,917 transcripts from normal lung tissues (24). In addition to fibulin-3, the expression of fibulin-1, fibulin-2 and fibulin-5 was also lower in lung cancer (Fig. 1A). Fibulin-5, whose expression was decreased by almost 10 fold, was the most significantly downregulated fibulin family member in lung cancer (Fig. 1A). Downregulation of fibulin-5 was confirmed by RT-PCR in lung tumors and lung cancer cell lines (Fig. 1B).
Fig. 1. Reduced expression of fibulin-5 in lung cancer.
(A) Expression of fibulin family members in SAGE databases containing 159,128 transcripts from 3 lung adenocarcinomas and 159,917 transcripts from 3 normal lung specimens. The expression (copies/cell) was normalized as in (41). (B) RT-PCR was used to analyze fibulin-5 expression in 5 matched pairs of normal (N)/tumor (T) lung tissues and 7 lung cancer cell lines. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a house-keeping gene, was used as an internal control. (C) Summary of fibulin-5 expression determined by immunohistochemistry in 95 lung tumors and 46 normal lung samples. Correlations between loss of fibulin-5 expression and tumor status and grade were analyzed by Fisher's exact test.
Fibulin-5 expression was further analyzed by immunohistochemistry in a tissue microarray containing 95 non-small cell lung cancer (NSCLC) and 46 normal lung specimens, including 32 matched tumor/normal pairs (Table S1). A highly significant (P<0.001, Fisher's exact test) difference between the normal and tumor tissues was observed: 73.9% (34/46) of normal lung specimens expressed fibulin-5, while only 22.1% (21/95) of NSCLC samples were positive for fibulin-5 (Fig. 1C, S1A and S1B). Among the 32 pairs, 17 (53.1%) tumors completely lost fibulin-5 expression compared to the matched normal tissues (Table S1). Fibulin-5 expression was detected in the cytoplasm of normal bronchial epithelial cells and fibulin-5-positive tumor cells (Fig. S1A and S1B). Importantly, loss of fibulin-5 expression correlated with tumor grade (P<0.05, chi-square exact test), with fibulin-5 staining detected in 50.0% (6/12) of grade-1 tumors, but in only 26.5% (9/34) of grade-2, and 16.3% (8/49) of grade-3/4 tumors (Fig. 1C and Table S1).
Silencing of fibulin-5 by promoter hypermethylation
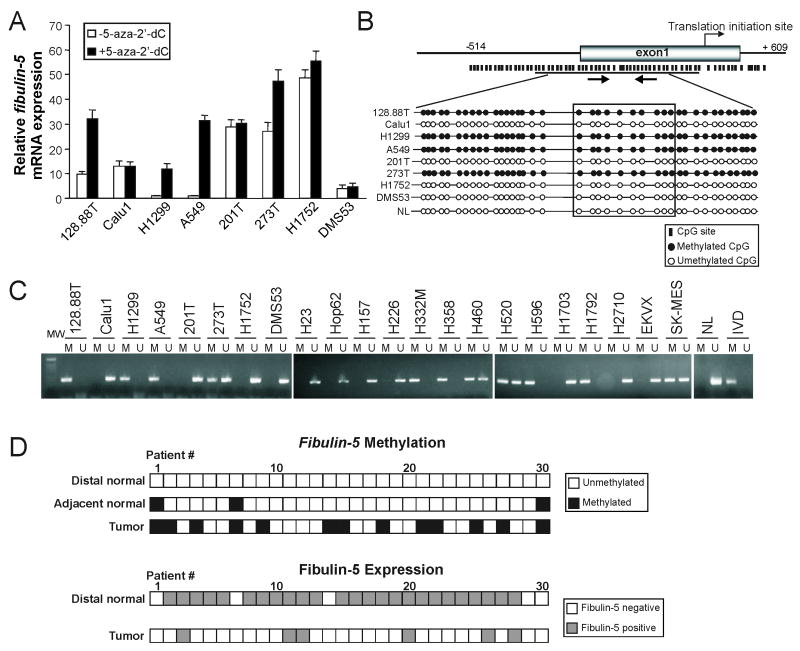
To determine whether downregulation of fibulin-5 is due to epigenetic silencing, eight lung cancer cell lines were treated with 5-aza-2′-dC, a pharmacological inhibitor of DNA methyltransferase. Fibulin-5 expression was significantly elevated in 4 cell lines following 5-aza-2′-dC treatment by real-time PCR (Fig. 2A). Bioinformatics analysis identified a CpG island in the 5′ promoter region of fibulin-5 (Fig. 2B). Bisulfite sequencing revealed that this CpG island was completely methylated in the 4 cell lines that were responsive to 5-aza-2′-dC treatment, but not in the other 4 that were insensitive to 5-aza-2′-dC (Fig. 2A, 2B and S2A), suggesting that promoter hypermethylation underlies downregulation of fibulin-5. Using a methylation specific PCR (MSP) assay, we found fibulin-5 promoter methylation in 11 out of 22 (50.0%) lung cancer cell lines (Fig. 2C).
