Abstract
Mitosis in higher eukaryotes is marked by the sequential assembly of two massive structures: the mitotic spindle and the nucleus. Nuclear assembly itself requires the precise formation of both nuclear membranes and nuclear pore complexes. Previously, importin alpha/beta and RanGTP were shown to act as dueling regulators to ensure that these assembly processes occur only in the vicinity of the mitotic chromosomes. We now find that the distantly related karyopherin, transportin, negatively regulates nuclear envelope fusion and nuclear pore assembly in Xenopus egg extracts. We show that transportin—and importin beta—initiate their regulation as early as the first known step of nuclear pore assembly: recruitment of the critical pore-targeting nucleoporin ELYS/MEL-28 to chromatin. Indeed, each karyopherin can interact directly with ELYS. We further define the nucleoporin subunit targets for transportin and importin beta and find them to be largely the same: ELYS, the Nup107/160 complex, Nup53, and the FG nucleoporins. Equally importantly, we find that transportin negatively regulates mitotic spindle assembly. These negative regulatory events are counteracted by RanGTP. We conclude that the interplay of the two negative regulators, transportin and importin beta, along with the positive regulator RanGTP, allows precise choreography of multiple cell cycle assembly events.
INTRODUCTION
During mitosis, the intracellular architecture of the higher eukaryotic cell undergoes constant and dramatic change. The chromosomes condense, the nuclear envelope breaks down, a mitotic spindle forms and separates the chromosomes, and finally a nuclear envelope is reassembled around each set of daughter chromosomes. The movement of the cell cycle from one stage of mitosis to the next involves the coordinated action of multiple kinases and phosphatases. Recently, however, new regulators have been identified that contribute not to the timing but to the spatial control of assembly of both the mitotic spindle and the nucleus: karyopherins and RanGTP.
Karyopherins are most well known for their function in interphase as the transport receptors for nuclear import and export (reviewed in Tran and Wente, 2006; Stewart, 2007). Nuclear proteins larger than ∼20–40 kDa are actively imported, whereas snRNAs, tRNAs, and other cargoes are exported through the nuclear pores by karyopherins. Karyopherins consist of a large family of importins (import receptors) and exportins (export receptors; reviewed in Weis, 2003; Pemberton and Paschal, 2005). The first and most studied import receptor of the karyopherin family was importin beta (Adam and Adam, 1994; Chi et al., 1995; Gorlich et al., 1995; Imamoto et al., 1995; Radu et al., 1995; Floer et al., 1997). Importin beta mediates the nuclear import of many proteins through the use of a specific adaptor, such as importin alpha (reviewed in Cook et al., 2007). Importin alpha recognizes cargoes containing classical or bipartite nuclear localization signals (NLSs), which are composed largely of basic amino acids (reviewed in Stewart, 2007). Importin beta can also bind to certain cargoes directly without the use of an adaptor. The second major import receptor discovered was transportin (or karyopherin beta-2), which is distantly related to importin beta (24% identical; Nakielny et al., 1996; Pollard et al., 1996; Bonifaci et al., 1997; Fridell et al., 1997; Siomi et al., 1997; Chook and Blobel, 1999). Transportin imports proteins without an adaptor. Transportin most commonly recognizes cargoes that contain a different consensus NLS, one with a terminal proline-tyrosine (PY) dipeptide preceded either by a hydrophobic or basic region, depending on the protein (Lee et al., 2006; Cansizoglu et al., 2007; Suel et al., 2008).
The directionality of transport is governed by a gradient of RanGTP, the GTP-bound form of the small GTPase Ran. The concentration of RanGTP is high in the nucleus and low in the cytoplasm (reviewed in Madrid and Weis, 2006), a result of the exclusive localization of the Ran-guanine exchange factor (GEF) on chromatin. Both importin beta and transportin release their cargoes upon encountering RanGTP in the nucleus.
From yeast to vertebrates, the basic architecture of the 60–120-MDa nuclear pore consists of a large central scaffold, eight cytoplasmic filaments, and a nuclear pore basket. Each nuclear pore contains ∼30 different proteins or nucleoporins in multiples copies and possesses eightfold symmetry (Rout et al., 2000; Cronshaw et al., 2002; Stoffler et al., 2003; Beck et al., 2007; Beck and Medalia, 2008; Brohawn et al., 2008; Debler et al., 2008; Jovanovic-Talisman et al., 2009; Lim et al., 2008). During transport, the karyopherins interact with a specific subset of nuclear pore proteins which have in common phenylalanine-glycine (FG) repeats (Gorlich and Kutay, 1999; Chook and Blobel, 2001; Macara, 2001; Denning et al., 2003; Fahrenkrog et al., 2004; Strawn et al., 2004; Peters, 2005; Madrid and Weis, 2006; Tran and Wente, 2006; Cook et al., 2007; Stewart, 2007; Lim et al., 2008). In organisms from Drosophila to vertebrates, the nuclear envelope disassembles and reforms during each cell cycle. Disassembly involves the complete disassembly of the nuclear pores into subunits and the simultaneous retraction of the nuclear membranes into the ER. Assembly of the nucleus at the end of mitosis requires all the components including nuclear membranes and nuclear pores to reassemble in a stepwise manner (reviewed in Burke and Ellenberg, 2002; Antonin et al., 2008; Kutay and Hetzer, 2008). Progress continues to be made on the order of assembly of the nucleoporins and/or nucleoporin subcomplexes into the pore, although this order remains approximate (Bodoor et al., 1999; Belgareh et al., 2001; Daigle et al., 2001; Walther et al., 2003a; Dultz et al., 2008; Rasala et al., 2008).
One valuable model system that has been used to study the regulated processes of mitotic spindle assembly and nuclear assembly, including nuclear pore assembly, is derived from extracts of Xenopus eggs. Mitotic Xenopus egg extracts can successfully recapitulate spindle assembly (see below). Similarly, interphase egg extracts have provided a powerful way to observe and manipulate nuclear assembly (Lohka and Masui, 1984; Newport, 1987; Smythe and Newport, 1991; Powers et al., 1995; Ullman and Forbes, 1995; Macaulay and Forbes, 1996; Goldberg et al., 1997; Hetzer et al., 2001; Harel et al., 2003a,b; Walther et al., 2003a,b; Chan and Forbes, 2006; Clarke and Zhang, 2008; Delmar et al., 2008). For nuclear assembly, fractionated Xenopus egg cytosol is combined with Xenopus egg membranes, sperm chromatin, and an energy-regenerating system. Notably, the fractionated Xenopus cytosol contains the soluble nucleoporins in ∼14 subcomplexes (Rasala et al., 2008) and these, poised for assembly, quickly assemble into nuclear pores. The vesiculated endoplasmic reticulum (ER) derived from lysing of the eggs serves as a source of membrane for nuclear membrane formation in vitro. In vivo, the reticular ER itself serves as the source of nuclear membrane; see Yang et al., (1997) and Anderson and Hetzer (2008) and references therein. Within ∼1 h of incubation at room temperature nuclei competent for nuclear import, and DNA replication is formed in vitro.
Intermediates in nuclear assembly have been identified in the Xenopus system using chemical and protein inhibitors (Newmeyer and Forbes, 1988; Finlay and Forbes, 1990; Macaulay and Forbes, 1996; Goldberg et al., 1997; Shumaker et al., 1998; Harel et al., 2003a; Hetzer et al., 2005) or by visualization using electron microscopy (Goldberg et al., 1997; Wiese et al., 1997). When chromatin, vesicles, and cytosol are mixed without any preincubation, electron microscopy reveals membrane vesicle binding to chromatin, followed by vesicle-vesicle fusion into patches of double membranes, nuclear pore assembly within the double membrane patches, and finally closure of any gaps in the nuclear envelope by additional fusion (Macaulay and Forbes, 1996; Burke and Ellenberg, 2002; Harel and Forbes, 2004; Baur et al., 2007; Rasala et al., 2008). However, the precise order and regulation of components in nuclear assembly remain an area of intense study (Antonin et al., 2008; D'Angelo and Hetzer, 2008; Rasala et al., 2008). Recently, the formation of nuclear pores was found to be initiated by the recruitment of a new pore-targeting protein, ELYS/MEL-28 to chromatin. ELYS recruits the Nup107-160 complex, the largest pore subcomplex, which contains 9–10 members (Li et al., 1995; Goldstein et al., 1996; Siniossoglou et al., 1996; Siniossoglou et al., 2000; Belgareh et al., 2001; Lutzmann et al., 2002; Harel et al., 2003b; Walther et al., 2003a; Loiodice et al., 2004; Zuccolo et al., 2007; Boehmer et al., 2008; Brohawn et al., 2008). Indeed, vertebrate ELYS was identified biochemically as a binding partner of the critical vertebrate Nup107-160 complex (Rasala et al., 2006; Franz et al., 2007; Gillespie et al., 2007). Strikingly, in the absence of ELYS, pore complexes do not assemble at the chromatin periphery, but instead form pore mimics in the ER; these cytoplasmic membrane stacks of pores are termed cytoplasmic annulate lamellae (AL; Rasala et al., 2006, 2008; Franz et al., 2007). Furthermore, the recruitment of ELYS and the Nup107-160 complex to chromatin was found to be a prerequisite for the recruitment of the integral membrane pore proteins POM121 and NDC1, followed by the bulk of the soluble nucleoporins, including the FG repeat nucleoporins (Rasala et al., 2008), to form the final mature nuclear pore perforating the double nuclear membranes. Interestingly, a fraction of ELYS moves with the Nup107-160 complex to kinetochores at mitosis, and mutation or RNAi of either causes cell cycle defects in yeast, Caenorhabditis elegans, zebrafish, and humans (Bai et al., 2004; Fernandez and Piano, 2006; Rasala et al., 2006; Davuluri et al., 2008; de Jong-Curtain et al., 2009).
Clearly, the formation of large cellular structures such as the mitotic spindle, nuclear membranes, and nuclear pores would be predicted to be the subject of careful and coordinated regulation. Two key regulators of all these assembly events have been identified, as stated above: RanGTP acts as a positive regulator, whereas importin beta acts as a negative regulator for spindle assembly (Kalab et al., 2002; Quimby and Dasso, 2003; Clarke and Zhang, 2008; Kalab and Heald, 2008) and for nuclear membrane and pore assembly (Ryan and Wente, 2002; Harel et al., 2003a; Ryan et al., 2003; Walther et al., 2003b; Clarke and Zhang, 2004; Harel and Forbes, 2004; Hetzer et al., 2005; D'Angelo et al., 2006; Ryan et al., 2007; Antonin et al., 2008). In the case of spindle assembly, importin beta, often in conjunction with importin alpha, binds to spindle assembly factors (SAFs) such as NuMA and TPX2 and sequesters them in an inactive form, preventing them from promoting microtubule assembly during mitosis—except in the area of the mitotic chromosomes (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Du et al., 2002; Schatz et al., 2003; Trieselmann et al., 2003; Ciciarello et al., 2004; Ems-McClung et al., 2004; Blower et al., 2005; Maresca et al., 2005; Albee et al., 2006; Ribbeck et al., 2006; Sillje et al., 2006; Tahara et al., 2008). RanGTP, produced only in the vicinity of the chromosomes, releases importin alpha and/or beta from nearby spindle assembly factors, allowing a spindle to form specifically around the mitotic chromosomes. Thus, importin beta and RanGTP act as spatial regulators or mitotic spindle assembly.
