Abstract
Recent studies indicate that regulation of cellular oxidative capacity through enhancing mitochondrial biogenesis may be beneficial for neuronal recovery and survival in human neurodegenerative disorders. The peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) has been shown to be a master regulator of mitochondrial biogenesis and cellular energy metabolism in muscle and liver. The aim of our study was to establish whether PGC-1α and PGC-1β control mitochondrial density also in neurons and if these coactivators could be up-regulated by deacetylation. The results demonstrate that PGC-1α and PGC-1β control mitochondrial capacity in an additive and independent manner. This effect was observed in all studied subtypes of neurons, in cortical, midbrain, and cerebellar granule neurons. We also observed that endogenous neuronal PGC-1α but not PGC-1β could be activated through its repressor domain by suppressing it. Results demonstrate also that overexpression of SIRT1 deacetylase or suppression of GCN5 acetyltransferase activates transcriptional activity of PGC-1α in neurons and increases mitochondrial density. These effects were mediated exclusively via PGC-1α, since overexpression of SIRT1 or suppression of GCN5 was ineffective where PGC-1α was suppressed by short hairpin RNA. Moreover, the results demonstrate that overexpression of PGC-1β or PGC-1α or activation of the latter by SIRT1 protected neurons from mutant α-synuclein- or mutant huntingtin-induced mitochondrial loss. These evidences demonstrate that activation or overexpression of the PGC-1 family of coactivators could be used to compensate for neuronal mitochondrial loss and suggest that therapeutic agents activating PGC-1 would be valuable for treating neurodegenerative diseases in which mitochondrial dysfunction and oxidative damage play an important pathogenic role.
Previous studies have shown that the PGC-1 family of coactivators, particularly PGC-1α, are potent stimulators of mitochondrial respiration and gene transcription in liver, heart, and skeletal muscle. It has been shown that PGC-1α acts by activating the nuclear respiratory factors NRF1 and NRF2 that in turn regulate expression of Tfam (mitochondrial transcription factor A), essential for replication, maintenance, and transcription of mitochondrial DNA. PGC-1α is also important for the expression of nuclear genes encoding respiratory chain subunits and other proteins that are required for proper mitochondrial functions (1–4).
Apart from gene expression, the activity of PGC-1α is influenced by posttranscriptional regulation by means of protein phosphorylation, acetylation, and methylation. PGC-1α is known to be regulated by p38 mitogen-activated protein kinase through the inhibition of the p160 Myb-binding protein (p160MBP)2 in brown fat cells and myotubes (5, 6). AMPK (AMP-activated protein kinase) phosphorylation of PGC-1α initiates many of the important gene-regulatory functions of AMPK in skeletal muscle (7). Acetylation status of PGC-1α is, on the other hand, regulated by the balanced action of the GCN5 acetyltransferase and the NAD+-dependent SIRT1 deacetylase. Activation of PGC-1α by deacetylation via SIRT1 has been shown to mediate the effects of PGC-1α on liver, fat, and muscle metabolism as well as on mitochondrial activity (8). The SIRT1 activator, resveratrol (3,4′,5-trihydroxystilbene), which is produced by different plants and is a constituent of red wine, has been shown to increase the activity of PGC-1α and to improve mitochondrial activity (9, 10). Also, it has been shown to have positive effects in the heart and in muscle metabolism, protecting against adverse effects of cell stress, metabolic disturbances, and high caloric intake (11, 12).
In contrast to the wealth of data on PGC-1 family factors in peripheral tissues, less is known about the role of these factors in brain plasticity and in neurodegeneration. Recent studies on PGC-1α-deficient mice show that the lack of PGC-1-mediated signaling led to neurodegeneration in parts of the brain. In addition, the PGC-1α knock-out mice showed an increased sensitivity to oxidative stressors, such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine and kainic acid, that induce degeneration of dopaminergic and glutamatergic neurons, respectively, in the brain (13). The aim of the current work was therefore to clarify whether and to what extent the PGC-1 coactivators could regulate mitochondrial capacity also in neurons and whether their posttranslational activation could be used to compensate for mitochondrial loss.
EXPERIMENTAL PROCEDURES
Primary Neuronal Culture
Primary cultures of rat cortical cells were prepared from neonatal Wistar rats. Briefly, cortices were dissected in ice-cold Krebs-Ringer solution (135 mm NaCl, 5 mm KCl, 1 mm MgSO4, 0.4 mm K2HPO2, 15 mm glucose, 20 mm HEPES, pH 7.4) containing 0.3% bovine serum albumin and trypsinized in 0.8% trypsin for 10 min at 37 °C. This was followed by trituration in a 0.008% DNase solution containing 0.05% soybean trypsin inhibitor. Neurons were resuspended in basal medium Eagle with Earle's salts (BME) containing 10% heat-inactivated fetal bovine serum, 25 mm KCl, 2 mm glutamine, and 100 μg/ml gentamicin and plated onto 35-mm glass bottom dishes (glass surface diameter 14 mm; MatTek, Ashland, MA) precoated with poly-l-lysine at a density of 106 cells/ml (2 ml of cell suspension/dish) or 170 μl/well into white microwell plates (96 F Nuclon Delta, Nunc; Thermo Fisher Scientific). 3 h later, medium was changed to NeurobasalTM-A medium containing B-27 supplement, 2 mm GlutaMAXTM-I, and 100 μg/ml gentamicin.