Fig. 2. Silencing of fibulin-5 in lung cancer by promoter hypermethylation.
(A) Fibulin-5 expression in 8 lung cancer cell lines with or without 5-aza-2′-dC treatment was determined by real-time RT-PCR. The results are the average of three independent experiments normalized to GAPDH levels. (B) CpG site distribution in the fibulin-5 promoter region and summary of bisulfite sequencing results for 8 lung cancer cell lines and normal lymphocytes (NL). The positions of MSP primers are indicated by arrows. (C) Fibulin-5 methylation status in 22 lung cancer cell lines was analyzed by MSP. M: amplification using primers specific for methylated DNA. U: amplification using primers specific for unmethylated DNA. NL and IVD (in vitro methylated DNA) were negative and positive controls, respectively. MW: molecular weight marker. (D) Thirty matched sets of samples (Table S2), including primary lung tumors (Tumor), pathologically normal lung tissues adjacent to the tumors (Adjacent normal), and normal lung tissues distal to the tumors (Distal normal), were analyzed for fibulin-5 methylation and expression by MSP and immunohistochemistry, respectively. Upper panel: Summary of MSP results. Lower panel: Summary of fibulin-5 expression in the distal normal and tumor samples.
The relationship between fibulin-5 downregulation and promoter methylation was further analyzed using 30 matched sets of frozen tissue samples. Each set included a tumor and histologically normal lung tissues adjacent and distal to the tumor (Table S2). While promoter methylation was found in 13 (43.3%) tumors, it was detected in only 3 (10%) adjacent normal, and in no (0%) distal normal samples (Fig. 2D and S2B). Immunohistochemistry confirmed the loss of fibulin-5 expression in all of the 13 tumors with promoter methylation (Fig. 2D and S2C). Among the 19 cases where tumors lost fibulin-5 expression compared to distal normal, promoter methylation was observed in 10 (52.6%) tumors, but in none of the corresponding normal specimens (Fig. 2D). In contrast, promoter methylation was not detected in any of the 6 cases where tumors expressed fibulin-5 (Fig. 2D). The correlation between loss of fibulin-5 expression and promoter methylation was statistically significant (P<0.05, Fisher's exact test). Together, these results indicate that promoter hypermethylation is the major, but not the only mechanism leading to fibulin-5 silencing in lung cancer.