For nuclear assembly, when excess importin beta is added, membrane vesicles are recruited to the chromatin, but fail to undergo the vesicle–vesicle fusion necessary to form the double nuclear membranes. This inhibition of fusion can be counteracted by the inclusion of increased RanGTP (Harel et al., 2003a; Walther et al., 2003b; Delmar et al., 2008). If excess importin beta is added to membrane-enclosed nuclear intermediates, it separately blocks nuclear pore assembly (Harel et al., 2003a; Walther et al., 2003b; Delmar et al., 2008). RanGTP reverses the inhibition of nuclear pore assembly, but only when untagged importin beta is used as the negative regulator (Delmar et al., 2008). Excess Ran alone causes excessive pore formation, both as nuclear annulate lamellae and as cytoplasmic annulate lamellae (Harel et al., 2003a; Walther et al., 2003b). These studies demonstrated a precise balance between the two regulators required for proper nuclear envelope assembly. For nuclear assembly, we believe that this regulatory system would be utilized in vivo to carefully regulate nuclear membrane fusion to occur where RanGTP is high, as on the surface of mitotic chromosomes, in order to prevent spatially or temporally undesirable fusion. Undesirable fusion would include the formation of intranuclear or cytoplasmic annulate lamellae, or the formation of disproportionate ratios of inner to outer nuclear membrane (Harel et al., 2003a; Walther et al., 2003b; Delmar et al., 2008). With respect to pore assembly, the importin beta negative regulatory system would prevent nuclear pores from forming in excess amounts on the nucleus or in inappropriate places as annulate lamellae elsewhere in the cell.
Analysis of the above studies poses a number of major questions: 1) Is the critical first step of nuclear pore assembly, the recruitment of ELYS to chromatin, regulated by importin beta? 2) Which downstream nucleoporin subunits are targets for importin beta regulation? 3) And perhaps most interestingly, do other transport receptors also regulate the steps of nuclear assembly—or spindle assembly?
In this study, we have found that transportin is indeed a regulator of cell cycle assembly events. Transportin negatively regulates nuclear membrane formation, nuclear pore assembly, and spindle assembly in the Xenopus nuclear in vitro system and does so in a Ran-sensitive manner. Focusing on the events of nuclear pore assembly, a search for the regulatory targets of transportin and importin beta in nuclear pore assembly was done and revealed largely the same soluble nucleoporin targets. Furthermore, both transportin and importin beta affect the critical first step of nuclear pore assembly, ELYS/MEL-28 binding to chromatin to initiate pore assembly.
MATERIALS AND METHODS
Recombinant Protein Cloning, Expression, Purification, and Antibodies
GST-human transportin (pGEX6P-Trn) was cloned by ligating a BamHI and XhoI fragment containing full-length transportin (Transportin 1; AAH40340) from pET28a-Trn (S. Shah and D. Forbes, unpublished data) into pGEX6P-3 vector (GE Healthcare, Piscataway, NJ). Cloning of GST-Xenopus importin beta was described in Delmar et al. (2008). Glutathione S-transferase (GST), GST-tagged Xenopus importin beta, and GST-tagged human transportin proteins were expressed in BL21 competent cells (EMD Chemicals, Gibbstown, NJ) by inducing with 0.1 mM IPTG and grown overnight at 17°C. Glutathione-Sepharose 4B beads (GE Healthcare) were used to purify the GST-tagged protein according to manufacturer's instructions. To obtain untagged importin beta and transportin, the GST-tagged protein was cleaved using GST-Precision Protease (GE Healthcare) and incubated at 4°C for 4 h. The cleaved protein was eluted and dialyzed into 5% glycerol in PBS (8 g/l NaCl, 2 g/l KCl, 1.44 g/l Na2HPO4, and 0.24 g/l KH2PO4, pH 7.4) and stored at −80°C. All the Xenopus importin beta and human transportin recombinant proteins added to the reactions are untagged, except in the case of pulldown experiments where GST-tagged importin beta and transportin were used. We note that GST-tagged and 6xHis-tagged transportin behaves indistinguishably from untagged transportin.
6xHis- and GST-tagged RanQ69L-GTP were expressed, purified, and loaded with GTP as previously described (Harel et al., 2003a). GST-ELYS fragments were expressed and purified as previously described (Rasala et al., 2008).
Antibodies used in this study included rabbit anti-gp210 (Harel et al., 2003a), anti-hNup133 and anti-xNup160 (Harel et al., 2003b), anti-xNup43 (Orjalo et al., 2006), anti-Nup93 (Miller and Forbes, 2000), anti-Nup53 and anti-xELYS (Rasala et al., 2008), anti-Orc2 (generous gift from Dr. John Newport, University of California, San Diego), anti-Xenopus importin beta (Rasala et al., 2006), and mouse anti-human transportin and anti-Nup62 (BD Biosciences, San Jose, CA).
Membrane Fusion and Nuclear Pore Assembly Assays
Xenopus high-speed cytosol and membranes, both from interphase Xenopus eggs, and Xenopus sperm chromatin were prepared as described in Harel et al. (2003a). Membrane fusion and nuclear pore assembly assays were performed as described previously (Harel et al., 2003a). Nuclear membrane assembly was analyzed (Figure 1A) using the fluorescent membrane dye 3,3-dihexyloxacarbocyanine iodide (DHCC; Eastman Kodak, Rochester, NY) and visualized with an Axiovert 200M confocal microscope (Carl Zeiss, Thornwood, NY) and a 63× objective. Images from the confocal microscope were recorded using a Coolsnap HQ (Photometrics, Tucson, AZ) camera and Metavue software (Molecular Devices, Downingtown, PA). Images were processed using ImageJ (available at http://rsb.info.nih.gov/ij/) and Adobe Photoshop (San Jose, CA). For dextran diffusion analysis of membrane integrity (Figure 1B), in vitro nuclear reconstitution reactions were carried out as above, except that at the beginning of the reaction additions were made as follows: GST, as a control (25 μM); GTPγS (2 mM); BAPTA (7.5 mM); RanQ69L-GTP (37.5 μM); GST-Trn (25 μM); or RanQ69L-GTP (37.5 μM) plus GST-Trn (25 μM). Assembly was allowed to proceed for 60 min, and then WGA (100 μM; EY Laboratories, San Mateo, CA) was added for 10 min to further ensure a tight seal of any nuclear pores present (although nuclear pores are already expected to exclude 70-kDa dextran). Reactions were stopped on ice, and then rhodamine-labeled dextran (70 kDa; 2.5 μg; Molecular Probes, Eugene, OR) was added to a 25-μl reaction, followed by 15-min incubation on ice. Reactions were diluted 1:1 and fixed with egg lysis buffer containing 7.4% paraformaldehyde. Dextran exclusion was visualized with an Axioskop2 microscope (Carl Zeiss) at a magnification of 100× using an oil objective (Carl Zeiss). Nuclear pore assembly (Figure 2) was visualized with the same microscope and a 63× objective as determined by staining for FG nucleoporins with mAb414 antibody (Covance, Berkeley, CA) which was directly labeled with Alexa 488 (Invitrogen, Eugene, OR) per manufacturer protocol. BAPTA [1,2-bis(2-aminophenoxy)ethane-N,N,N,N-tetraacetic acid; EMD Chemicals] was added at 8 μM.
Figure 1.
Excess transportin blocks nuclear membrane fusion in a Ran-regulated manner. (A) Nuclear formation from Xenopus egg extract was performed at room temperature for 1 h with the addition of PBS buffer (control), 2 mM GTPγS (GTPγS), 37.5 μM RanQ69L-GTP (Ran), 25 μM transportin (Trn), or 25 μM transportin and 37.5 μM RanQ69L-GTP (Trn+Ran). Membrane fusion was observed without fixation using confocal microscopy and by staining nuclei with the membrane dye DHCC (green). DNA was stained with DAPI (blue). Sections of membrane stain were magnified three times (3×) and represented to the right of the merged images. Discontinuities in the DHCC staining indicate regions with little or no membrane fusion. Representative images are shown. Bar, 10 μm. (B) To determine membrane integrity, six in vitro nuclear reconstitution reactions were set up and supplemented with the following additions: GST as a control (25 μM), GTPγS (2 mM), BAPTA (7.5 mM), RanQ69L-GTP (37.5 μM), GST-Trn (25 μM), or RanQ69L-GTP (37.5 μM) plus GST-Trn (25 μM). After 60 min of assembly, the nuclei were treated as described in Materials and Methods. The entry or exclusion of rhodamine-labeled 70-kDa dextran (70-kDa Rhod-Dex) was visualized using fluorescence microscopy. The membrane integrity of the same nuclei was also assessed as visualized with differential interference contrast microscopy in the right panels (DIC). Bar, 10 μm.
Figure 2.
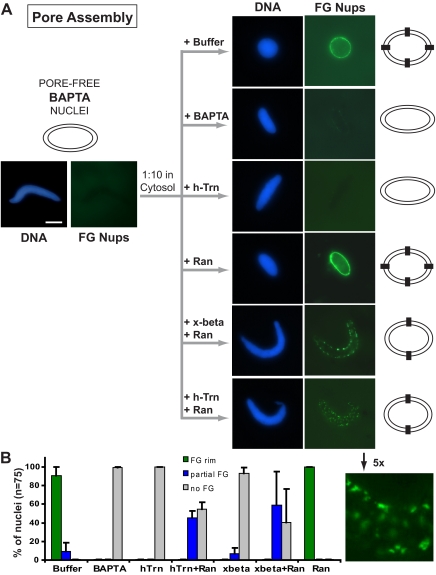
Excess transportin blocks nuclear pore assembly. (A) Pore-free BAPTA nuclei intermediates were assembled by adding 8 μM BAPTA in the nuclear formation reaction at t = 0 min. At t = 60 min, BAPTA-arrested nuclei were diluted 1:10 in fresh cytosol in the presence or absence of recombinant proteins. To assess nuclear pore formation, Alexa-488 directly labeled antibody against FG nucleoporins (mAb414) was added to the nuclei at t = 90 min for an additional 20 min before visualization by fluorescence microscopy. As expected, no FG pore staining was observed in the starting BAPTA nuclei (left panels). When the BAPTA nuclei were then diluted 1:10 into fresh cytosol to which a small amount of control buffer was added (+buffer), or to which 30 μM RanQ69L-GTP (+Ran) was added, nuclear pores formed, as detected by anti-FG staining. If, however, BAPTA (8 μM) were included in the fresh cytosol, pore assembly was prevented (+BAPTA). Strikingly, human transportin (20 μM; +h-Trn) blocked nuclear pore formation, as did Xenopus importin beta (20 μM; data not shown; Delmar et al., 2008). In contrast, significant, albeit not full, FG staining was observed when 20 μM transportin was added in conjunction with 30 μM RanQ69L-GTP (+h-Trn+Ran) or when 20 μM Xenopus importin beta was added with 30 μM RanQ69L-GTP (+x-beta+Ran). A 5× magnification of a representative Trn+Ran nucleus is shown, with the signal of the FG Nup staining brightened to show detail. DNA was stained with DAPI (blue). Representative images are shown. Bar, 10 μm. (B) Quantitation of data in A. Seventy-five nuclei per experiment were counted under each condition, and the percentage of nuclei that contained strong, partial, or no FG-nucleoporin staining was plotted. Error bars, SD calculated over three independent experiments.