For preparation of primary cultures of cerebellar granule cells, the cerebelli from 8-day-old Wistar rats were dissociated by trypsinization in 0.25% trypsin at 35 °C for 15 min, followed by trituration in a 0.004% DNase solution containing 0.05% soybean trypsin inhibitor. Cells were resuspended in BME containing 10% fetal bovine serum, 25 mm KCl, 2 mm glutamine, and 100 μg/ml gentamicin. Neurons were plated onto 35-mm glass bottom dishes precoated with poly-l-lysine at a density of 1.3 × 106 cells/ml. 10 μm cytosine arabinoside was added 24 h after plating to prevent the proliferation of glial cells.
Primary cultures of mesencephalic neurons were prepared from mesencephali of mouse embryos at embryonic day 15. Midbrain anlages were dissected, cleaned from meninges, triturated mechanically through a 1-ml pipette tip in BME containing 10% fetal bovine serum, and plated as droplets containing 0.2 × 105 cells directly on poly-l-lysine-precoated glass bottom dishes. 1 h after plating, 2 ml of NeurobasalTM-B medium containing B-27 supplement, 2 mm GlutaMAXTM-I, and 100 μg/ml gentamicin was added. All of the cultures were grown in a humidified 5% CO2, 95% air incubator at 37 °C.
Plasmids
Plasmids overexpressing PGC-1α (Addgene plasmid 10974), Gal4-PGC-1α (plasmid 8892), PGC-1β (plasmid 1031), p160MBP (plasmid 41), p160C (plasmid 46), p67MBP (plasmid 42), SIRT1 (plasmid 10962), SIRT1 G261A (plasmid 10963), or PGC-1α promoter-luciferase (plasmid 8887) were from ADDGENE (Cambridge, MA). GFP and mitochondrial pDsRED2 were from Clontech. Plasmids expressing short hairpin RNA-s against PGC-1α, PGC-1β, and GCN5 were from SABiosciences (Frederick, MD). Plasmids expressing dominant negative or constitutively active AMPK isoforms were generous gifts from Dr. D. Carling (14). Luciferase reporter constructs of murine COX8a and COX6c promoters (15) were gifts from Dr. M. T. Wong-Riley, and luciferase reporter constructs of mitofusion promoter were from Dr. A. Zorzano (16). 120Q Htt-overexpressing plasmid (17) was a gift from Dr. L. Hasholt, and A53T α-synuclein-expressing plasmid was from Dr. S. Kõks. Expression vectors were confirmed in HEK293 cells using Western blot. shRNA-encoding plasmids were validated using reverse transcription-PCR according to the manufacturer's protocols (supplemental Fig. S1).
Transfection
Cultures were transiently transfected on the second day (on the first day in the case of cerebellar granule cells) in vitro with LipofectamineTM 2000. Briefly, conditioned medium was collected, and 100 μl of Opti-MEM® I medium containing 2% LipofectamineTM 2000 and 1–2 μg of total DNA was added directly to the cells on the glass bottom surface of dishes and incubated for 3–4 h in a humidified 5% CO2, 95% air incubator. In multiwell plates, 50 μl of Opti-MEM® I medium with 2% LipofectamineTM 2000 and 0.6–0.7 μg of DNA were added to each well. For primary cerebellar granule cells, conditioned medium was added at the completion of incubation, and for cortical and mesencephalic cultures conditioned medium was mixed 1:1 with fresh NeurobasalTM medium. The transfected DNAs were allowed to express and accumulate respective proteins, and experiments were performed on the fifth day in vitro unless specified otherwise. Controls were transfected as treated groups with an equal amount of non-interfering DNA. All of the culture media, supplements, and transfection reagents were obtained from Invitrogen.
Image Acquisition and Analysis
For mitochondrial density measurements in axons, the neuronal cultures were transfected with GFP, mitochondrial pDsRed2, and plasmids of interest. 3 days later, 10 fluorescence images were randomly captured from each dish using an Olympus IX70 inverted microscope equipped with WLSM PlanApo ×40/0.90 water immersion objective and Olympus DP70 CCD camera. Morphometric analysis was performed using MicroImage software (Media Cybernetics, Bethesda, MD). For mitochondrial density (mitochondrial length/axonal length), at least 40 axons per group were analyzed (1 axon/image).