Suppression of lung cancer cell invasion and MMP-7 expression by fibulin-5
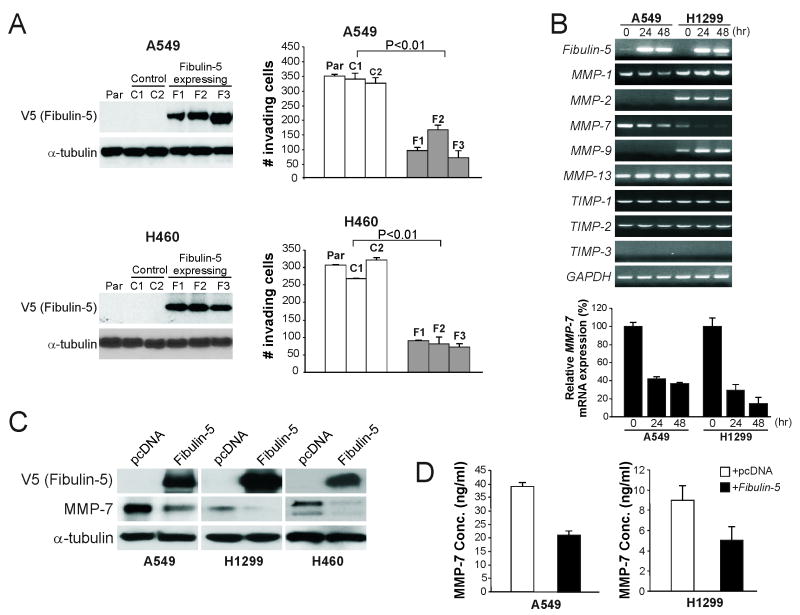
Based on the biochemical properties and the RGD-containing feature of fibulin-5 (14), we hypothesized that it is involved in regulating cancer cell invasion. To test this hypothesis, A549, H460 and H1299 lung cancer cells, which contain promoter methylation and lack fibulin-5 expression (Fig. 2A-C), were used to establish stable fibulin-5-expressing cell lines by transfection (Fig. 3A and S3A). Analysis of cell invasion by Matrigel assays revealed that fibulin-5 expression significantly suppressed invasion of all three cell lines (P<0.01, Fisher's exact test) (Fig. 3A and S3). However, fibulin-5 expression did not significantly affect cell growth determined by soft agar and 5-bromo-2-deoxyuridine (BrdU) incorporation assays (Fig. S4A and S4B), or spontaneous and drug-induced apoptosis (Fig. S4C). Transfection of fibulin-5 into H1299 cells altered the expression of fibulin-2 and fibulin-6, but not other fibulins (Fig. S4D).
Fig. 3. Fibulin-5 suppressed lung cancer cell invasion and MMP-7 expression.
(A) Suppression of lung cancer cell invasion by fibulin-5. Left panel: Parental (Par), fibulin-5-negative (C1 and C2), and stable fibulin-5-expressing (F1-3) A549 and H460 lung cancer cell lines were analyzed for fibulin-5 (V5) expression by Western blotting. α-tubulin was the loading control. Right panel: Matrigel invasion assay was used to analyze invasion of A549 and H460 cell lines with or without fibulin-5 expression. The results are the average of 3 independent experiments. (B) A549 and H1299 cells were transfected with fibulin-5 or control pcDNA vector. Upper panel: Indicated MMPs and TIMPs were analyzed by RT-PCR at 24 and 48 hr after transfection. Lower panel: MMP-7 expression was confirmed by real-time RT-PCR with GAPDH as the internal control. The results were normalized to the cells without fibulin-5 transfection (0 hr), which were defined as 100%. (C) Indicated lung cancer cell lines were transfected with V5-tagged fibulin-5 or control pcDNA vector. Fibulin-5 and MMP-7 were analyzed by Western blotting 36 hr following transfection. (D) Levels of secreted MMP-7 in the culture media of A549 and H1299 cells were measured by ELISA at 36 hr after fibulin-5 transfection.
To determine how fibulin-5 inhibits lung cancer cell invasion, genes encoding several proteolytic enzymes involved in degrading the basement membrane, including three TIMPs and five MMPs, were analyzed by RT-PCR in A549 and H1299 cells following fibulin-5 transfection. Only the expression of MMP-7 was consistently downregulated by 60-80% following fibulin-5 transfection (Fig. 3B). Concordantly, fibulin-5 expression suppressed MMP-7 protein expression (Fig. 3C), the level of secreted MMP-7 (Fig. 3D), and the activity of an MMP-7 luciferase reporter (Fig. S5A).