Annulate Lamellae Assembly
Xenopus cytosol (40 μl), membranes (5 μl), glycogen (5 μl), and the indicated amount of recombinant proteins were incubated together for 90 min at room temperature. Ten microliters of the reaction were diluted in 190 μl ELB (10 mM HEPES, pH 7.6, 50 mM KCl, and 2.5 mM MgCl2) and overlaid onto a 300-μl sucrose cushion (0.5 M) in ELB. The samples were spun at 25,000 rpm for 20 min at 2°C in a TL-100 tabletop ultracentrifuge (Beckman Coulter, Fullerton, CA). The membrane pellet was collected, rinsed with ELB, and resuspended in 100 μl 1× SDS-PAGE sample loading buffer. One tenth of the volume was loaded for SDS-PAGE and processed for immunoblotting.
For GST pulldowns, recombinant GST, GST-Xenopus importin beta, and GST-human transportin were incubated with glutathione-Sepharose beads (GE Healthcare) without cross-linking. After blocking with 20 mg/ml bovine serum albumin (BSA) in PBS for 30 min, Xenopus egg cytosol was spun for an additional 150,000 rpm for 10 min at 4°C to remove residual membrane contamination and then added to the beads and incubated for 2 h at 4°C (25 μL cytosol in 500 μL PBS). After washing the beads with PBS, the bound proteins were eluted with 0.1 M glycine, pH 2.5, and neutralized with 1 M Tris, pH 8.0. One fifth of each reaction was loaded for SDS-PAGE and processed for immunoblotting.
Chromatin Binding for Immunofluorescence Microscopy
Xenopus cytosol, sperm chromatin, and recombinant proteins were incubated together for 20 min at room temperature. The reactions were diluted in 800 μl ELB, overlaid on a 300 μl 25% sucrose cushion in ELB, and centrifuged at 750 rpm (100 × g) for 15 min onto poly-l-lysine–treated coverslips. Coverslips were then fixed in 4% formaldehyde in PBS for 10 min at room temperature and processed for immunofluorescence microscopy. Nuclei were visualized with an Axioskop 2 microscope (63× objective; Carl Zeiss).
Anchored Chromatin Assay for Immunoblotting
Xenopus cytosol was heat inactivated at 100°C for 3 min and then spun at 14,000 rpm for 20 min to collect the supernatant. Twenty-five microliters of heat-inactivated cytosol, which contains primarily the protein nucleoplasmin, was used to decondense 500,000 Xenopus sperm chromatin at room temperature for ∼15 min, and the state of chromatin decondensation was monitored by DAPI staining and fluorescence microscopy. The decondensed sperm chromatin were diluted in 300 μl ELB (10 mM HEPES, pH 7.6, 50 mM KCl, and 2.5 mM MgCl2) and allowed to bind to poly-l-lysine–coated coverslips by gravity for 2 h at room temperature. The chromatin covered coverslips were blocked with 4% BSA in ELB. The blocked chromatin coated coverslips were then incubated with cytosol or cytosol in the presence of 20 μM Xenopus importin beta, 20 μM human transportin, or 30 μM RanQ69L-GTP recombinant protein for 20 min at room temperature. After three washes in ELBK (10 mM HEPES, pH 7.6, 100 mM KCl, and 2.5 mM MgCl2) the chromatin was lysed in 30 μl 1× SDS-PAGE sample loading buffer, and the bound proteins were subjected to SDS-PAGE and immunoblotting.
ELYS Direct Binding Assay
To assess direct binding, 1.0 μM untagged Xenopus importin beta or human transportin was incubated with 1 μM GST, GST-RanQ69L, GST-ELYS AT-hook+, GST-ELYS ΔAT-hook, or GST-ELYS short AT-hook+ in 1× binding buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 0.2% BSA, 1 μg/ml μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml protease inhibitor TPCK, 1 mM benzamidine, and 1 mM PMSF) with or without 5 μM RanQ69L-GTP on ice for 1 h. Meanwhile, glutathione-Sepharose beads (GE Healthcare) were washed and blocked with 20 mg/ml BSA (Sigma-Aldrich, St. Louis, MO) for 30 min at room temperature in 1× binding buffer. Ten microliters of the glutathione beads were added to the recombinant proteins in each reaction and incubated for additional 30 min at 4°C in a total volume of 100 μl, before washing three times with 1× binding buffer (without BSA) and two times with PBS. The bound proteins were eluted with 27 μl 0.1 M glycine, pH 2.5, followed by neutralization with 3 μl 1 M Tris, pH 8.0, and addition to 10 μl 4× SDS-PAGE sample loading buffer. Samples were boiled and loaded onto 12% SDS-PAGE gels, which were silver-stained, stained with Coomassie blue, or transferred to PVDF for immunoblotting.
Spindle Assembly Experiments
Frogs for interphase extract preparation were induced to ovulate with injection of 500 U of human chorionic gonadotropin (Sigma-Aldrich) and put into individual buckets containing 100 mM NaCl for 18 h. The frogs laid eggs overnight at 18°C, and the eggs were collected the next morning. After removing most of the NaCl solution from the best batches, eggs were dejellied with a 2% cysteine solution, pH 7.7. Activated and dead eggs were constantly removed using a transfer pipette (Fisher Scientific, Pittsburg, PA). When dejellied, eggs were washed extensively three times with XB buffer (100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 50 mM sucrose, pH 7.7). Using a cut transfer pipette, eggs were transferred in 2-ml polypropylene tubes (Beckman) and crushed by centrifugation using a TL55S rotor in a Beckman TL100 ultracentrifuge for 15 min at 15,000 rpm. The cytoplasmic layer was removed with a 18-guage needle on a 3-ml syringe, gently drawn out, and supplemented with cytochalasin B (50 μg/ml) and a protease inhibitor cocktail (10 μg/ml aprotinin and 10 μg/ml leupeptin). The cytoplasmic extract was then centrifuged a second time for 15 min at 15,000 rpm. The light layer was recovered and used as the interphase extract.
The mitotic extract (cytostatic factor or CSF extract) was made exactly as the interphase extract, except for the use of the buffer CSF-XB (100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 50 mM sucrose, and 7 mM EGTA, pH 7.7) instead of XB.
For spindle assembly, ∼6.5 × 104 sperm chromatin DNA (3000 SpC/μl) were added to 20 μl of interphase extract, as well as ATP energy mix (10 mM phosphocreatine, 80 μg/ml creatine kinase, 1 mM ATP, 2 mM MgCl2, and 100 μM EGTA). At t = 90 min after the DNA addition, the extract now containing assembled nuclei was supplemented with rhodamine-tubulin (final concentration 20 μg/ml) and 20 μl of mitotic extract to induce entry into mitosis. Where indicated, GST-transportin was added to a final concentration of 20 μM and RanQ69L-GTP to a final concentration of 5 μM. At t = 15 or 60 min after mitotic extract addition, aliquots were withdrawn and fixed with 1 μl of fixation buffer (48% glycerol, 11% formaldehyde, and 10 mM HEPES, pH 7.5) supplemented with 5 μg/ml Hoechst DNA dye. Spindle formation was monitored using a Zeiss Axioskop 2 microscope using a 63× objective.
RESULTS
Transportin Blocks Nuclear Membrane Formation In Vitro in a Ran-reversible Manner
Importin beta has been shown to act as a negative regulator of both nuclear membrane and nuclear pore assembly (Harel et al., 2003a; Walther et al., 2003b; Delmar et al., 2008). We asked whether another nuclear import receptor, transportin, could regulate nuclear envelope assembly in the Xenopus laevis in vitro nuclear assembly system. In a control reaction, when chromatin was mixed with Xenopus egg cytosol and membranes, completely fused nuclear membranes were observed to encircle the chromatin by 60 min, as indicated by a smooth membrane stain surrounding each nucleus (Figure 1A, Ctrl). The addition of GTPγS, a known inhibitor of vesicle–vesicle fusion, inhibited nuclear membrane formation as evidenced by the discontinuous membrane profile, indicative of unfused membrane vesicles (Figure 1A, GTPγS; Newport and Dunphy, 1992; Macaulay and Forbes, 1996). Importin beta gave, as expected, a similarly discontinuous membrane profile around the chromatin (data not shown; Harel et al., 2003a; Delmar et al., 2008). When excess transportin was added, unfused membranes were also observed (Figure 1A, Trn). Importantly, this membrane fusion defect was counteracted by the positive regulator Ran in the form of RanQ69L-GTP (a form of Ran that cannot hydrolyze GTP; Hughes et al., 1998; Figure 1A, compare Trn+Ran with Trn), as previously seen with importin beta (Harel et al., 2003a; Delmar et al., 2008). We also measured membrane fusion (or lack thereof) by determining whether the structures formed around chromatin after 60 min of reconstitution were able to exclude 70-kDa rhodamine-labeled dextran; 70-kDa dextran is known to be unable to diffuse through nuclear pores in completely formed nuclear membranes, but can diffuse into nuclei if there are gaps in the nuclear membranes or if nuclear membrane assembly never proceeds beyond the vesicle-binding stage. We found that when either transportin or GTPγS was added, then exclusion of rhodamine 70-kDa dextran was not observed (Figure 1B, red). Observation of a discontinuous nuclear rim by differential interference contrast (DIC) microscopy confirmed this lack of fusion when transportin or GTPγS were added (Figure 1B, gray). When Ran was added with transportin, dextran exclusion and a continuous rim were now observed. Taken together, these results showing a discontinuous rim by DIC microscopy (Figure 1B) and by DHCC membrane labeling (Figure 1A), as well as an absence of dextran exclusion (Figure 1B, red) all indicated that an intact nuclear membrane does not form in the presence of excess transportin. We conclude that transportin negatively regulates nuclear membrane fusion and does so in a Ran-reversible manner.