For mitochondrial density measurements in cell body, the neuronal cultures were transfected with GFP, mitochondrial pDsRed2, and plasmids of interest. 3 days later, 512 × 512-pixel sections were acquired by an LSM 510 META Zeiss confocal microscope equipped with a Plan-Apo ×100/1.4 oil immersion objective. Voxels were collected at 43–74-nm lateral and 100-nm axial intervals. Raw images were three-dimensionally deconvolved and reconstructed by the AutoDeblur and Autovisualize X software package (Media Cybernetics). Reconstructed images were then subjected to stereologic analysis using the Computer-assisted Stereology Toolbox software (Olympus, Ballerup, Denmark). The mitochondrial volume as well as the volume of cell body was estimated using the Cavalieri principle. A grid of points was superimposed on the images, after which the points that overlaid the fluorescent signal were counted. Mitochondrial density in cell body was calculated by dividing the mitochondrial volume with the volume of cell body. Seven or eight cell bodies were analyzed in each group.
Estimation of Autophagy
For autophagy measurements, the cortical cultures were transfected with EGFP-LC3, mitochondrial pDsRed2, and plasmids of interest. 3 days later, fluorescence images were randomly captured using LSM 510 META Zeiss using CI Plan-Neofluar ×63/1.33 water immersion objective. The temperature was maintained at 37 °C using a climate chamber. Distribution and colocalization of GFP-LC3 punctates (for autophagosome visualization) and pDsRed2 (for mitochondrial visualization) were analyzed using MicroImage software. Altogether, 42 axons/group were analyzed (1 axon/image).
Luciferase Reporter Gene Assays
For luciferase assays, cortical neurons were transiently transfected on the second day after plating, as described above, using 0.6–0.7 μg of total DNAs in the desired combination of luciferase reporter plasmids or Gal4 fusions as well as SIRT1 or p160MBP and p67MBP plasmids. All of these transfections included 0.1 μg of Renilla luciferase (pRL-CMV) plasmid. Luciferase assays were performed 24 h after transfection using Dual-Glo Luciferase Assay reagent (Promega, Madison, WI) according to the manufacturer's instructions. The promoter activity of COX subunits (COX6c and COX8a) and PGC-1α as well as posttranscriptional activation of PGC-1α was determined as relative firefly luciferase luminescence normalized to Renilla luciferase signal measured by a MicroBeta® TriLux luminescence counter (PerkinElmer Life Sciences). In all luminescence experiments, eight independent samples were analyzed.
ATP Levels
Cortical neurons were transfected with plasmids expressing firefly luciferase, Renilla luciferase, and plasmids of interest. For measurement of firefly luciferase activity, we incubated cells for 10 min with 25 μm 1-(4,5-dimethoxy-2-nitrophenyl)diazoethane-caged luciferin at 37 °C and measured luminescence by MicroBeta® TriLux. For measurement of Renilla luciferase activity, the neurons were lysed, and luminescence was measured using Dual-Glo luciferase assay reagent. Firefly luciferase activity from living cells normalized per Renilla luciferase activity from lysed cells was used as an estimate of ATP level in transfected cells. In all experiments, 16 independent samples were analyzed.
Statistics
One-way ANOVA followed by Newman-Keuls post hoc test or t test were used to compare the difference from the control group, and two-way ANOVA was used to analyze the interaction between treatments.
RESULTS
PGC-1α and PGC-1β Control Mitochondrial Density
Overexpression of PGC-1α in cortical neurons led to a 13% increase in mitochondrial density in axons, and PGC-1α suppression by shRNAs decreased mitochondrial density up to 17% (Fig. 1, A–C). Similarly, overexpression of PGC-1β increased mitochondrial density in axons 19%, and PGC-1β suppression by shRNAs decreased mitochondrial density up to 25% (Fig. 1D). Similar trends were observed also in midbrain and cerebellar granule neurons (supplemental Fig. S2). Fig. 1, E–H, demonstrates that PGC-1α and PGC-1β control mitochondrial density also in the neuronal body; suppression of either PGC-1α or PGC-1β decreased significantly mitochondrial mass in the cortical neuron body, whereas overexpression of PGC-1α and PGC-1β did not increase mitochondrial density over control.
FIGURE 1.
Effects of PGC-1 α and PGC-1 β on mitochondrial density in primary cortical neurons. Cortical neurons were transfected with plasmids expressing wild type PGC-1 coactivators or their shRNAs, GFP, and mitochondria-targeted pDsRED2 at day 2 after plating. A (expressing wild type PGC-1α) and B (expressing shRNA suppressing PGC-1α) demonstrate substantial differences in axonal mitochondrial density 3 days later. Further morphometric analysis demonstrates that axonal mitochondrial density is controlled both by PGC-1α (C) and PGC-1β (D). To estimate mitochondrial density in the neuron's body, we performed three-dimensional scan with a confocal laser microscope, followed by three-dimensional reconstruction of mitochondrial network in neuronal body. E (expressing wild type PGC-1α) and F (expressing shRNA suppressing PGC-1α) demonstrate also a clear difference in mitochondrial density in the neuronal body. Stereological analysis of three-dimensional reconstructed mitochondrial networks demonstrates a decrease of density in response of PGC-1α (G) and PGC-1β (H) suppression. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control (one-way ANOVA followed by Newman-Keuls post hoc test).