Fibulin-5 inhibits lung cancer cell invasion by downregulating MMP-7
The role of MMP-7 in lung cancer cell invasion was then investigated. Transfection of MMP-7 into fibulin-5-expressing A549 cells restored their invasiveness (Fig. 4A). Conversely, knockdown of MMP-7 by two independent siRNA significantly decreased the invasiveness of both A549 and H1299 cells (Fig. 4B). To determine if fibulin-5 silencing alone is sufficient to drive MMP-7 expression and cell invasion, siRNA was used to knock down fibulin-5 in H1752 cells (Fig. 4C), which expressed a relatively high level of fibulin-5 with no detectable fibulin-5 promoter methylation (Fig. 2A and 2B). Transfection with two independent fibulin-5 siRNA, but not the control siRNA, led to increased H1752 cell invasion as well as elevated MMP-7 expression (Fig. 4C), suggesting that fibulin-5 downregulation alone is sufficient for stimulating lung cancer cell invasion through upregulation of MMP-7.
Fig. 4. Fibulin-5 inhibited lung cancer cell invasion by suppressing MMP-7.
(A) Fibulin-5-expressing A549 cell lines F1 and F2 were transfected with HA-tagged MMP-7 or the control vector. Left panel: Transfected MMP-7 expression was analyzed by HA Western blotting 36 hr after MMP-7 transfection. Right panel: Invasion of A549 F1 and F2 cells was analyzed by Matrigel assay 36 hr after transfection. (B) A549 and H1299 cells were transfected with control siRNA, or two independent MMP-7 siRNA. Left panel: MMP-7 expression was analyzed by Western blotting 36 hr after siRNA transfection. Right panel: Cell invasion was analyzed by Matrigel assay 36 hr after siRNA transfection. (C) H1752 cells were transfected with control siRNA, or two independent fibulin-5 siRNA. Left panel: Fibulin5 and MMP-7 were analyzed by Western blotting 48 hr after transfection. Right panel: Matrigel assay was used to analyze cell invasion 24 hr after siRNA transfection. (D) Summary of fibulin-5 and MMP-7 expression in 95 NSCLC samples (Table S1) analyzed by immunohistochemistry. The inverse correlation between fibulin-5 and MMP-7 expression was statistically significant (two-tailed chi-square test).
MMP-7 expression was then analyzed by immunohistochemistry using the aforementioned tissue microarray (Table S1), in comparison with fibulin-5 expression (Fig. 4D and S1C). No MMP-7 immunoreactivity was detected in the 46 normal lung specimens (data not shown), consistent with the previous finding that MMP-7 is not expressed in normal lung (25). In contrast, 39 out of 95 (41.1%) NSCLC samples were positive for MMP-7 (Fig. 4D and Table S1). Remarkably, a statistically significant (P<0.05, two-tailed chi-square test) inverse correlation between fibulin-5 and MMP-7 expression was found, with 54.7% (52/95) of tumors expressing either MMP-7 or fibulin-5, but only 5.5% (5/95) expressing both proteins (Fig. 4D). Together, these results suggest that fibulin-5 suppresses lung cancer cell invasion by inhibiting the expression of MMP-7.
RGD-dependent inhibition of MMP-7 by fibulin-5 through the ERK pathway
To investigate the mechanism of MMP-7 regulation by fibulin-5, we determined the functional role of its RGD motif. Several deletion mutants of fibulin-5 were constructed and expressed in A549 cells (Fig. 5A). A deletion fragment containing the amino-terminal 1/3 of fibulin-5 (N136) including the RGD motif retained most of the activities in suppressing MMP-7 protein expression and its reporter activities (Fig. 5B and S5B). In contrast, all deletion mutants without the RGD motif, including a micro deletion of the RGD residues only, lost over 80% of the activities in suppressing MMP-7 expression and the level of secreted MMP-7 (Fig. 5B), suggesting that the inhibition of MMP-7 by fibulin-5 is mediated by the RGD motif through integrin signaling.
Fig. 5. RGD-dependent suppression of MMP-7 by fibulin-5 via the ERK pathway.