Transportin Negatively Regulates Nuclear Pore Assembly
To determine whether transportin separately blocks mature nuclear pore assembly, we formed nuclear intermediates that contained complete nuclear membranes, but which lacked nuclear pores. This was done by performing nuclear assembly in the presence of the calcium chelator BAPTA (Macaulay and Forbes, 1996; Harel et al., 2003a; Delmar et al., 2008). BAPTA nuclear intermediates contain a fused nuclear envelope, but no mature nuclear pores, as previously shown by electron microscopy and a lack of detectable FG nucleoporins by immunofluorescence with mAb414 (Macaulay and Forbes, 1996; Harel et al., 2003a; Delmar et al., 2008). When such BAPTA nuclear intermediates are diluted 1:10 into fresh cytosol lacking BAPTA, nuclear pore assembly quickly ensues (Macaulay and Forbes, 1996; Harel et al., 2003a; Delmar et al., 2008). This rescue was typically apparent from the acquisition of FG-nucleoporin staining by the rescued nuclei as shown in Figure 2A (+Buffer). If the added Xenopus cytosol instead contained a second addition of BAPTA, rescue was not observed (Figure 2A, +BAPTA). However, if BAPTA nuclei were diluted into cytosol containing excess transportin at either 10 or 20 μM concentrations, the incorporation of FG-nucleoporins was substantially reduced by 10 μM transportin (Supplemental Figure S1) and was completely blocked by 20 μM transportin (Figure 2A, +h-Trn; Supplemental Figure S1). We found that the ability of transportin to block nuclear pore assembly was partially reversed by RanQ69L-GTP: FG-staining pores were now observed, albeit fewer in number (Figure 2A, +h-Trn+Ran; see also 5× high-magnification inset) than in the control (+buffer). This partial rescue of pore assembly by Ran-Q69L was also observed when importin beta was the inhibitory agent (Figure 2A, +x-beta +Ran). When RanQ69L-GTP was added alone, it did not have any negative effect on nuclear pore assembly (Figure 2A, +Ran). To quantitate these effects, seventy-five nuclei for each condition in each of three different experiments were counted for a strong FG nucleoporin rim, a partial FG rim, or the absence of FG staining. The quantitation, as shown in Figure 2B, clearly confirms that transportin blocks nuclear pore assembly and that this block is partially reversed by Ran-GTP.
Annulate Lamellae Assembly Is Negatively Regulated by Transportin
AL are stacked membranes within the ER membrane network that contain cytoplasmic mimics of intact nuclear pore complexes (Merisko, 1989; Dabauvalle et al., 1991; Meier et al., 1995). In most ways these AL pores are identical to nuclear pores, but they are assembled differently in that they do not require the pore-targeting protein ELYS or chromatin in order to form (Kessel, 1989; Merisko, 1989; Dabauvalle et al., 1991; Meier et al., 1995; Miller and Forbes, 2000; Rasala et al., 2008). A previous study found that 10 μM transportin did not block AL pore assembly (Walther et al., 2003b). However, in that electron microscopic experiment, transportin was only tested in the presence of 5 μM RanQ69L-GTP and thus was likely lower in effective concentration. We observed in our experiments above that 20 μM transportin was required to completely block nuclear pore assembly. [We note that our assembly extracts are about twice as concentrated as those used in Walther et al. (2003b)]. We next set out to ask whether transportin (20 μM, with no added RanQ69L-GTP) could block in vitro AL pore assembly, as determined by immunoblotting of membrane pellets for nucleoporins. To form ALs, Xenopus egg cytosol was mixed with Xenopus membranes in the absence of chromatin and incubated for 90 min. The AL were isolated by centrifugation and subjected to immunoblot analysis, and the incorporation of multiple key nucleoporins into ALs was assessed. When ALs were formed under control conditions, all the individual nucleoporins tested including Nup133, Nup43, Nup93, and Nup53 were detected as present, indicating the formation of AL pores (Figure 3C, lane 1). When we added 20 μM importin beta to an AL assembly reaction, as expected from previous work, it blocked the incorporation of these soluble nucleoporins into membranes (Figure 3C, lane 2). Addition of 20 μM transportin also clearly blocked AL assembly (Figure 3C, lane 4). Indeed, importin beta and transportin inhibited the incorporation of all tested nucleoporins, which in addition to the ones shown in Figure 3C, included Nup214, Nup155, Nup153, Nup98, and Nup62 (data not shown). The block to AL formation by importin beta and transportin was significantly reversed by the addition of RanQ69L-GTP (Figure 3C, lanes 3 and 5). These results demonstrate that transportin is mirroring its affect on nuclear pore assembly and negatively regulating AL pore assembly.
Figure 3.
Nucleoporin targets of importin beta and transportin. (A) GST, GST-importin beta, or GST-transportin was added to Xenopus cytosol and incubated for 2 h at 4°C. The bound proteins were analyzed by immunoblotting. Immunoblotting controls were shown in lane 1 (Cytosol). GST alone did not interact with any of the tested nucleoporins (lane 2). FG-nucleoporins (Nup358, Nup214, Nup153, Nup98, Nup62, Nup50, and Nup53) interact with both importin beta and transportin as expected (lanes 3 and 4). Members of the Nup107-160 complex (Nup160, Nup133, and Nup85), the Nup107-160 complex–associated proteins ELYS and centrin also bind both importin beta and transportin. Nup155 and Nup93 do not associate with importin beta or transportin. In the Nup205 immunoblot, a higher nonspecific band was observed. (B) GST or GST-transportin pulldowns were performed and bound proteins were analyzed as in A, except that 10 μM RanQ69L-GTP was added in the reactions in lanes 3 and 5 (GST+RanQ69L, and GST-Trn+RanQ69L). (C) Annulate lamellae pore assembly is regulated by transportin in a Ran-mediated manner. Xenopus cytosol and membranes were incubated with or without recombinant proteins for 90 min. AL membranes were isolated by centrifugation and the associated nucleoporins were detected by immunoblotting. When AL is formed, all tested nucleoporins (gp210, Nup133, Nup43, Nup93, and Nup53) are present (lane 1). However, when excess importin beta or transportin (20 μM) are added, these nucleoporins no longer accumulate on AL membranes (lanes 2 and 4). The importin beta and transportin block to AL assembly can be largely reversed by addition of excess RanQ69L-GTP (30 μM; lanes 3 and 5). Excess RanQ69L-GTP alone does not affect AL pore assembly (lane 6). AL was not formed when only cytosol (lane 7) or membrane (lane 8) was added to sperm chromatin. Equal amounts of membranes were collected under each condition, as indicated by the pore membrane protein gp210 (top row).
A Subset of Nucleoporins Binds to Both Transportin and Importin Beta
We next wanted to determine the nucleoporin subunit targets of transportin and importin beta regulation of pore assembly. Both direct and indirect interactions between transportin or importin beta and nucleoporins could affect pore assembly. During import in intact cells, both karyopherins are known to bind to FG-containing nucleoporins that are essential for nucleocytoplasmic transport (Madrid and Weis, 2006; Terry et al., 2007; Lim et al., 2008). One hypothesis would be that transportin and importin beta regulate pore assembly by sequestering the essential FG-nucleoporins, except in the region of the chromosomes where RanGTP is high. A broader hypothesis might predict that all nucleoporin subunits are targets of regulation. Lastly, it is conceivable that transportin and importin beta might have overlapping and nonoverlapping nucleoporin targets that can be regulated differentially.
To address the potential nucleoporin subunit targets of receptor inhibition in the Xenopus system in a comprehensive manner, we performed GST pulldowns from Xenopus cytosol using GST, GST-importin beta, or GST-transportin as bait. We probed for the different pore subunits by immunoblotting using antibodies to 12 of the 14 soluble nuclear pore complex subunits with the exception of Aladin and Tpr (Rasala et al., 2008). We found that both importin beta and transportin clearly interacted with the FG-nucleoporins Nup358, Nup214, Nup153, Nup98, Nup62, and Nup50 (Figure 3, A, lanes 3 and 4, and B, lane 4), as expected (Moroianu et al., 1995; Shah and Forbes, 1998; Shah et al., 1998; Yaseen and Blobel, 1999; Fontoura et al., 2000; Ben-Efraim and Gerace, 2001; Lindsay et al., 2002). Nup53, which contains a low number of FGs (Marelli et al., 1998; Devos et al., 2006) also associated with importin beta and transportin (Figure 3A, lanes 3 and 4).
The major subunit of the nuclear pore, the Nup107-160 complex, exists in two complexes: one containing nine core nucleoporins (Nup107, Nup160, Nup133, Nup96, Nup85, Nup43, Nup37, Sec13, and Seh1) and a second complex that also contains the pore-targeting nucleoporin ELYS/MEL-28. A previous study has shown that an ELYS/Nup107-160 complex immunoprecipitated by anti-ELYS antibody from human cell lysates does not contain importin beta (Rasala et al., 2006). In contrast, immunoprecipitates of ELYS/Nup107-160 complex from Xenopus egg extracts with anti-ELYS antibodies contain importin beta (Franz et al., 2007 and our unpublished observations). When GST-importin beta was used here to perform a pulldown assay from Xenopus egg cytosol, all of the tested members of the Nup107-160 complex were observed (Figure 3A, lane 3). The complex was also observed in GST-transportin pulldowns (Figure 3, A, lane 4, and B, lane 4). Strikingly, the two newly discovered Nup107-160 complex-associated proteins, ELYS and centrin (Rasala et al., 2006; Franz et al., 2007; Gillespie et al., 2007; Resendes et al., 2008), were also found to bind to both GST-importin beta and GST-transportin pulldowns (Figure 3A, lanes 3 and 4).
Not all nucleoporin subunits were found to bind to importin beta and transportin: members of the key Nup93/188/205 scaffold subunit (Grandi et al., 1997; Hawryluk-Gara et al., 2005), as well as another subunit of the pore scaffold, Nup155 (Aitchison et al., 1995; Franz et al., 2005), did not show interaction with importin beta or transportin (Figure 3A, lanes 3 and 4).
The above results show that exogenously added importin beta and transportin can bind to a specific and similar subset of nucleoporin complexes in Xenopus egg cytosol. To ask whether importin beta or transportin is found naturally in endogenous nucleoporin subcomplexes, we performed immunoprecipitation with several different anti-Nup antibodies and probed for the presence of importin beta and transportin. We found that endogenous importin beta and transportin were each present in immunoprecipitates with anti-Nup62, anti-Nup50, and anti-Nup98 antibodies (Supplemental Figure S2, A and B). Significantly, these nucleoporins are all known to be present in Xenopus extracts in separate Nup subcomplexes (see, for example, Rasala et al., 2008). We conclude from this sampling of subcomplexes that importin beta and transportin are clearly present in endogenous Nup subcomplexes in vivo.
We looked further at the effect of Ran-GTP on the nucleoporin targets of transportin. The interaction between GST-transportin and the large Nup107-160 complex (Nup160, Nup133, and Nup43) was abolished by the presence of RanQ69L-GTP (Figure 3B, lane 5). Similarly, the binding of transportin to the FG-nucleoporins Nup214 and Nup98 was also prevented by the presence of RanQ69L-GTP (Figure 3B, lane 5), and the interaction with Nup62 was significantly decreased. In contrast, the interaction between transportin and the FG-nucleoporins Nup358, Nup153, and Nup50 remained the same or increased in the presence of RanQ69L-GTP (Figure 3B, lane 5). Interestingly, these latter three proteins are unique among all nucleoporins in that they have the ability to bind RanGTP on their own (Wu et al., 1995; Yokoyama et al., 1995; Saitoh et al., 1996; Nakielny et al., 1999; Lindsay et al., 2002).
Together, the above data demonstrate that importin beta and transportin interact with a near identical and specific subset of nucleoporins. This suggests a model where importin beta and transportin regulate nuclear pore assembly by a similar mechanism. It is interesting that neither importin beta nor transportin interact with the Nup93/188/205 subcomplex or with Nup155. We conclude that the recruitment of these latter nucleoporins into the forming nuclear pore is not likely to be regulated by importin beta or transportin. Thus, it appears that transportin and importin beta regulate not all, but a subset, of nucleoporins and that this subset is substantially similar between the two karyopherins.