Observed changes were accompanied by changes in expressional level; overexpression of PGC-1α and PGC-1β activated transcription of downstream reporter genes containing promoter areas from nuclear mitochondrial genes known to be activated by nuclear respiratory factor-1. Luciferase reporter constructs of murine COX8a or COX6c promoters (15) were cotransfected with PGC-1α and PGC-1β expression plasmids. Luciferase activity performed 24 h later demonstrated (Fig. 2, A and B) that overexpression of PGC-1α and PGC-1β increase relative luminescence, suggesting increased expression of COX8a and COX6c fusion proteins. PGC-1α overexpression led also to an 85 ± 8% expression increase of luciferase reporter construct of the mitofusin-2 promoter known to be directly regulated by PGC1α (16).
FIGURE 2.
Effects of PGC-1 α and PGC-1 β on the COX8a and COX6c promoter activity. Cortical neurons were cotransfected with plasmids expressing COX8a or COX6c promoter-luciferase reporter, Renilla luciferase, and PGC-1α or PGC-1β constructs at day 1–2 after plating. Overexpression of PGC-1α (A) or PGC-1β (B) led to increased luciferase activity performed 24 h later, suggesting increased expression of these reporter genes. Results are normalized to Renilla luciferase signal (**, p < 0.01; ***, p < 0.001 versus control, t test).
Next, we estimated the relative ATP levels in PGC-1α- and PGC-1β-overexpressing neurons. Neurons were cotransfected with firefly and Renilla luciferase-expressing plasmids, and firefly luciferase activity was measured 24 h later in living cells using 1-(4,5-dimethoxy-2-nitrophenyl)diazoethane-caged luciferin as a substrate. Results normalized to Renilla luciferase activity measured after the lysis of cells demonstrate a 31 and 39% increase in PGC-1α- or PGC-1β-overexpressing neurons, respectively (Fig. 3).
FIGURE 3.
Effects of PGC-1 α and PGC-1 β on the ATP level. Cortical neurons were cotransfected with plasmids expressing firefly luciferase, Renilla luciferase and PGC-1α or PGC-1β at day 2 after plating. Firefly luciferase activity was measured 24 h later, using 1-(4,5-dimethoxy-2-nitrophenyl)diazoethane-caged luciferin as a substrate, after which the neurons were lysed and Renilla luciferase was measured. Overexpression of PGC-1α and PGC-1β led to an increased firefly to Renilla luciferase activity ratio. **, p < 0.01; ***, p < 0.001 versus control (one-way ANOVA followed by Newman-Keuls post hoc test).
We also tested the possibility that overexpression of PGC-1α and PGC-1β increases mitochondrial density via decreasing mitochondrial degradation by autophagy. PGC-1α or PGC-1β was cotransfected with EGFP-LC3 plus mitochondrial pDsRED2, and the number of autophagosomes per axonal segment was estimated. However, there were no major changes in macroautophagy or in mitochondrial removal by macroautophagy in PGC-1-overexpressing neurons (Fig. 4).
FIGURE 4.
Effects of PGC-1 α and PGC-1 β on the autophagy. Cortical neurons were transfected with plasmids expressing EGFP-LC3, mitochondria-targeted pDsRed2, and PGC-1α or PGC-1β at day 2 after plating. 3 days later, the LC3-positive dots were visualized. A, EGFP-LC3-expressing autophagosome (left), mitochondria-targeted pDsRed2 expressing mitochondria (middle), and superimposed channels to demonstrate colocalization of signals (right and below). The number of autophagosomes (B and D) as well as the number of mitochondria engulfed by autophagosomes per mm of axonal length (C and E) was unchanged in PGC-1α- or PGC-1β-overexpressing neurons. Rapamycin used as a positive control increased the number of LC3-positive mitochondria.
PGC-1α and PGC-1β control mitochondrial density also in other neuron types. Overexpression of PGC-1α and PGC-1β in cerebellar neurons increased mitochondrial density 30% (p < 0.001) and 20% (p < 0.001), respectively, whereas suppression of PGC-1α and PGC-1β led to a 20% (p < 0.01) or 34% (p < 0.001) decrease, respectively. Overexpression of PGC-1α and PGC-1β in midbrain neurons increased mitochondrial density 19% (p < 0.001) and 17% (p < 0.001), respectively, whereas suppression of PGC-1α and PGC-1β led to a 17% (p < 0.001) or 14% (p < 0.01) decrease, respectively.