(A) Left panel: Schematic diagrams of V5-tagged wild-type (WT) and deletion mutants of fibulin-5. Right panel: Expression of the WT and mutant fibulin-5 in A549 cells was detected by Western blotting 36 hr after transfection. (B) A549 cells were transfected with WT or the indicated mutant fibulin-5 constructs. Upper panel: MMP-7 expression was analyzed by Western blotting 36 hr after transfection. Lower panel: The levels of secreted MMP-7 were determined by ELISA 36 hr after transfection. (C) Upper panel: Indicated kinases were probed by Western blotting 24 hr after A549 and H1299 cells were transfected with the control or WT fibulin-5. Lower panel: ERK activation was analyzed by Western blotting 24 hr after transfecting A549 cells with WT fibulin-5 or the RGD deletion mutant (ΔRGD). (D) A549 cells were treated with the ERK inhibitor PD98059 (50 μM) for 12 or 24 hr. ERK activation and MMP-7 expression were determined by Western blotting.
To further delineate the mechanism by which fibulin-5 inhibits MMP-7 expression, several kinases downstream of integrin signaling, including ERK, p38, and JNK, were analyzed. Phosphorylation of ERK, but not that of p38 and JNK, was markedly reduced following fibulin-5 transfection (Fig. 5C). The inhibition of ERK phosphorylation by fibulin-5 was RGD-dependent (Fig. 5C). ERK inhibitor PD98059 blunted MMP-7 expression in A549 and H1299 cells (Fig. 5D). Furthermore, fibulin-5 also suppressed serum- or epidermal growth factor (EGF)-stimulated ERK and FAK phosphorylation, as well as the expression of the integrin downstream effectors ILK and PINCH (Fig. S5C and S5D). These observations suggest that fibulin-5 functions as an inhibitor of ERK signaling via its RGD motif to inhibit MMP-7 expression and lung cancer cell invasion.
Potent inhibition of H460 tumor metastasis by fibulin-5
The previously described NCI-H460 metastasis model was then used to determine whether fibulin-5 can suppress tumor metastasis (26). Parental and fibulin-5-expressing H460 cells, which had similar growth rate (Fig. S6A), were injected intravenously (i.v.) into nude mice. After 5-7 weeks, the mice receiving the parental H460 cells had a number of lung metastasis nodules (Fig. 6A, S6B) and lymph node metastasis (Table S3). In contrast, mice injected with fibulin-5-expressing H460 cells had much fewer lung metastasis nodules (Fig. 6A and 6B), and no lymph node metastasis (Table S3). The mice receiving the parental H460 cells also had significantly lower body weights compared with those injected with fibulin-5-expressing H460 cells (Table S3). Furthermore, the parental H460 lung tumors expressed much higher levels of MMP-7 and phosphorylated ERK compared with those expressing fibulin-5 (Fig. 6C). These results suggest that fibulin-5 functions as a suppressor of lung cancer metastasis by inhibiting MMP-7 expression and ERK signaling.
Fig. 6. Inhibition of lung cancer metastasis by fibulin-5 in mice.
(A) Parental and fibulin-5-expressing H460 cells were injected (106 cells/injection) i.v. by tail vein into BALB/c nude mice. Representative pictures of fixed lungs at 7 weeks after injection were shown, with H&E staining pictures on the right. Arrows indicate metastasis nodules. (B) Metastasis nodules in the parental and fibulin-5-expressing H460 tumors were quantified. The difference between the groups with and without fibulin-5 expression was analyzed by Fisher's exact test. (C) Expression of MMP-7 and phospho-ERK in representative metastatic tumors was analyzed by immunohistochemistry (200×), with upper right showing enlarged pictures.