Transportin and Importin Beta Regulate Chromatin Binding of ELYS in the First Step of Nuclear Pore Assembly
The first known step of nuclear pore complex assembly involves recruitment of the pore-targeting nucleoporin ELYS/MEL-28 to AT-rich regions of chromatin. Importantly, this recruitment can occur either in the absence or presence of the Nup107-160 complex in Xenopus egg extracts (Rasala et al., 2006, 2008; Franz et al., 2007; Gillespie et al., 2007). However, the Nup107/160 complex cannot bind to chromatin in the absence of ELYS (Franz et al., 2007). ELYS thus initiates nuclear pore assembly. Because we have found that both transportin and importin beta interact with ELYS and the Nup107-160 complex (Figure 3A), it is feasible that these transport receptors regulate the initial step of nuclear pore assembly. We tested this hypothesis using a chromatin-binding assay. For this assay, Xenopus cytosol and sperm chromatin were mixed in solution with or without the recombinant receptor proteins and incubated for 20 min at room temperature. The chromatin templates were centrifuged onto polylysine-coated coverslips and processed for immunofluorescence. In the control, where cytosol was added to chromatin without any added importin beta or transportin, the chromatin binding proteins ELYS and Orc2 both bound to the chromatin (Figure 4A, +Buffer). In contrast, ELYS no longer associated with chromatin if excess importin beta was present in the reaction (Figure 4A, +beta). ELYS was also greatly reduced on chromatin in the presence of transportin (Figure 4A, +Trn). The binding of Orc2 to chromatin was unaffected by either of these karyopherins (Figure 4A, + beta; +Trn). The inhibition of ELYS binding to chromatin by transportin was substantially prevented if recombinant RanQ69L-GTP was included (Figure 4A, +Trn+Ran). A similar effect was observed with the simultaneous addition of importin beta and Ran-GTP (Figure 4A, +beta+Ran). These effects on ELYS binding were quantitated in four different experiments and are shown in Figure 4B. We conclude that importin beta and transportin negatively regulate the binding of ELYS to chromatin, as judged by this immunofluorescence assay.
Figure 4.
Transportin and importin beta regulate the initial step in nuclear pore assembly, ELYS recruitment. (A) Xenopus cytosol, sperm chromatin, and recombinant proteins were incubated together for 20 min at room temperature, before centrifugation of the chromatin onto coverslips and processing for immunofluorescence with anti-ELYS and anti-ORC2 antibodies. Representative images are shown. When Xenopus cytosol was incubated with a chromatin source in the absence of membranes, ELYS bound to chromatin (+Buffer). However, when 20 μM importin beta or 20 μM transportin were added to the reaction, ELYS no longer bound to chromatin (+beta, and +Trn). Both blocks to chromatin binding of ELYS were substantially prevented by the inclusion of 30 μM RanQ69L-GTP (+beta+Ran, and +Trn+Ran). The binding to chromatin of the known chromatin-binding protein, Orc2, was not significantly changed upon addition of an excess of any of the recombinant proteins. DNA was stained with DAPI. Bar, 10 μm. (B) Quantitation of the data from four experiments performed as in A are plotted. For each experiment, 50 areas per condition were quantitated. Specifically, 10 sections of 10 × 10 pixels in each of five nuclei per condition were measured for pixel brightness using ImageJ software. These values were averaged per condition and normalized to the average pixel brightness value obtained for the control. SDs were calculated over the four experiments.
The binding of ELYS to chromatin was also tested biochemically, using instead an anchored chromatin assay. In this assay, decondensed sperm chromatin was bound to poly-l-lysine coverslips, then incubated with Xenopus cytosol. The unbound cytosolic proteins were washed away, whereas the chromatin-bound proteins were collected in SDS sample buffer, subjected to SDS-PAGE, and tested for the presence of ELYS and the Nup107-160 complex by immunoblotting. In the absence of any added recombinant receptor proteins, ELYS and members of the Nup107-160 complex bound to chromatin, as expected (Figure 5, lane 5; ELYS, Nup160, Nup133, and Nup43). We have previously shown that AT-rich DNA sequences capture ELYS (Rasala et al., 2006, 2008; Franz et al., 2007; Gillespie et al., 2007). ELYS then recruits the Nup107-160 complex (Rasala et al., 2006, 2008; Franz et al., 2007; Gillespie et al., 2007). We found that association of the Nup107-160 complex with chromatin was greatly reduced in the presence of importin beta (Figure 5, lane 6) or transportin (Figure 5, lane 9). The Orc2 signal present on the chromatin remained unchanged whether or not importin beta or transportin were added (Figure 5, cf. lanes 6 and 9 with lane 5). The block of ELYS and Nup107-160 complex binding to chromatin could, in large part, be prevented by the addition of RanQ69L-GTP (Figure 5, lanes 7 and 10). Taken together, the data indicate that importin beta and transportin can negatively regulate the recruitment of the pore targeting protein ELYS to chromatin and do so in a Ran-regulated manner.
Figure 5.

Transportin and importin beta block ELYS from binding to anchored chromatin. Chromatin was decondensed and allowed to settle onto poly-l-lysine–coated coverslips. The anchored chromatin was incubated with Xenopus egg cytosol in the presence and absence of added recombinant protein for 20 min at room temperature. Proteins bound to chromatin were isolated and analyzed by immunoblotting. An immunoblotting control of total Xenopus egg cytosol is shown in lane 1. Negative controls included cytosol incubated with either recombinant importin beta or transportin but without chromatin (lanes 2 and 3), or decondensed chromatin templates with no further addition (lane 4). The Nup107-160 complex (represented by Nup160, Nup133, and Nup43) and ELYS bind to chromatin (lane 5), whereas Nup62 does not bind to chromatin. Orc2, a chromatin-binding protein, was included as a positive control for chromatin binding. When 20 μM importin beta or transportin was added, the binding of ELYS and the Nup107-160 complex to chromatin was largely abolished (lanes 6 and 9, respectively). This block could be significantly reversed by inclusion of 30 μM RanQ69L-GTP (lanes 7 and 10). RanQ69L-GTP alone did not adversely affect the binding of ELYS and the Nup107-160 complex to the chromatin (lane 8).
Transportin and Importin Beta Bind to ELYS Directly
As shown above, transportin and importin beta negatively regulate nuclear pore assembly, prevent ELYS from binding to chromatin, and interact with ELYS in pulldown assays from Xenopus egg cytosol. To determine whether importin beta or transportin can bind to ELYS directly, we performed a direct binding assay using different regions of the C-terminus of ELYS which are known to associate with chromatin (Rasala et al., 2008). The ELYS fragments used were GST-tagged versions of ELYS AT-hook+ (aa 2281-2408), ELYS ΔAT-hook (aa 2359-2408), and ELYS-short AT-hook+ (aa 2281-2359; Figure 6A). All three fragments bind to chromatin, although GST-ELYS AT-hook+ binds somewhat more strongly, as it possesses two chromatin binding domains (Rasala et al., 2008; and data not shown). The GST-tagged versions of each of these ELYS fragments were incubated with untagged importin beta or untagged transportin in the absence of any cytosolic proteins. The bound fraction was analyzed by silver stain to detect importin beta (Figure 6B). GST-RanQ69L-GTP, a positive control, clearly bound importin beta (Figure 6B, lane 1). GST alone, a negative control, did not bind importin beta (Figure 6B, lane 2). We found that both ELYS AT-hook+ and ELYS ΔAT-hook directly bound importin beta (Figure 6B, lanes 3 and 4, respectively). However, ELYS-short AT-hook+ did not bind to importin beta (Figure 6B, lane 5).
Figure 6.
Transportin and importin beta directly bind to ELYS. (A) Schematics of the Xenopus ELYS and three GST-tagged Xenopus ELYS C-terminal fragments used in the direct binding assay. ELYS AT-hook+ contains two chromatin-binding domains (CBD 1 and CBD 2) including an AT-hook motif (black bar) in CBD 1. ELYS ΔAT-hook contains the last 50 amino acids of the C-terminus of ELYS (yellow and red) and lacks the AT-hook. ELYS short AT-hook+ (CBD 1) includes the AT-hook but lacks CBD 2. The sequence of the final 128 amino acids of Xenopus ELYS is shown at the right with the AT-hook boxed, the initial portion of CBD 2 in yellow, and the basic region of CBD 2 in red (see text). (B and C) GST-RanQ69L-GTP, GST alone, or one of three GST-ELYS fragments (GST-ELYS AT-hook+, GST-ELYS ΔAT-hook, or GST-ELYS short AT-hook+) were incubated with 1.0 μM untagged recombinant importin beta (B) or transportin (C) in the presence of BSA but the absence of any Xenopus nuclear or cytoplasmic proteins. The GST-tagged proteins and bound protein were isolated on glutathione-Sepharose beads and subjected to gel electrophoresis and silver stain analysis in B or Western blotting and Coomassie staining in C. The gel in D was Coomassie stained, although E is an immunoblot using anti-transportin antibody. GST-RanQ69L-GTP served as a positive control for importin beta and transportin direct binding (lane 1 in B–D; lane 2 in E), whereas GST served as a negative control (lane 2 in B–D; lane 1 in E). Importin beta and transportin were found to directly bind to GST-ELYS AT-hook+ and GST-ELYS ΔAT-hook, but not to GST-ELYS short AT-hook+ (see lanes 3–5 in B and C; lanes 3–8 in D and E), indicating that binding requires a region in the extreme C-terminus (aa 2359–2408) of ELYS (yellow + red). Comparable amounts of GST recombinant proteins were recovered and loaded on the gels as determined by silver or Coomassie staining; these are marked with hatchmarks (B–D) on the right of each panel. (D and E) Experiments were performed as in B and C except with the addition of RanQ69L-GTP (5 μM) in parallel reactions (lanes 4, 6, and 8).
We further found that both ELYS AT-hook+ and ELYS ΔAT-hook bound to transportin directly (Figure 6C, lanes 3 and 4), whereas GST-ELYS-short AT-hook+ did not bind transportin (Figure 6C, lane 5), as determined by immunoblotting. RanGTP substantially prevented the binding of transportin to ELYS AT-hook+ and ELYS ΔAT-hook (Figure 6E, lanes 8 and 6, respectively), a significant reversal also observed with importin beta and Ran (Figure 6D, lanes 4 and 6). These data indicate that transportin and importin beta can bind directly to the extreme C-terminus of ELYS in a region that does not include the AT-hook.
Transportin Blocks Spindle Assembly in a Ran-sensitive Manner
Importin beta has been shown in multiple studies using mitotic Xenopus egg extracts to regulate spindle assembly. Mitotic spindles readily assemble in vitro around chromatin added to such an extract (see Kalab and Heald, 2008; Clarke and Zhang, 2008 for review). It has been proposed that the location of spindle assembly specifically in the area of mitotic chromosomes in cells is orchestrated by importin beta and its adaptor protein importin alpha, which act to inhibit spindle assembly factors elsewhere (Clarke and Zhang, 2008; Kalab and Heald, 2008). RanGTP, produced by its chromatin-bound RanGEF, releases spindle assembly factors from inhibition in the vicinity of chromosomes; thus Ran acts as a positive regulator of spindle assembly and importin alpha and/or beta act as negative regulators. When excess exogenous Ran is added to Xenopus mitotic cytosol, abundant asters then are induced to form throughout the cytosol, even in the complete absence of chromatin (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999).
