Abstract
Regulation of translation factor activity plays a major role in protein synthesis-dependent forms of synaptic plasticity. We examined translational control across the critical period of Arc synthesis underlying consolidation of long term potentiation (LTP) in the dentate gyrus of intact, anesthetized rats. LTP induction by high frequency stimulation (HFS) evoked phosphorylation of the cap-binding protein eukaryotic initiation factor 4E (eIF4E) and dephosphorylation of eIF2α on a protracted time course matching the time-window of Arc translation. Local infusion of the ERK inhibitor U0126 inhibited LTP maintenance and Arc protein expression, blocked changes in eIF4E and eIF2α phosphorylation state, and prevented initiation complex (eIF4F) formation. Surprisingly, inhibition of the mTOR protein complex 1 (mTORC1) with rapamycin did not impair LTP maintenance or Arc synthesis nor did it inhibit eIF4F formation or phosphorylation of eIF4E. Rapamycin nonetheless blocked mTOR signaling to p70 S6 kinase and ribosomal protein S6 and inhibited synthesis of components of the translational machinery. Using immunohistochemistry and in situ hybridization, we show that Arc protein expression depends on dual, ERK-dependent transcription and translation. Arc translation is selectively blocked by pharmacological inhibition of mitogen-activated protein kinase-interacting kinase (MNK), the kinase coupling ERK to eIF4E phosphorylation. Furthermore, MNK signaling was required for eIF4F formation. These results support a dominant role for ERK-MNK signaling in control of translational initiation and Arc synthesis during LTP consolidation in the dentate gyrus. In contrast, mTORC1 signaling is activated but nonessential for Arc synthesis and LTP. The work, thus, identifies translational control mechanisms uniquely tuned to Arc-dependent LTP consolidation in live rats.
Introduction
The adult mammalian brain is known to express diverse forms of activity-dependent synaptic plasticity (1, 2). Bursts of synaptic activity can induce short term changes in synaptic strength, but more stable modifications typically require modulation of gene expression at the transcriptional and post-transcriptional levels (3, 4). Through post-transcriptional regulation, synaptic activity may dictate the time and place of neuronal protein synthesis.
Regulated phosphorylation of translation factors and other ribosome-associated proteins is a major mechanism for controlling the activity of the translational machinery (5, 6). Translation control studies of LTP2 have concentrated mainly on the Schaffer-collateral input to hippocampal CA1 pyramidal cells. Studies employing knock-out mice and pharmacological inhibitors support a role for eukaryotic initiation factor 4E (eIF4E) and eIF2α in consolidation of LTP in the CA1 region and long term memory (7–10). The function of the cap-binding protein eIF4E during translational initiation is controlled by eIF4E-binding proteins (4E-BPs), which inhibit initiation complex (eIF4F) formation by competing with the scaffolding protein eIF4G for a shared binding site on eIF4E. Activation of the mammalian target of rapamycin protein complex 1 (mTORC1) facilitates initiation through phosphorylation and inhibition of 4E-BP. Phosphorylation of eIF4E on Ser209 requires ERK signaling and is usually (but not always) correlated with enhanced translation of mRNA subpopulations (11–15). Thus, the prevailing model is that ERK and mTORC1 work synergistically in regulation of eIF4E and translational initiation in long term synaptic plasticity. In addition, LTP in the CA1 region is associated with mTORC1-dependent phosphorylation of ribosomal protein S6 (rpS6), a component of the 40 S ribosomal subunit, and enhanced synthesis of several translation factors which may function to boost translational capacity (16–18).
Neuronal activity also modulates the elongation step of protein synthesis through regulation of eukaryotic elongation factor 2 (eEF2) (19, 20). eEF2 is a GTP-binding protein that mediates translocation of peptidyl-tRNAs from the A-site to the P-site on the ribosome. Phosphorylation of eEF2 on Thr56 inhibits eEF2-ribosome binding and arrests peptide chain elongation (21, 22). Paradoxically, eEF2 is phosphorylated in protein synthesis-dependent forms of LTP and long term depression (23–25), yet synthesis of several plasticity-related proteins, including Arc, is maintained.
Currently, little is known about the dynamic, coordinate regulation of diverse translation factors (eIF4E, eIF2α, rpS6, and eEF2) implicated in long term synaptic plasticity. The most detailed data is from Schaffer collateral-CA1 synapses, yet it is becoming increasing clear that excitatory synapses in different brain regions exhibit qualitatively distinct forms of synaptic plasticity that engage distinct molecular mechanisms. In a recent study in the dentate gyrus of intact rats, LTP consolidation was reported to require a remarkably prolonged period of Arc translation lasting between 2 and 4 h after the induction of LTP by patterned high frequency stimulation (26). Capitalizing on this model, the present study demonstrates a unique, time-dependent regulation of translation factor activity and synthesis mediated by NMDAR, ERK, and mTORC1 signaling. The results couple ERK signaling through MAP kinase-interacting kinase (MNK) to translational initiation and Arc-dependent LTP consolidation. Surprisingly, rapamycin does not affect LTP maintenance, eIF4E activity, or Arc expression, yet it blocks mTORC1 signaling to rpS6 and translation factor synthesis. Furthermore, our findings dissociate eEF2 phosphorylation from Arc synthesis in dentate gyrus LTP.