Discussion
Similar to tumor initiation, invasion and metastasis are also driven by genetic and epigenetic alterations (27). Our results demonstrate that fibulin-5 functions as a suppressor of lung cancer invasion, and epigenetic inactivation of fibulin-5 contributes to lung cancer progression. However, deregulation in fibulin-5 does not appear to be a major factor in tumor initiation, as fibulin-5 silencing was mostly found in high-grade lung tumors. Previous studies also described downregulation of fibulin-5 mostly in advanced malignancies (11). Knockout of fibulin-5 in mice is not sufficient to affect tumor incidence (20). Our data revealed downregulation of multiple fibulin family members in lung cancer. This consistent pattern seems to be distinguished from the context-specific expression of fibulin family members in other tumor types (10, 11).
Fibulin-5 is downregulated in lung cancer largely by promoter hypermethylation. Interestingly, fibulin-5 hypermethylation was found in several histologically normal tissues adjacent to the tumors (Fig. 2D). This might be due to an epigenetic field effect and/or infiltrating tumor cells described in other studies (28). Additional mechanisms, such as expression of the oncoproteins c-Myc and VEGF, may also lead to fibulin-5 silencing (29, 30). Fibulin-5 is localized on 14q32.1, a tumor suppressor-containing region (31). Mutations or homozygous deletions of fibulin-5 have yet to be found.
The effects of fibulin-5 on lung cancer are mediated by the inhibition of MMP-7, which is upregulated in lung cancer (32, 33), and plays an important role in cancer invasion (4, 6). MMP-7 overexpression is associated with poor prognosis of NSCLC (34). The frequent change of fibulin-5, along with the inverse correlation between fibulin-5 and MMP-7 expression, suggests that epigenetic silencing of fibulin-5 might be a major mechanism underlying MMP-7 overexpression in NSCLC. Fibulin-5 can also modulate the expression of MMP-2, MMP-3, TIMP-1, and TIMP-3 in suppressing the angiogenesis of fibrosarcoma cells (17), indicating a broad relevance in regulating the activities of the MMP/TIMP families. The antiangiogenic activity may also be a contributing factor for its inhibitory effect on lung cancer invasion.
Our results suggest a model in which fibulin-5 inhibits ERK activity through RGD-mediated integrin signaling, leading to transcriptional repression of MMP-7 and suppression of cell invasion. Integrin signaling is not only critical for cell migration and invasion, but is also involved in the regulation of MMP and TIMP activities (5). The RGD motif of fibulin-5 binds to integrins αvβ3, αvβ5, and α9β1 (16). It remains to be determined which integrin subunit(s) mediates the inhibitory effect of fibulin-5 on MMP-7 in the lung. It is also unclear how MMP-7 expression is downregulated through the ERK pathway. Several binding sites of the ETS transcription factors, which have been implicated in regulating MMP expression (35), were found in the promoter region of MMP-7 (data not shown). The roles of ETS and other transcription factors in the inhibition of MMP-7 by fibulin-5 need to be further investigated.
Tumor metastasis is stimulated upon inactivation of a group of metastasis suppressors, such as NM23, KAI1, and MKK4 (36). These genes are downregulated in metastatic tumors and possess the activity of reducing the metastatic propensity of cancer cells in animal tumor models. Identifying additional metastasis suppressors is critical for understanding the mechanisms of tumor metastasis. Our data strongly suggest that fibulin-5 functions as a metastasis suppressor. Several additional lines of evidence support this notion. During mouse development, fibulin-5 is widely expressed in the so called “epithelial-mesenchymal transition” (EMT) region (14). EMT is a critical process in cancer metastasis, during which epithelial cells acquire phenotypes of motile fibroblasts. Emerging evidence indicates that ERK/FAK-mediated integrin signaling and tumor-associated MMPs stimulate EMT (37, 38). Fibulin-5 expression is regulated by TGF-β (11), which plays an important role in cancer metastasis and EMT (39). Therefore, epigenetic silencing of fibulin-5 and subsequent activation of MMP-7 may be events involved in EMT. However, this speculation seems to contradict a recent finding that fibulin-5 can promote EMT in breast tumors (40), reinforcing the context specificity involved in the regulation of fibulin family proteins.