Here we asked whether the import receptor transportin would show a similar ability to regulate spindle assembly. We reasoned that this could very well be the case since, like importin beta, transportin negatively inhibits both nuclear membrane assembly and nuclear pore assembly (Figures 1–5).
To assess effects of transportin on mitotic spindle assembly, nuclei were assembled by adding sperm chromatin to a Xenopus interphase egg extract (20 μl) and allowing them to assemble into nuclei for 90 min (Figure 7A, top left panel, 0 min). During this time, it has been demonstrated that the DNA present faithfully replicates (Newport, 1987; Tutter and Walter, 2006). Mitotic egg extract (20 μl) was then added to convert the reaction to a mitotic state. Rhodamine-labeled tubulin was simultaneously added to allow the monitoring of mitotic spindle (or aster) assembly. Transportin (20 μM) was added with or without RanQ69L-GTP (5 μM) in parallel reactions to test for effects on spindle assembly. At 15 min after the addition of mitotic cytosol, essentially all of nuclei in the reactions had been converted to condensed chromatin figures that had an attached rhodamine-labeled half spindle, as is typical of this time point in spindle assembly assays in Xenopus mitotic extracts (Mitchison et al., 2004; Maresca and Heald, 2006; Orjalo et al., 2006; Figure 7A, 15 min). We noted that when transportin was present the half spindles appeared somewhat smaller. At 30 min, small bipolar spindles were beginning to form in both the control and transportin reactions (data not shown). By 60 min, the control showed abundant robust bipolar spindles organized around condensed chromatin (66%; Figure 7A, 60 min). However, the chromatin in reactions with added transportin were for the most part now devoid of associated microtubules (89%; Figure 7A, 60 min). In contrast, when RanQ69L-GTP was included with the transportin, robust (40%) and weak (22%) bipolar spindles formed (Figure 7A, 60 min). Addition of RanGTP alone caused large numbers of asters to form independent of chromatin (52%) as well as the formation of multipolar spindles in the vicinity of chromatin (38%). The data from three such experiments has been quantitated in Figure 7B. The 60-min Control results are shown in blue, transportin results are in red, and transportin plus RanGTP results shown in yellow. We conclude that transportin, like importin beta, is an inhibitor of mitotic spindle assembly in vitro and this inhibition can be counteracted by RanGTP.
Figure 7.
Transportin negatively regulates mitotic spindle assembly. (A) Xenopus interphase egg extract was supplemented with sperm chromatin and energy mix to allow nuclear formation and DNA replication for 90 min (see Materials and Methods). Spindle assembly was then induced and followed by the addition of 20 μl of Xenopus mitotic extract (CSF extract) and rhodamine-labeled tubulin. Human GST-transportin (20 μM; + Trn) or RanQ69L-GTP (5 μM; + Ran) were added where noted. A representative nucleus visualized at t = 0 min after mitotic extract addition corresponds to 90 min after adding sperm chromatin to the Xenopus interphase extract (top left picture). After adding mitotic extract, aliquots were withdrawn at 15 min (left panels) and 60 min (right panels), prefixed with fixation buffer, and the structures formed were counted for Figure 7B. DNA was visualized with 5 μg/ml Hoechst DNA dye. Bar, 10 μm. (B) Quantitation was done to enumerate the in vitro spindles or defective spindles observed after no addition, transportin addition, and/or RanGTP addition. The graph represents the quantitation of the 60-min time points derived from three different experiments done as in A. For each condition, ∼60–80 structures were counted and classified into the categories shown on the x-axis: robust or weak bipolar spindles, robust or weak half spindle, multipolar spindles, asters, or DNA with no microtubules (MTs) at all. All the structures counted contained DNA except in the case of asters (*); added RanGTP caused the formation of ∼60% asters (no associated DNA) and 29% multipolar spindles (contained DNA).
DISCUSSION
Mitosis is a complex stepwise process that requires regulation at multiple points. Here, we show that transportin is a negative regulator of spindle assembly, nuclear membrane formation, and nuclear pore assembly (Figures 1, 2, and 7). Moreover, transportin also negatively regulates assembly of the cytoplasmic mimic of nuclear pores, annulate lamellae (Figure 3C). The nucleoporin targets of transportin and importin beta involve many, but not all nucleoporins. The targets include the FG-nucleoporins and the large critical Nup107-160 complex, but do not include the Nup93-188-205 or Nup155 pore scaffold subunits (Figure 3, A and B). These data imply that the recruitment of certain nucleoporins is regulated by transportin and importin beta (through direct or indirect interactions), but that the recruitment of other Nups is not. When tested, RanGTP reversed the binding of transportin to key Nup subunits (Figures 3, A and B, and 6), consistent with the model that RanGTP promotes pore assembly by releasing nucleoporins from karyopherin sequestration (Harel et al., 2003a; Hetzer et al., 2005; Antonin et al., 2008; Delmar et al., 2008; Lim et al., 2008).
We provide evidence for the first time that both transportin and importin beta can negatively regulate the earliest known step in the initiation of pore assembly, the recruitment to chromatin of the pore-targeting protein ELYS/MEL-28 (Figures 4 and 5). We further find that transportin and importin beta can bind directly to the extreme C-terminus of ELYS in a chromatin-binding region that contains an NLS-like region, but not the ELYS AT-hook (Figure 6A, CBD2). A simple model would be that the direct binding of transportin or importin beta to this NLS-like sequence at the C-terminus of ELYS would be sufficient—in the cellular context—to prevent full-length ELYS from binding to chromatin; alternatively, it is equally possible that full-length ELYS has multiple chromatin-binding domains that bind transportin and/or importin beta during the regulation of ELYS that controls future pore assembly. Prevention of ELYS from binding to chromatin has been previously shown to block the recruitment of the Nup107-160 complex to chromatin and to prevent all further pore assembly (Rasala et al., 2006, 2008; Franz et al., 2007; Gillespie et al., 2007). Thus, negative regulation of ELYS/chromatin binding down-regulates nuclear pore assembly (Figure 8). We hypothesize that transportin and importin beta act in a multiplex manner by also binding to nucleoporin targets other than ELYS, for example, the FG-Nups and the Nup107-160 complex (Figure 3, A and B), in order to ensure that neither pore assembly nor assembly of abortive nucleoporin aggregates occur in inappropriate regions of the cell. These inappropriate regions would include intranuclear annulate lamellae, cytoplasmic annulate lamellae, and smaller soluble aggregates of Nups in the cytoplasm. RanGTP appears to play a counteracting role to the karyopherin regulators in both the initial and downstream steps of pore assembly.
Figure 8.
Transportin and importin beta regulate multiple mitotic assembly events: from spindle assembly to pore assembly. On induction of mitosis, sequential assembly of a number of large cellular structures is required. After prophase, a mitotic spindle must be assembled around the metaphase chromosomes. Transportin (Trn) negatively regulates this assembly event (Figure 7), as was previously shown for importin beta. The next large assembly event is that of the nucleus. In early telophase, the initial step of nuclear pore assembly involves the binding of ELYS (red ovals) to chromatin (blue half circles). Here, we demonstrate that transportin and importin beta both negatively regulate the binding of ELYS to chromatin (Figures 4 and 5). This inhibition is counteracted by RanGTP. ELYS next recruits the Nup107-160 pore subunit (yellow Y-shapes). During this period, membrane vesicles (white circles) and/or sheets are also recruited to the chromatin and then fuse to form a double nuclear membranes (curved lines). The fusion of membrane vesicles to form a nuclear envelope is negatively regulated by transportin (Figure 1) and importin beta (Harel et al., 2003a; Delmar et al., 2008) in a RanGTP-sensitive manner (Figure 1). After vesicle–vesicle fusion at the chromatin to form double membrane patches, the bulk of nucleoporins are recruited into the membrane to form mature nuclear pores (green nuclear pore). This latter process is negatively regulated by transportin (Figure 2) and importin beta (Harel et al., 2003a; Walther et al., 2003b; Delmar et al., 2008) and positively regulated by RanGTP. The transportin and importin beta hexagons outlined in red represent findings presented here for the first time.
Dueling Regulators
Ran reversal of importin beta inhibition is, for the most part, a hallmark of the studies of importin beta regulation to date. When tagged human importin beta was shown to block mitotic spindle assembly and nuclear membrane assembly in Xenopus extracts, both processes were reversed by RanGTP (Nachury et al., 2001; Wiese et al., 2001; Harel et al., 2003a; Ems-McClung et al., 2004; Blower et al., 2005; Albee et al., 2006; Ribbeck et al., 2006; Tahara et al., 2008). An exception to this was the inhibition of nuclear pore assembly by tagged human importin beta, where RanGTP did not reverse the inhibition (Harel et al., 2003a). It was later found, however, that when an untagged version of either human or Xenopus importin beta was used to inhibit nuclear pore assembly, the inhibition was RanGTP-reversible (Delmar et al., 2008). In the present study, we used untagged human transportin, but found that its block to pore assembly was only partially reversed by excess RanGTP. One possibility is that human transportin may be less sensitive to RanGTP than importin beta is, specifically in the area of nuclear pore assembly. An alternate explanation is that there may be a separate effector of transportin in addition to the GTPase Ran involved in pore assembly. Potentially consistent with this, we did observe that although RanGTP was able to release transportin from certain nucleoporins (the FG nucleoporins Nup98 and Nup214 and the Nup107-160 complex), RanGTP left intact transportin's binding to the FG nucleoporins Nup358, Nup153, and Nup50 (Figure 3B), perhaps arguing for the existence of a second positive regulator for transportin. It is known that these latter proteins are unique among nucleoporins in that they contain known RanGTP-binding domains (Wu et al., 1995; Yokoyama et al., 1995; Saitoh et al., 1996; Nakielny et al., 1999; Lindsay et al., 2002). It is possible that their regulation by transportin is reversed via a different mechanism.
Multiple Points of Regulation in Pore Assembly
In mammalian cells, the order of assembly of the Nups has been very broadly categorized using GFP-tagged nucleoporins into early, intermediate, and late-assembling nucleoporins (Bodoor et al., 1999; Dultz et al., 2008). Assuming this order is followed in Xenopus cells as seems likely (Walther et al., 2001; Harel et al., 2003b; Walther et al., 2003b; Rasala et al., 2006, 2008), we conclude that transportin and importin beta bind to nucleoporin subunit targets in each of the temporal stages of pore assembly. Transportin and importin beta bind to ELYS and the Nup107-160 complex, which assemble early; Nup98, which has an intermediate assembly time (Dultz et al., 2008); and Nup50 and Nup214, which assemble late, as well as Nup153 which has both early- and late-assembling populations (Bodoor et al., 1999; Dultz et al., 2008). Thus, it appears that there could be sequential points of regulation by transportin and importin beta during nuclear pore assembly.