It should also be noted that the effects of PGC-1α and PGC-1β were additive and independent. In these experiments, PGC-1α increased mitochondrial density in cortical neurons 16% and PGC-1β 19%, but when overexpressed together, the mitochondrial density increased 30% (p = 0.53 for interaction). Similarly, suppression of PGC-1α and PGC-1β by the respective shRNAs was additive and independent; suppression of PGC-1α decreased mitochondrial density 29%, suppression of PGC-1β 23%, and both together 51% (p = 0.77 for interaction) (supplemental Fig. S3).
Overexpression of SIRT1 and Suppression of GCN5 Increase Mitochondrial Density
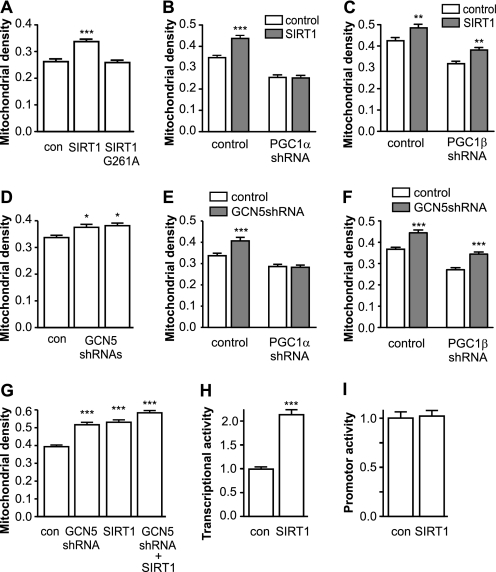
Fig. 5A demonstrates that overexpression of wild type SIRT1 increased mitochondrial density in cortical neurons up to 30%, whereas overexpression of inactive G261A-mutated SIRT1 had no effect. Fig. 5, B and C, demonstrates that this effect was mediated exclusively by PGC-1α, since the positive effect of SIRT1 on mitochondrial density disappeared in the absence of PGC-1α (p = 0.0002 for interaction) but not in the absence of PGC-1β (p = 0.89 for interaction). A similar effect was observed when suppressing GCN5 acetyltransferase. Fig. 5D demonstrates that GCN5 suppression by specific short hairpin RNA-s leads to significant increase in mitochondrial density. Similarly to SIRT1, this effect disappeared when PGC-1α was suppressed (p = 0.0033 for interaction; Fig. 5E) but sustained when PGC-1β was suppressed (p = 0.86 for interaction; Fig. 5F). It should be noted that the effects of SIRT1 overexpression and GCN5 suppression were dependent (p = 0.0084 for interaction; Fig. 5G).
FIGURE 5.
Effects of SIRT1 and GCN5 shRNA. A–G, cortical neurons were transfected with plasmids expressing GFP and mitochondrial pDsRED2 and with different plasmids interfering with PGC-1 at day 2 after plating. A demonstrates that overexpression of SIRT1 but not its inactive mutant, SIRT G261A, increases mitochondrial density. Effect of SIRT1 disappeared in the presence of shRNA suppressing PGC-1α (B) (p < 0.001 for interaction, two-way ANOVA) but not PGC-1β (C) (p = 0.89). D demonstrates the positive effects of GCN5 suppressing shRNA that disappeared in the presence of PGC-1α shRNA (E) (p = 0.003) but not in the presence of PGC-1β shRNAs (F) (p = 0.009). G demonstrates that although combination of SIRT1 and GCN5 shRNA increased mitochondrial density more that each treatment separately; both treatments were still dependent (p = 0.008 for interaction). H and I, cortical neurons were cotransfected with Gal4-PGC-1α fusion protein, GAL4-UAS-luciferase reporter, and SIRT1 or with PGC-1α promoter reporter and SIRT1 plasmids. H demonstrates that overexpression of SIRT1 increases PGC-1α transcriptional activity whereas the activity of PGC-1α promoter reporter (I) remains unchanged (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control; one-way ANOVA followed by Newman-Keuls post hoc test was used to compare the difference from control group, and two-way ANOVA was used to analyze the interaction between treatments).
Our next step was to find out whether or not the effect of SIRT1 was related with increased transcriptional activity of PGC-1α or its expression. Cortical neurons were cotransfected with a fusion protein connecting the yeast Gal4 DNA binding domain with full-length PGC-1α, along with an appropriate Gal4-UAS-luciferase reporter, and SIRT1. Luciferase assays were conducted 24 h later and revealed that SIRT1 increased Gal4-PGC1α transcriptional activity more than 2-fold (Fig. 5H). In another set of experiments cortical neurons were cotransfected with reporter gene plasmid containing mouse PGC-1α promoter area and SIRT1. Luciferase activity performed 24 h later, showed no difference between control and SIRT1-overexpressing cortical neurons (Fig. 5I).