Supplementary Material
Acknowledgments
This work is supported in part by NIH grants CA106348 and CA121105, the Career Investigator Award from the American Lung Association (ALA), the American Cancer Society Grant RSG-07-156-01-CNE, the Developmental Research Program, the Career Development Award (L.Z.), the Postdoctoral Fellowship (W.Y.) from the Lung Cancer Specialized Program of Research Excellence at the University of Pittsburgh Cancer Institute (CA090440), the Flight Attendant Medical Research Institute (FAMRI), the Alliance for Cancer Gene Therapy (ACGT), the NIH grant CA129829 (J.Y.), the NIH grant GM065188 (C.W.), and the National Basic Research Program of China 2009CB521702 (W.Y.). L.Z. is a V Foundation scholar.
Abbreviations
- 5-aza-2′-dC
5-aza-2′-deoxycytidine
- ECM
extracellular matrix
- EGF
epidermal growth factor
- ERK
extracellular signal-regulated kinase
- FAK
focal adhesion kinase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IVD
in vitro methylated DNA
- MSP
methylation specific PCR
- NL
normal lymphocytes
- NSCLC
non-small cell lung cancer
- MMP
matrix metalloproteinase
- RT-PCR
reverse transcriptase-PCR
- SAGE
serial analysis of gene expression
- siRNA
small interfering RNA
- TIMP
tissue inhibitor of metalloproteinase
References
- 1.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 2.Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12:89–96. doi: 10.1006/scbi.2001.0416. [DOI] [PubMed] [Google Scholar]
- 3.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–14. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–8. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 5.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 6.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–52. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 8.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: physiological and disease perspectives. EMBO Rep. 2003;4:1127–31. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4:479–89. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–40. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Schiemann WP, Blobe GC, Kalume DE, Pandey A, Lodish HF. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J Biol Chem. 2002;277:27367–77. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- 12.Wlazlinski A, Engers R, Hoffmann MJ, et al. Downregulation of several fibulin genes in prostate cancer. Prostate. 2007;67:1770–80. doi: 10.1002/pros.20667. [DOI] [PubMed] [Google Scholar]
- 13.Yi CH, Smith DJ, West WW, Hollingsworth MA. Loss of fibulin-2 expression is associated with breast cancer progression. Am J Pathol. 2007;170:1535–45. doi: 10.2353/ajpath.2007.060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albig AR, Schiemann WP. Fibulin-5 function during tumorigenesis. Future Oncol. 2005;1:23–35. doi: 10.1517/14796694.1.1.23. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura T, Ruiz-Lozano P, Lindner V, et al. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J Biol Chem. 1999;274:22476–83. doi: 10.1074/jbc.274.32.22476. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura T, Lozano PR, Ikeda Y, et al. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415:171–5. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- 17.Albig AR, Neil JR, Schiemann WP. Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res. 2006;66:2621–9. doi: 10.1158/0008-5472.CAN-04-4096. [DOI] [PubMed] [Google Scholar]
- 18.Kuang PP, Goldstein RH, Liu Y, Rishikof DC, Jean JC, Joyce-Brady M. Coordinate expression of fibulin-5/DANCE and elastin during lung injury repair. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1147–52. doi: 10.1152/ajplung.00098.2003. [DOI] [PubMed] [Google Scholar]
- 19.Merklinger SL, Wagner RA, Spiekerkoetter E, et al. Increased fibulin-5 and elastin in S100A4/Mts1 mice with pulmonary hypertension. Circ Res. 2005;97:596–604. doi: 10.1161/01.RES.0000182425.49768.8a. [DOI] [PubMed] [Google Scholar]