Multiple Regulators
Why is more than one nuclear import receptor used to regulate nuclear assembly? One explanation would be that, although transportin and importin beta bind to a similar subset of nucleoporins, they may bind with different affinities. The nucleoporins could then be differentially regulated in a more controlled manner. Another possibility is that the availability or concentration of importin beta accessible for regulation in vivo might be quite different from that of transportin, providing another mechanism for fine tuning the regulation of steps in nuclear assembly. Lastly, we do not exclude the possibility that additional import receptors may regulate nuclear assembly in a similar way.
Regulation of the Initial Step in Nuclear Pore Assembly
The binding of ELYS to chromatin is the initial step in pore assembly. We show this to be regulated by both transportin and importin beta (Figures 4 and 5). In the absence of the recruitment of ELYS to chromatin, pore complexes no longer form in the nuclear envelope, but instead form as annulate lamellae pores in the cytoplasm (Rasala et al., 2006, 2008; Franz et al., 2007). We would predict that if an excess of ELYS were allowed to bind to chromatin, then excessive nuclear pores would form that would lead to defects in nuclear assembly including, for example, aberrant assembly of intranuclear annulate lamellae as previously observed in the presence of excess Ran (Harel et al., 2003a) and to defects in pore spacing or nuclear lamina formation. A disproportion of nuclear pore numbers could also adversely affect the novel functions recently recognized to be mediated by the nuclear pore complex, such as transcriptional activation of certain genes (Casolari et al., 2004; Cabal et al., 2006; Taddei et al., 2006; Brown et al., 2008) and DNA break repair (Nagai et al., 2008). In consequence, we believe it is likely that pore number would be tightly regulated. Interestingly, most vertebrate cultured cell lines have a very similar number of nuclear pores (∼2000–5000), whereas inactive B cells have many fewer nuclear pores (∼400; Maul, 1977). In reality, little is known of the mechanism of pore number regulation. Regulation of the targeting of ELYS to chromatin by transportin and importin beta could be one such mode of regulation.
Karyopherin-binding Sites on the C-Terminus of ELYS
The C-terminal 50 aa of Xenopus ELYS binds both importin beta and transportin directly (aa 2359-2408, Figure 6A, yellow/red). Sequence analysis reveals that the last 18 amino acids contain 10 positively charged residues in two clusters (RRTRRRIIAKPVTRRKMRCOOH; Figure 6A, red). Importin beta is well known to bind to positively charged regions in proteins ranging from its adaptor, importin alpha, to cargoes that importin beta binds directly, such as HIV Rev and ribosomal proteins L23a and S7 (Gorlich and Kutay, 1999). Thus, each basic cluster may bind importin beta directly. The most recently reported transportin NLS consensus sequences contain a PY dipeptide preceded by various alternate upstream amino acid sequences, one of which contains 5–8 basic amino acids (basic-enriched5–8-X8–10-PY; Lee et al., 2006). Although this abundant class of transportin NLSs was preselected by computer analysis on the basis of containing a PY element, the protein HuR, a known transportin cargo, contains a PG in place of PY in its NLS (Lee et al., 2006). Also, of two known cargoes for the S. cerevisiae transportin homologue kap104, Hrp1 contains a PY, whereas Nab2 has a PY-type NLS where a PL replaces the PY (aa 200-250). Each is essential for kap104 binding of these cargoes (Siomi et al., 1998; Truant et al., 1998; Lee and Aitchison, 1999; Marfatia et al., 2003; Lange et al., 2008). Here we found that the C-terminus of Xenopus ELYS directly binds transportin, even though it does not contain a PY; it does, however, contain a PV dipeptide that maps between the two basic clusters at the ELYS C-terminus (Figure 6A, red). It is possible that this PV serves in place of the PY. Indeed, the ELYS sequence RRTRRRIIAKPVTRRKMRCOOH resembles somewhat the predicted transportin NLS of the human metal response element-binding transcription factor 2 (KKGKKKSVGRPPGPYTRKM; Lee et al., 2006).
For vertebrate ELYS, transportin and importin beta have both been shown to bind to the last 50 amino acids (Figure 6), although they may also bind elsewhere on ELYS. If it is found that they bind to the identical sequence or to adjacent sequences within the positively charged ELYS C-terminus, it may be that they contribute to the negative regulation of ELYS binding to chromatin in an interchangeable manner. Alternatively, one of the karyopherins may be present in greater abundance in vivo or be higher in its binding affinity and thus act as the dominant regulator of that particular site. It is also highly possible that importin beta and transportin bind to additional regions of ELYS and that multiple binding events are required for controlling ELYS. In any case, their goal presumably would be to achieve negative regulation of ELYS except in areas of high RanGTP, such as at the surface of chromatin.
Multiplex Regulation of Spindle Assembly
Lastly, we demonstrated that transportin has the ability to negatively regulate an entirely different cell cycle structure, the mitotic spindle, and does so in a Ran-sensitive manner (Figure 7). Our data thus support the idea that, like importin beta, transportin has diverse functions during different parts of the cell cycle, namely as an import receptor during interphase and as a negative regulator of mitotic spindle and nuclear assembly during mitosis. Importin beta was demonstrated to regulate mitotic spindle assembly by binding and sequestering SAFs in areas away from the mitotic chromosomes (reviewed in Harel and Forbes, 2004; Funabiki, 2005; Kalab and Heald, 2008). RanGTP, produced on the mitotic chromosomes by the RanGEF RCC1, acts as a positive regulator for mitotic spindle formation by triggering dissociation of importin beta from its SAF binding partners (Kalab and Heald, 2008). An interesting question will be whether transportin binds to the same SAFs as importin alpha and beta or different ones. At present, at least 10 SAFs have been identified that are direct targets of importin beta and/or alpha (including NuMA, TPX2, and the kinesin/MAP XCTK2; Clarke and Zhang, 2008; Kalab and Heald, 2008). A cursory analysis of these SAFs reveals that several contain a PY dipeptide (D. Forbes, unpublished data), but whether these are authentic transportin binding sites is not yet known. Significantly, the S. cerevisiae kap104 mutant kap104-E604K has been found to speed up mitosis in vivo, consistent with a negative regulatory role for transportin in mitosis (Asakawa and Toh-e, 2002). However, because yeast retain an intact nucleus during their closed mitosis, this has also been suggested to result from a defect in the import role of kap104.
An interesting question is whether other karyopherins similarly sequester SAFs. At present it is known that the export karyopherin Crm1 acts during mitosis but in a different manner, targeting to kinetochores and aiding in the binding of a set of proteins important for kinetochore function (Arnaoutov et al., 2005; Arnaoutov and Dasso, 2005; Knauer et al., 2006). Because export receptors are stabilized by Ran, one might expect that only import receptors could duel with RanGTP to regulate assembly of the large mitotic cellular structures around the chromatin. However, export receptors could also regulate these assembly events if they used an entirely different way to reverse target sequestration such as, for example, being dissociated by RanGAP.
Summary
In conclusion, we have demonstrated that transportin is a negative regulator of nuclear membrane formation and pore assembly in the Xenopus in vitro system and that this regulatory complexity extends to spindle assembly. Focusing on nuclear pore assembly, we showed that both transportin and importin beta negatively regulate the initial step in pore assembly, ELYS binding to chromatin. RanGTP acts in an opposite manner from that of transportin to balance the effects of this new negative regulator. Both transportin and importin beta bind directly to the C-terminus of ELYS, a region critical for ELYS recruitment to chromatin. The precise choreography of regulation provided by the new regulatory karyopherin, transportin, reveals yet another level of complexity to the regulation of mitotic spindle and nuclear assembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Forbes lab for helpful discussions and Michelle Gaylord for excellent technical assistance. The work described here was supported by National Institutes of Health Grant R01-GM033279 to D.J.F.
Abbreviations used:
- AL
annulate lamellae
- FG
phenylalanine-glycine
- GST
glutathione-S-transferase
- NLS
nuclear localization signal
- PY
proline-tyrosine
- SAF
spindle assembly factors
- Trn
transportin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0152) on July 29, 2009.
REFERENCES
- Adam E. J., Adam S. A. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Rout M. P., Marelli M., Blobel G., Wozniak R. W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albee A. J., Tao W., Wiese C. Phosphorylation of maskin by Aurora-A is regulated by RanGTP and importin beta. J. Biol. Chem. 2006;281:38293–38301. doi: 10.1074/jbc.M607203200. [DOI] [PubMed] [Google Scholar]
- Anderson D. J., Hetzer M. W. Shaping the endoplasmic reticulum into the nuclear envelope. J. Cell Sci. 2008;121:137–142. doi: 10.1242/jcs.005777. [DOI] [PubMed] [Google Scholar]
- Antonin W., Ellenberg J., Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- Asakawa K., Toh-e A. A defect of Kap104 alleviates the requirement of mitotic exit network gene functions in Saccharomyces cerevisiae. Genetics. 2002;162:1545–1556. doi: 10.1093/genetics/162.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S. W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. The fission yeast Nup107–120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell. Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur T., Ramadan K., Schlundt A., Kartenbeck J., Meyer H. H. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J. Cell Sci. 2007;120:2895–2903. doi: 10.1242/jcs.010181. [DOI] [PubMed] [Google Scholar]
- Beck M., Lucic V., Forster F., Baumeister W., Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- Beck M., Medalia O. Structural and functional insights into nucleocytoplasmic transport. Histol. Histopathol. 2008;23:1025–1033. doi: 10.14670/HH-23.1025. [DOI] [PubMed] [Google Scholar]
- Belgareh N., et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I., Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W. H., Bastos R., Lohka M., Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 1999;112(Pt 13):2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Boehmer T., Jeudy S., Berke I. C., Schwartz T. U. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol. Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N., Moroianu J., Radu A., Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Leksa N. C., Spear E. D., Rajashankar K. R., Schwartz T. U. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. R., Kennedy C. J., Delmar V. A., Forbes D. J., Silver P. A. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Ellenberg J. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Cabal G. G., et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Cansizoglu A. E., Lee B. J., Zhang Z. C., Fontoura B. M., Chook Y. M. Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Casolari J. M., Brown C. R., Komili S., West J., Hieronymus H., Silver P. A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chan R. C., Forbes D. J. In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. Methods Mol. Biol. 2006;322:289–300. doi: 10.1007/978-1-59745-000-3_20. [DOI] [PubMed] [Google Scholar]
- Chi N. C., Adam E. J., Adam S. A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y. M., Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Chook Y. M., Blobel G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Ciciarello M., Mangiacasale R., Thibier C., Guarguaglini G., Marchetti E., Di Fiore B., Lavia P. Importin beta is transported to spindle poles during mitosis and regulates Ran-dependent spindle assembly factors in mammalian cells. J. Cell Sci. 2004;117:6511–6522. doi: 10.1242/jcs.01569. [DOI] [PubMed] [Google Scholar]
- Clarke P. R., Zhang C. Spatial and temporal control of nuclear envelope assembly by Ran GTPase. Symp. Soc. Exp. Biol. 2004;56:193–204. [PubMed] [Google Scholar]
- Clarke P. R., Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M. A., Anderson D. J., Richard E., Hetzer M. W. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- D'Angelo M. A., Hetzer M. W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle M. C., Loos K., Merkert H., Scheer U. Spontaneous assembly of pore complex-containing membranes (“annulate lamellae”) in Xenopus egg extract in the absence of chromatin. J. Cell Biol. 1991;112:1073–1082. doi: 10.1083/jcb.112.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G., Gong W., Yusuff S., Lorent K., Muthumani M., Dolan A. C., Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4:e1000240. doi: 10.1371/journal.pgen.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong-Curtain T. A., et al. Abnormal nuclear pore formation triggers apoptosis in the intestinal epithelium of elys-deficient Zebra fish. Gastroenterology. 2009;136:193–204. doi: 10.1053/j.gastro.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler E. W., Ma Y., Seo H. S., Hsia K. C., Noriega T. R., Blobel G., Hoelz A. A fence-like coat for the nuclear pore membrane. Mol. Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Delmar V. A., Chan R. C., Forbes D. J. Xenopus importin beta validates human importin beta as a cell cycle negative regulator. BMC Cell Biol. 2008;9:14. doi: 10.1186/1471-2121-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B. T., Rout M. P., Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Taylor L., Compton D. A., Macara I. G. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S. C., Zheng Y., Walczak C. E. Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Koser J., Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Fernandez A. G., Piano F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr. Biol. 2006;16:1757–1763. doi: 10.1016/j.cub.2006.07.071. [DOI] [PubMed] [Google Scholar]