p160 Myb-binding Protein Represses PGC-1α Activity but Independently of SIRT1
We next tested whether the effect of SIRT1 could be related to the PGC-1α repressor, p160 Myb-binding protein (p160MBP), which interacts with the negative regulatory domain of PGC-1α and reduces its transcriptional activity. Fig. 6A demonstrates that suppression of p160MBP leads to a considerable, 20% increase in mitochondrial density. On the other hand, overexpression of p160MBP or its N-terminal fragment of 67-kDa p67MBP suppressed mitochondrial density about 20%. In experiments performed with an N-terminal deletion mutant of p160MBP (which expresses amino acids 580–1330), no effect on mitochondrial density was observed. Experiments also demonstrate that p160MBP suppresses PGC-1α transcriptional activity but not expression. Cortical neurons were cotransfected with Gal4-PGC1α, Gal4-UAS-luciferase reporter, and p160MBP or p67MBP. Luciferase assays revealed that p160MBP or p67MBP suppressed Gal4-PGC1α transcriptional activity around 25% (Fig. 6B). At the same time, no effect of p160MBP or p67MBP on reporter gene plasmid containing mouse PGC-1α promoter area was observed (Fig. 6C). The effects of p160MBP were mediated only by PGC-1α; effects of p160MBP suppression/overexpression disappeared when PGC-1α was suppressed by shRNA against PGC-1α (Fig. 6D). The experiments depicted in Fig. 6E demonstrate also that effects of p160MBP and SIRT1 were additive and independent. Another set of experiments demonstrated that the effects of SIRT1 were also independent from AMPK, since the effect of SIRT was present also in neurons expressing negative dominant forms of AMPK1 and -2 (p = 0.54 for interaction).
FIGURE 6.
Effect of p160MBP. A demonstrates that overexpression of p160MBP and p67MBP but not the deletion mutant of p160MBP decreases mitochondrial density, whereas p160MBP shRNA increases it. Experiments with Gal4-PGC-1α (B) and PGC-1 promoter reporter (C) confirm that p160MBP and p67MBP regulate only PGC-1α transcriptional activity and not the expression. Effects of p160MBP overexpression/suppression disappeared in the presence of shRNA suppressing PGC-1α (D) (p < 0.011 for interaction, two-way ANOVA). Effects of p160MBP overexpression/suppression and SIRT1 were additive (E) (p = 0.45 for interaction). (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control; one-way ANOVA followed by Newman-Keuls post hoc test).
PGC-1α, PGC-1β, and SIRT1 Overexpression and GCN5 Suppression Protect A53T α-Synuclein or 120Q Huntingtin-overexpressing Cortical Neurons from Mitochondrial Loss
Our next aim was to see whether PGC-1α and PGC-1β could reverse mitochondrial loss observed in models of Huntington and Parkinson disease. Both overexpression of mutant A53T α-synuclein and 120Q huntingtin in cortical neurons induced considerable, 15–25% mitochondrial loss. However, when A53T α-synuclein or 120Q huntingtin was cotransfected with PGC-1α or PGC-1β (Fig. 7, A and B), no mitochondrial loss was observed. Similarly, overexpression of SIRT1 or suppression of GCN5 protected A53T α-synuclein- and 120Q huntingtin-overexpressing cortical neurons from mitochondrial loss (Fig. 7, C and D). The protective effects of SIRT1 and GCN5 suppression were mediated by PGC-1α, since no protective effects were observed in an experiment where PGC-1α was suppressed by respective shRNA (data not shown).
FIGURE 7.
PGC-1 α, PGC-1 β, and SIRT1 overexpression and GCN5 suppression protect against mitochondrial loss. Cortical neurons were transfected with plasmids expressing A53T-mutated α-synuclein or 120Q huntingtin, GFP, or mitochondrial pDsRED2 and with different plasmids modulating expression or activity of PGC-1 coactivators. A and B demonstrate that PGC-1α and PGC-1β protect against A53T-mutated α-synuclein or 120Q huntingtin-induced mitochondrial loss, respectively. C and D demonstrate that also SIRT1 and GCN5 shRNA protect against A53T-mutated α-synuclein or 120Q huntingtin-induced mitochondrial loss, respectively. E and F demonstrate that overexpression of PGC-1α and SIRT1 protect also against A53T α-synuclein or 120Q huntingtin-induced neuronal death, respectively (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control; one-way ANOVA followed by Newman-Keuls post hoc test).
PGC-1α or SIRT1 Overexpression Protect A53T α-Synuclein or 120Q Huntingtin-overexpressing Cortical Neurons from Degeneration
In final experiments, we tested whether overexpression of PGC-1α or SIRT1 could protect the neurons against A53T α-synuclein- or 120Q huntingtin-induced degeneration. Fig. 7, E and F, demonstrates that both PGC1α and SIRT1 increased the number of surviving neurons in A53T α- synuclein- or 120Q huntingtin-transfected cultures.