EXPERIMENTAL PROCEDURES
Electrophysiology and Intrahippocampal Infusion
Experiments were performed on 220 adult male Sprague-Dawley rats. The electrophysiological methods have been detailed elsewhere (27, 28). Briefly, rats were anesthetized with urethane, and electrodes were positioned for selective unilateral stimulation of the medial perforant fibers in the angular bundle and extracellular recording of evoked field potentials in the hilar region of the dentate gyrus (illustrated in Fig. 1A). Recordings were done with borosilicate glass micropipettes (tip size 1–5 μm) filled with 1 m NaCl (input impedance 1–1.5 megaohms). Drugs were infused with a second micropipette (tip size 10–15 μm) connected via a polyethylene (PE50) tube to a 5-μl Hamilton syringe (Reno, NV) and infusion pump. The two micropipettes were clamped together on a micromanipulator with a vertical tip separation of 700 μm. The tip of the infusion cannula was located in deep stratum lacunosum-moleculare of field cornu ammonis 1 (CA1), ∼300 μm from the nearest medial perforant path-granule synapses in the upper blade of the dorsal dentate gyrus. Test pulses were applied at 0.033 Hz throughout the experiment except during the period of HFS. Responses were allowed to stabilize, and 20 min of base-line recording was obtained. After base-line recording, drugs were locally infused at 0.06 μl/min, with volumes ranging from 0.3 to 2 μl depending on the drug. After completing the infusion, test responses were collected for a further 45 min followed by application of HFS. HFS was given in three sessions with 5 min between each. Each session consisted of four 400-Hz stimulus trains (8 pulses/burst) with a 10-s interval between each train. The total HFS duration was 10.5 min, and the total pulse number was 128 pulses. After HFS, evoked responses were collected for periods of 15 min, 2 h, or 4 h.
FIGURE 1.
LTP-specific modulation of eIF4E and eIF2α phosphorylation state in the dentate gyrus in vivo. A, experiments were performed in live anesthetized rats. The schematic shows the hippocampus on the left and right side of the forebrain. B, electrodes were positioned for selective unilateral stimulation of medial perforant fibers in the angular bundle and extracellular recording of evoked field potentials in the hilar region of the dentate gyrus (DG). Drugs were infused through a micropipette positioned immediately above the dentate gyrus. C, left, time course plot showing changes in medial perforant path-granule cell-evoked fEPSPs. Values are the means (±S.E.) expressed in percentage of base line. Test pulses were applied at a 0.033 Hz. AP5 (0.3 μl, 5 min, 50 mm, n = 15) or vehicle PBS were infused during the period, indicated by the black bar. HFS (indicated by arrows) was applied in three series of 400-Hz bursts separated by 5 min (10.5-min total duration). A third treatment group received AP5 infusion and low frequency test stimulation (LFS) only. n = 5 for all treatment groups and time points. Right, sample field potential traces (mean of 5 sweeps) recorded at the times indicated (1 = pre-infusion, 2 = pre-HFS, 3 = post-HFS). Calibration: 5 mV, 2ms. D and E, quantification of Western blot assays of dentate gyrus homogenate samples collected at 15 min, 2 h, and 4 h post-HFS in vehicle and AP5-infused rats. Bar graphs show percentage change (±S.E.) in the expression of phosphorylated (p) eIF4E- Ser209 (D) and eIF2α-Ser51 (E) in the treated dentate gyrus relative to the contralateral control dentate gyrus. Phosphoprotein levels were normalized to total protein levels for the respective translation factors. Total protein levels were normalized to GAPDH loading controls (shown in Fig. 6). n = 5 in all groups. *, p < 0.05 (significantly different from control). F, representative immunoblots of phosphoprotein expression in treated (+) and contralateral control (−) dentate gyrus.
Drugs and Antibodies
2-Amino-5-phosphonopentanoic acid (AP5, 50 mm prepared in 1× PBS; Tocris), anisomycin (86.7 mm in 1 n NaOH diluted in saline; Sigma), actinomycin D (5 mg/ml in saline; Sigma), U0126 (100 μm in 1× PBS with 1% dimethyl sulfoxide (DMSO); Promega), rapamycin (100 μm in 1× PBS with 0.1% DMSO) and CGP57380 (2 mm in 1× PBS with 0.1% DMSO; Sigma).