- Finlay D. R., Forbes D. J. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G., Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J. Biol. Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fontoura B. M., Blobel G., Yaseen N. R. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin beta 2-mediated nuclear import. J. Biol. Chem. 2000;275:31289–31296. doi: 10.1074/jbc.M004651200. [DOI] [PubMed] [Google Scholar]
- Franz C., Askjaer P., Antonin W., Iglesias C. L., Haselmann U., Schelder M., de Marco A., Wilm M., Antony C., Mattaj I. W. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I. W., Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell R. A., Truant R., Thorne L., Benson R. E., Cullen B. R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J. Cell Sci. 1997;110(Pt 11):1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Funabiki H. Two birds with one stone—dealing with nuclear transport and spindle assembly. Cell. 2005;121:157–158. doi: 10.1016/j.cell.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gillespie P. J., Khoudoli G. A., Stewart G., Swedlow J. R., Blow J. J. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. W., Wiese C., Allen T. D., Wilson K. L. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J. Cell Sci. 1997;110(Pt 4):409–420. doi: 10.1242/jcs.110.4.409. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., Snay C. A., Heath C. V., Cole C. N. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kostka S., Kraft R., Dingwall C., Laskey R. A., Hartmann E., Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Grandi P., Dang T., Pane N., Shevchenko A., Mann M., Forbes D., Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Harel A., Chan R. C., Lachish-Zalait A., Zimmerman E., Elbaum M., Forbes D. J. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell. 2003a;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Forbes D. J. Importin beta: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D. J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003b;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara L. A., Shibuya E. K., Wozniak R. W. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol. Biol. Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M., Meyer H. H., Walther T. C., Bilbao-Cortes D., Warren G., Mattaj I. W. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- Hetzer M. W., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hughes M., Zhang C., Avis J. M., Hutchison C. J., Clarke P. R. The role of the ran GTPase in nuclear assembly and DNA replication: characterisation of the effects of Ran mutants. J. Cell Sci. 1998;111(Pt 20):3017–3026. doi: 10.1242/jcs.111.20.3017. [DOI] [PubMed] [Google Scholar]
- Imamoto N., Shimamoto T., Kose S., Takao T., Tachibana T., Matsubae M., Sekimoto T., Shimonishi Y., Yoneda Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- Jovanovic-Talisman T., Tetenbaum-Novatt J., McKenney A. S., Zilman A., Peters R., Rout M. P., Chait B. T. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R. T., Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis K., Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kessel R. G. The annulate lamellae—from obscurity to spotlight. Electron Microsc. Rev. 1989;2:257–348. doi: 10.1016/0892-0354(89)90003-8. [DOI] [PubMed] [Google Scholar]
- Knauer S. K., Bier C., Habtemichael N., Stauber R. H. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 2006;7:1259–1265. doi: 10.1038/sj.embor.7400824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Hetzer M. W. Reorganization of the nuclear envelope during open mitosis. Curr. Opin. Cell Biol. 2008;20:669–677. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Mills R. E., Devine S. E., Corbett A. H. A PY-NLS nuclear targeting signal is required for nuclear localization and function of the Saccharomyces cerevisiae mRNA-binding protein Hrp1. J. Biol. Chem. 2008;283:12926–12934. doi: 10.1074/jbc.M800898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. J., Cansizoglu A. E., Suel K. E., Louis T. H., Zhang Z., Chook Y. M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. C., Aitchison J. D. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J. Biol. Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- Li O., Heath C. V., Amberg D. C., Dockendorff T. C., Copeland C. S., Snyder M., Cole C. N. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell. 1995;6:401–417. doi: 10.1091/mbc.6.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R.Y., Aebi U., Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M. E., Plafker K., Smith A. E., Clurman B. E., Macara I. G. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell. 2002;110:349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- Lohka M. J., Masui Y. Roles of cytosol and cytoplasmic particles in nuclear envelope assembly and sperm pronuclear formation in cell-free preparations from amphibian eggs. J. Cell Biol. 1984;98:1222–1230. doi: 10.1083/jcb.98.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I. G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C., Forbes D. J. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J. Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A. S., Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J. D., Wozniak R. W. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T. J., Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- Maresca T. J., Niederstrasser H., Weis K., Heald R. Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr. Biol. 2005;15:1755–1761. doi: 10.1016/j.cub.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Marfatia K. A., Crafton E. B., Green D. M., Corbett A. H. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- Maul G. G. The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int. Rev. Cytol. Suppl. 1977;6:75–186. [PubMed] [Google Scholar]
- Meier E., Miller B. R., Forbes D. J. Nuclear pore complex assembly studied with a biochemical assay for annulate lamellae formation. J. Cell Biol. 1995;129:1459–1472. doi: 10.1083/jcb.129.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merisko E. M. Annulate lamellae: an organelle in search of a function. Tissue Cell. 1989;21:343–354. doi: 10.1016/0040-8166(89)90049-9. [DOI] [PubMed] [Google Scholar]
- Miller B. R., Forbes D. J. Purification of the vertebrate nuclear pore complex by biochemical criteria. Traffic. 2000;1:941–951. [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Maddox P., Groen A., Cameron L., Perlman Z., Ohi R., Desai A., Salmon E. D., Kapoor T. M. Bipolarization and poleward flux correlate during Xenopus extract spindle assembly. Mol. Biol. Cell. 2004;15:5603–5615. doi: 10.1091/mbc.E04-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J., Hijikata M., Blobel G., Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., Brown G. W., Varela E., Hediger F., Gasser S. M., Krogan N. J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Shaikh S., Burke B., Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Siomi M. C., Siomi H., Michael W. M., Pollard V., Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp. Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- Newmeyer D. D., Forbes D. J. Nuclear import can be separated into distinct steps in vitro: nuclear pore binding and translocation. Cell. 1988;52:641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Newport J., Dunphy W. Characterization of the membrane binding and fusion events during nuclear envelope assembly using purified components. J. Cell Biol. 1992;116:295–306. doi: 10.1083/jcb.116.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Orjalo A. V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S. G., Fontoura B., Briggs S., Dasso M., Forbes D. J. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- Pollard V. W., Michael W. M., Nakielny S., Siomi M. C., Wang F., Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Powers M. A., Macaulay C., Masiarz F. R., Forbes D. J. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby B. B., Dasso M. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 2003;15:338–344. doi: 10.1016/s0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- Radu A., Blobel G., Moore M. S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Orjalo A. V., Shen Z., Briggs S., Forbes D. J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Ramos C., Harel A., Forbes D. J. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendes K. K., Rasala B. A., Forbes D. J. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol. Cell. Biol. 2008;28:1755–1769. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell. 2006;17:2646–2660. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., McCaffery J. M., Wente S. R. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Wente S. R. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 2002;3:17. doi: 10.1186/1471-2156-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Zhou Y., Wente S. R. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Cooke C. A., Burgess W. H., Earnshaw W. C., Dasso M. Direct and indirect association of the small GTPase ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevis egg extracts. Mol. Biol. Cell. 1996;7:1319–1334. doi: 10.1091/mbc.7.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz C. A., Santarella R., Hoenger A., Karsenti E., Mattaj I. W., Gruss O. J., Carazo-Salas R. E. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Forbes D. J. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich S., Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D. K., Vann L. R., Goldberg M. W., Allen T. D., Wilson K. L. TPEN, a Zn2+/Fe2+ chelator with low affinity for Ca2+, inhibits lamin assembly, destabilizes nuclear architecture and may independently protect nuclei from apoptosis in vitro. Cell Calcium. 1998;23:151–164. doi: 10.1016/s0143-4160(98)90114-2. [DOI] [PubMed] [Google Scholar]
- Sillje H. H., Nagel S., Korner R., Nigg E. A. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S., Lutzmann M., Santos-Rosa H., Leonard K., Mueller S., Aebi U., Hurt E. Structure and assembly of the Nup84p complex. J. Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E. C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Siomi M. C., Eder P. S., Kataoka N., Wan L., Liu Q., Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Fromont M., Rain J. C., Wan L., Wang F., Legrain P., Dreyfuss G. Functional conservation of the transportin nuclear import pathway in divergent organisms. Mol. Cell. Biol. 1998;18:4141–4148. doi: 10.1128/mcb.18.7.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C., Newport J. W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Stoffler D., Feja B., Fahrenkrog B., Walz J., Typke D., Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J. Mol. Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Suel K. E., Gu H., Chook Y. M. Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 2008;6:e137. doi: 10.1371/journal.pbio.0060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S. M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tahara K., et al. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J. Cell Biol. 2008;180:493–506. doi: 10.1083/jcb.200708003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Shows E. B., Wente S. R. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Trieselmann N., Armstrong S., Rauw J., Wilde A. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 2003;116:4791–4798. doi: 10.1242/jcs.00798. [DOI] [PubMed] [Google Scholar]
- Truant R., Fridell R. A., Benson R. E., Bogerd H., Cullen B. R. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol. Cell. Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutter A. V., Walter J. C. Chromosomal DNA replication in a soluble cell-free system derived from Xenopus eggs. Methods Mol. Biol. 2006;322:121–137. doi: 10.1007/978-1-59745-000-3_9. [DOI] [PubMed] [Google Scholar]
- Ullman K. S., Forbes D. J. RNA polymerase III transcription in synthetic nuclei assembled in vitro from defined DNA templates. Mol. Cell. Biol. 1995;15:4873–4883. doi: 10.1128/mcb.15.9.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003a;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I. W., Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003b;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Fornerod M., Pickersgill H., Goldberg M., Allen T. D., Mattaj I. W. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wiese C., Goldberg M. W., Allen T. D., Wilson K. L. Nuclear envelope assembly in Xenopus extracts visualized by scanning EM reveals a transport-dependent ‘envelope smoothing’ event. J. Cell Sci. 1997;110(Pt 13):1489–1502. doi: 10.1242/jcs.110.13.1489. [DOI] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wu J., Matunis M. J., Kraemer D., Blobel G., Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yang L., Guan T., Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen N. R., Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc. Natl. Acad. Sci. USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N., et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Zuccolo M., et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.