DISCUSSION
This work reports the first evidence that the PGC-1 family of coactivators (both PGC-1α and PGC-1β) controls mitochondrial density in primary neurons. This effect was observed in all studied subtypes of neurons, in cortical, midbrain, and cerebellar granule neurons. Consistent with findings in non-neuronal cells (18), we observed that PGC-1α and PGC-1β control mitochondrial capacity in an additive and independent manner.
We also observed that endogenous neuronal PGC-1α but not PGC-1β could be activated through its repressor domain (amino acids 200–400). In non-neuronal cells, this domain is suppressed by p160MBP, and it has been suggested that undocking of p160MBP from PGC-1α leads to the activation of the latter (6). As an example, p38 mitogen-activated protein kinase phosphorylates PGC-1α at three residues in the repression domain (19), leading to the inability of the phospho-PGC-1α repressor domain to bind the p160MBP. Results presented here indicate that PGC-1α is suppressed by p160MBP or its N-terminal fragment p67MBP, also in neurons. Moreover, PGC-1α seems to be under the control of p160MBP since suppression of p160MBP by shRNA led to considerable increase in mitochondrial density. This effect of p160MBP on mitochondrial density was exclusively mediated via PGC-1α. It has been also demonstrated that the repressor domain of PGC-1α interacts with SIRT1, shown to deacetylate PGC-1α in at least 13 lysines in different domains of the protein (4). The present work demonstrates that SIRT1 activates transcriptional activity of PGC-1α in neurons and thus increases the mitochondrial density. A similar effect was observed when we suppressed GCN5 acetyltransferase. Effects of SIRT1 overexpression and GCN5 suppression were dependent, suggesting that deacetylation rather than some other effects of SIRT1 or GCN5 suppression is responsible for increased mitochondrial density. Moreover, these effects were mediated exclusively via PGC-1α, since overexpression of SIRT1 or suppression of GCN5 was ineffective where PGC-1α was suppressed by shRNA. We also tested whether the effect of SIRT1 could be related with undocking of p160MBP or p67MBP or with AMPK but found no interaction. These experiments suggest the effect of SIRT overexpression to be direct rather than working through the p160MBP/p38 mitogen-activated protein kinase or AMPK kinase pathways, as suggested for 293T cells (20, 21). Again, these results support the earlier results by Puigserver et al. (19) showing that SIRT1 is activating (4) and GCN5 is inactivating (22) the PGC-1α in non-neuronal cells.
Taken together, this evidence suggests that activation or overexpression of PGC-1α or PGC-1β could be used to compensate for neuronal mitochondrial loss. Mitochondrial deficit is one of the main features of all major age-related neurodegenerative diseases (reviewed recently by Reeve et al. (23)). Lines of evidence suggest a pathogenic role of oxidative damage and mitochondrial dysfunction in causing Parkinson disease. α-Synuclein-transfected cells show a significant 30% decrease in mitochondrial activity (24), and transgenic mice expressing A53T α-synuclein develop mitochondrial pathology (25, 26). Consistent deficits in the subunits and activity of mitochondrial complex I of the electron transport chain in blood platelets and in substantia nigra of Parkinson disease patients is a prominent phenomenon (27). Here we show that overexpression of PGC-1α or PGC-1β reverses fully the mitochondrial loss induced by A53T α-synuclein. Similarly, overexpression of SIRT1 or suppression of GCN5 protected neurons from A53T α-synuclein-induced mitochondrial loss. The protective effect of SIRT1 or suppression of GCN5 disappeared when PGC-1α was suppressed by specific shRNAs, suggesting that such a protective effect was exclusively mediated by PGC-1α.
Also, Huntington disease patients exhibit well described metabolic defects (28, 29) that have been linked to mitochondrial dysfunction (reviewed in Refs. 30 and 31). Cui et al. (32) demonstrated that mutant huntingtin causes disruption of mitochondrial function by inhibiting expression of PGC-1α. Mutant huntingtin repressed PGC-1α transcription by associating with the promoter and interfering with the CREB/TAF4-dependent transcriptional pathway critical for the regulation of PGC-1α gene expression. They also demonstrated that lentivirus-mediated delivery of PGC-1α in the striatum provided neuroprotection in the transgenic Huntington disease mice. Here we demonstrate that also overexpression of SIRT1 or suppression of GCN5 protected neurons from mutant huntingtin-induced mitochondrial loss in primary cortical neurons.
Taken together, our results demonstrate that PGC-1α and PGC-1β control mitochondrial capacity in neurons and that neuronal PGC-1α could be activated by SIRT1 deacetylase or suppressing GCN5 acetyltransferase. Moreover, all of these treatments protected neurons from mutant α-synuclein- or mutant huntingtin-induced mitochondrial loss. These results support the view that therapeutic agents activating the PGC-1 family of coactivators would be valuable for treating Huntington disease and Parkinson disease as well as other neurodegenerative diseases in which mitochondrial dysfunction and oxidative damage play an important pathogenic role, such as Alzheimer disease and amyotrophic lateral sclerosis (33). The next step of that work should thus be to find pharmacological activators for activation of PGC-1 family coactivators.