Antibodies used for Western blotting were as follows (all were obtained from Cell Signaling Technology, Beverly, MA, unless otherwise indicated): Arc (1:400; C-7 Santa Cruz), phospho-eIF2α-Ser51 (1:1000), eIF2α (1:1000), eEF1A (1:1000; Upstate Biotechnology), phospho-eIF4E Ser209 (1:500), eIF4E (1:1000), phospho-eEF2 Thr56(1:1000), eEF2 (1:1000), GAPDH (1:400; Sigma mouse monoclonal G8795), phospho-rpS6 Ser240/244 (1:1000), rpS6 (1:1000), phospho-p70S6K Thr421/Ser424 (1:1000) (Sigma), phospho-p70S6K1-Thr389) (1:1000), p70S6K1 antibody (1:1000), phospho-mTOR Ser2448 (1:1000), anti-mTOR (1:1000), 4E-BP2 (1:1000), phospho-MNK1 Thr197/202 (1:500), MNK1 (1:1000), and eIF4G1 (1:1000) (from the Sonenberg laboratory).
SDS-PAGE and Western Blotting
At the end of electrophysiological recording rats were decapitated, and the dentate gyrus was rapidly dissected on ice and homogenized as previously described (26). Samples were boiled in sample buffer (Bio-Rad) and resolved on 10% or 8% SDS-PAGE minigels. Proteins were transferred to polyvinylidene difluoride membranes (Amersham Biosciences) which were then blocked, probed with antibodies, and developed using chemiluminescence reagents (ECL, Amersham Biosciences). The blots were scanned using Gel DOC EQ (Bio-Rad), and bands intensity were quantified using analytical software (Quantity one 1D analysis software; Bio-Rad). Blots treated with phospho-specific antibody were stripped with 100 mm 2-mercaptoethanol, 2% SDS, and 62.5 mm Tris-HCL, pH 6.7, at 50 °C for 30 min, washed, blocked, and reprobed with antibody recognizing total protein. Optical density values were expressed per unit of protein (GAPDH) applied to the gel lane. The phosphoproteins were normalized relative to the total protein on the same lane. Total proteins were normalized to GAPDH. Values from the treated dentate gyrus were expressed in percent of the values from contralateral control dentate gyrus. Significant differences between the treated and non-treated dentate gyrus were determined using Student's t test for dependent samples. Group comparison was done by analysis of variance. The p value for significance was 0.05.
Cap Analogue Pulldown Assay
Microdissected dentate gyri were homogenized on ice in homogenization buffer (50 mm Tris, pH 7.5, 150 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride), and protein concentration was determined using the Pierce BCA protein assay reagent (Thermo Scientific, Pierce). 250 μg of tissue lysates were incubated with 7-methyl-GTP-Sepharose 4B beads (GE Healthcare) at 4 °C for 12 h with moderate shaking to precipitate eIF4E and the associated proteins. The beads were pelleted, washed, and resuspended in 40 μl of 2× SDS-PAGE loading buffer and denatured at 100 °C for 5 min. Equivalent proportions of precipitated proteins in each group were separated on a 7–15% gradient gel and analyzed by immunoblotting. In some experiments an m7GDP-agarose resin was used as indicated. Precipitated material was washed 3 times and then eluted with 40 μl of 2× sample buffer followed by SDS-PAGE.
Immunohistochemical Staining
The brains were fixed and processed for staining as described (22). Briefly, sections were blocked and then incubated with primary antibodies recognizing phospho-rpS6-Ser240/244 (1:1000) and Arc (1:400) overnight at 4 °C. After primary antibody incubation the sections were washed and incubated in respective biotinylated secondary antibodies (1: 200) (Vector Laboratories, Burlingame, CA) at room temperature for 2 h. The sections were washed and incubated in Vector ABC reagents (Vector Laboratories) for 1 h, and final color was developed using 3,3′-diaminobenzidine with nickel as the chromogen. Sections were mounted on slides, dehydrated through alcohols to xylene, and covered with dibutyl phthalate xylene.