- 20.Yanagisawa H, Davis EC, Starcher BC, et al. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415:168–71. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- 21.Yue W, Dacic S, Sun Q, et al. Frequent inactivation of RAMP2, EFEMP1 and Dutt1 in lung cancer by promoter hypermethylation. Clin Cancer Res. 2007;13:4336–44. doi: 10.1158/1078-0432.CCR-07-0015. [DOI] [PubMed] [Google Scholar]
- 22.Ming L, Wang P, Bank A, Yu J, Zhang L. PUMA dissociates Bax and BCL-XL to induce apoptosis in colon cancer cells. J Biol Chem. 2006;281:16034–42. doi: 10.1074/jbc.M513587200. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, Fukuda T, Li Y, Zha X, Qin J, Wu C. Molecular dissection of PINCH-1 reveals a mechanism of coupling and uncoupling of cell shape modulation and survival. J Biol Chem. 2005;280:27631–7. doi: 10.1074/jbc.M504189200. [DOI] [PubMed] [Google Scholar]
- 24.Nacht M, Dracheva T, Gao Y, et al. Molecular characteristics of non-small cell lung cancer. Proc Natl Acad Sci U S A. 2001;98:15203–8. doi: 10.1073/pnas.261414598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo F, Kaminski N, Eugui E, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci U S A. 2002;99:6292–7. doi: 10.1073/pnas.092134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soon LL, Yie TA, Shvarts A, Levine AJ, Su F, Tchou-Wong KM. Overexpression of WISP-1 down-regulated motility and invasion of lung cancer cells through inhibition of Rac activation. J Biol Chem. 2003;278:11465–70. doi: 10.1074/jbc.M210945200. [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 28.Dammann R, Strunnikova M, Schagdarsurengin U, et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–36. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277:36921–30. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 30.Albig AR, Schiemann WP. Fibulin-5 antagonizes vascular endothelial growth factor (VEGF) signaling and angiogenic sprouting by endothelial cells. DNA Cell Biol. 2004;23:367–79. doi: 10.1089/104454904323145254. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y, Ko JM, Lung HL, Lo PH, Stanbridge EJ, Lung ML. Monochromosome transfer provides functional evidence for growth-suppressive genes on chromosome 14 in nasopharyngeal carcinoma. Genes Chromosomes Cancer. 2003;37:359–68. doi: 10.1002/gcc.10228. [DOI] [PubMed] [Google Scholar]
- 32.Safranek J, Holubec L, Jr, Topolcan O, et al. Expression of mRNA MMP-7 and mRNA TIMP-1 in non-small cell lung cancer. Anticancer Res. 2007;27:2953–6. [PubMed] [Google Scholar]
- 33.Lin TS, Chiou SH, Wang LS, et al. Expression spectra of matrix metalloproteinases in metastatic non-small cell lung cancer. Oncol Rep. 2004;12:717–23. [PubMed] [Google Scholar]
- 34.Liu D, Nakano J, Ishikawa S, et al. Overexpression of matrix metalloproteinase-7 (MMP-7) correlates with tumor proliferation, and a poor prognosis in non-small cell lung cancer. Lung Cancer. 2007;58:384–91. doi: 10.1016/j.lungcan.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Horiuchi S, Yamamoto H, Min Y, Adachi Y, Itoh F, Imai K. Association of ets-related transcriptional factor E1AF expression with tumour progression and overexpression of MMP-1 and matrilysin in human colorectal cancer. J Pathol. 2003;200:568–76. doi: 10.1002/path.1387. [DOI] [PubMed] [Google Scholar]
- 36.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 37.Giehl K, Menke A. Microenvironmental regulation of E-cadherin-mediated adherens junctions. Front Biosci. 2008;13:3975–85. doi: 10.2741/2985. [DOI] [PubMed] [Google Scholar]
- 38.Radisky DC, LaBarge MA. Epithelial-mesenchymal transition and the stem cell phenotype. Cell Stem Cell. 2008;2:511–2. doi: 10.1016/j.stem.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Minn AJ, Kang Y, Serganova I, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YH, Albig AR, Regner M, Schiemann BJ, Schiemann WP. Fibulin-5 initiates epithelial-mesenchymal transition (EMT) and enhances EMT induced by TGF-beta in mammary epithelial cells via a MMP-dependent mechanism. Carcinogenesis. 2008;29:2243–51. doi: 10.1093/carcin/bgn199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue W, Sun Q, Dacic S, et al. Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis. 2008;29:84–92. doi: 10.1093/carcin/bgm267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.