Supplementary Material
Acknowledgments
We thank Dr. A. Zharkovsky for continuous support and Dr. Carling, Dr. L. Hasholt, Dr. S. Kõks, Dr. A. Zorzano, and Dr. M. T. Wong-Riley for providing the plasmids used in this study.
This work was supported by Estonian Science Foundation Grants 6227 and 7175, European Community Projects BRAIN BIOENERGETICS and ESTBIOREG, and the European Regional Development Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- MBP
- Myb-binding protein
- BME
- basal medium Eagle with Earle's salts
- GFP
- green fluorescent protein
- ANOVA
- analysis of variance
- shRNA
- short hairpin RNA
- EGFP
- enhanced GFP
- AMPK
- AMP-activated protein kinase.
REFERENCES
- 1.Finck B. N., Kelly D. P. (2006) J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handschin C., Spiegelman B. M. (2006) Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 3.Feige J. N., Auwerx J. (2007) Trends Cell Biol. 17, 292–301 [DOI] [PubMed] [Google Scholar]
- 4.Rodgers J. T., Puigserver P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tcherepanova I., Puigserver P., Norris J. D., Spiegelman B. M., McDonnell D. P. (2000) J. Biol. Chem. 275, 16302–16308 [DOI] [PubMed] [Google Scholar]
- 6.Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J., Jäeger S., Erdjument-Bromage H., Tempst P., Spiegelman B. M. (2004) Genes Dev. 18, 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers J. T., Lerin C., Gerhart-Hines Z., Puigserver P. (2008) FEBS Lett. 582, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 10.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 11.Baur J. A., Sinclair D. A. (2006) Nat. Rev. Drug Discov. 5, 493–506 [DOI] [PubMed] [Google Scholar]
- 12.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 13.St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., Spiegelman B. M. (2006) Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- 14.Woods A., Azzout-Marniche D., Foretz M., Stein S. C., Lemarchand P., Ferré P., Foufelle F., Carling D. (2000) Mol. Cell. Biol. 20, 6704–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhar S. S., Ongwijitwat S., Wong-Riley M. T. (2008) J. Biol. Chem. 283, 3120–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soriano F. X., Liesa M., Bach D., Chan D. C., Palacín M., Zorzano A. (2006) Diabetes 55, 1783–1791 [DOI] [PubMed] [Google Scholar]
- 17.Hasholt L., Abell K., Nørremølle A., Nellemann C., Fenger K., Sørensen S. A. (2003) J. Gene Med. 5, 528–538 [DOI] [PubMed] [Google Scholar]
- 18.Srivastava S., Barrett J. N., Moraes C. T. (2007) Hum. Mol. Genet. 16, 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J. C., Zhang C. Y., Krauss S., Mootha V. K., Lowell B. B., Spiegelman B. M. (2001) Mol Cell 8, 971–982 [DOI] [PubMed] [Google Scholar]
- 20.Lan F., Cacicedo J. M., Ruderman N., Ido Y. J. (2008) J. Biol. Chem. 283, 27628–27635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeve A. K., Krishnan K. J., Turnbull D. M. (2008) Biotechnol. J. 3, 750–756 [DOI] [PubMed] [Google Scholar]
- 24.Hsu L. J., Sagara Y., Arroyo A., Rockenstein E., Sisk A., Mallory M., Wong J., Takenouchi T., Hashimoto M., Masliah E. (2000) Am. J. Pathol. 157, 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stichel C. C., Zhu X. R., Bader V., Linnartz B., Schmidt S., Lübbert H. (2007) Hum. Mol. Genet. 16, 2377–2393 [DOI] [PubMed] [Google Scholar]
- 26.Martin L. J., Pan Y., Price A. C., Sterling W., Copeland N. G., Jenkins N. A., Price D. L., Lee M. K. (2006) J. Neurosci. 26, 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney P. M., Xie J., Capaldi R. A., Bennett J. P., Jr. (2006) J. Neurosci. 26, 5256–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leenders K. L., Frackowiak R. S., Quinn N., Marsden C. D. (1986) Mov. Disord. 1, 69–77 [DOI] [PubMed] [Google Scholar]
- 29.Jenkins B. G., Koroshetz W. J., Beal M. F., Rosen B. R. (1993) Neurology 43, 2689–2695 [DOI] [PubMed] [Google Scholar]
- 30.Lin M. T., Beal M. F. (2006) Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 31.Browne S. E., Beal M. F. (2004) Neurochem. Res. 29, 531–546 [DOI] [PubMed] [Google Scholar]
- 32.Cui L., Jeong H., Borovecki F., Parkhurst C. N., Tanese N., Krainc D. (2006) Cell 127, 59–69 [DOI] [PubMed] [Google Scholar]
- 33.McGill J. K., Beal M. F. (2006) Cell 127, 465–468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.