In Situ Hybridization
RNA probes were prepared as previously described (23). Fixed floating sections were washed in PBS for 5 min, treated with pre-warmed proteinase K (10 μg/ml) for 5 min at 37 °C, and post-fixed (5 min with 4% paraformaldehyde/PBS) at room temperature. After post-fixation, sections were incubated 3 min in 0.1 m triethanolamine (TEA, pH 8.0) and treated with 0.25% acetic anhydride in 0.1 m TEA, pH 8.0, for 10 min, washed twice in 2× SSC (1× SSC = 0.15 m NaCl and 0.015 m sodium citrate), and blocked for 10 min in prehybridization buffer. Hybridization with an Arc riboprobe was carried out in a humidified chamber at 60 °C overnight. Sections were washed twice with 2× SSC at room temperature for 20 min, once with 50% formamide in 2× SSC at 65 °C, and twice in 2× SSC at 37 °C. Unbound probe was removed by incubation with 20 μg/ml RNase A at 37 °C for 30 min, and the RNase A was sequentially inactivated by incubating the sections in RNase A buffer for at 65 °C for 30 min. After blocking in 2% blocking reagent for 2 h at room temperature, AP-coupled anti-digoxigenin antibody (1:2000, Roche Applied Science) was applied overnight at 4 °C. Visualization was done with the chromogenic substrates nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science) according to the supplier's instructions. Sections incubated with a control, sense probe for Arc did not show any staining.
RESULTS
LTP-specific Modulation of eIF4E and eIF2a Phosphorylation State
LTP at medial perforant path-granule cell synapses of the dentate gyrus requires Arc synthesis and is associated with rapid Arc transcription, transport of mRNA to dendrites, and enhanced protein expression (26, 29, 30). Local infusions with Arc antisense oligodeoxynucleotides revealed that LTP consolidation requires sustained Arc synthesis during a time-window that starts within 15 min of HFS and lasts between 2 and 4 h (26). The present study examines translation control mechanisms across the critical period of Arc-dependent LTP consolidation. Stable LTP of the field excitatory postsynaptic potential (fEPSP) slope was induced by HFS of the medial perforant pathway (Fig. 1). The total duration of the HFS paradigm was 10.5 min (3 sessions of 400-Hz stimulation with 5 min between sessions). Dentate gyrus tissue was collected at 15 min, 2 h, and 4 h after completing HFS, and Western blot analysis of homogenate samples was used to determine changes in the expression of phosphorylated translation factor. In all figures levels of phosphorylated translation factors are normalized to total levels of the respective translation factor proteins.
We first examined the possible changes in the phosphorylation state of initiation factors eIF4E and eIF2α. Cap-dependent translation starts with recognition of the m7GpppN cap structure on the 5′-end of the mRNA by eIF4E, resulting in recruitment of an initiation complex that includes the 40 S ribosomal subunit. This complex scans the mRNA to the start site, where 60 S subunit joining occurs, and the elongation step of protein synthesis commences. Phosphorylation of eIF4E on Ser209 is generally correlated with enhanced rates of translation, whereas dephosphorylation is associated with decreased translation (31). Phosphorylation of eIF4E promotes its dissociation from the cap, which could serve to speed ribosomal scanning or promote further rounds of initiation (32). As shown in Fig. 1D, HFS led to a rapid increase in Ser209 phosphorylation of eIF4E that declined during the course of LTP maintenance. Phospho-eIF4E levels in homogenate samples from HFS-treated dentate gyrus were significantly elevated 65.3 ± 20.7% above levels in the contralateral control dentate gyrus at 15 min post-HFS and 30 ± 1.1% above control at 2 h post-HFS but were not significantly elevated at 4 h.
eIF2α is a heterotrimeric GTP-binding protein required for recruitment of tRNAiMet to the 40 S subunit. GTP is hydrolyzed during translation initiation, and regeneration of active eIF2α-GTP requires the guanine-nucleotide exchange factor eIF2B. When phosphorylated on Ser51, eIF2α competitively inhibits eIF2B, thus blocking recycling of eIF2 and inhibiting translation (5, 8). As shown in Fig. 1E, HFS resulted in rapid dephosphorylation of eIF2α-Ser51, indicating rapid activation of initiation. Activation of eIF2α followed a decremental time course that paralleled the effects on phospho-eIF4E, although levels of phosph-eIF2α remained significantly decreased at 4 h post-HFS. We then asked whether these changes are specific to NMDAR-dependent LTP induction. Rats were briefly infused with the NMDAR antagonist AP5 (0.3 μl, 5 min, 50 mm, 15 nmol) or vehicle (PBS) 45 min before HFS. The tip of the infusion pipette was located in stratum lacunosum-moleculare of CA1, some 300 μm from the nearest medial perforant path synapses in the upper blade of the dorsal dentate gyrus. fEPSPs were reduced briefly after AP5 but resumed the stable base line by 25 min before HFS. AP5 had no effect on fEPSPs evoked by low frequency test stimulation over 2 h of recording (Fig. 1B) and no effect on initiation factor phosphorylation in tissue collected 15 min, 2 h, and 4 h after AP5 infusion (not shown). As expected, LTP induction was blocked in AP5-treated rats (Fig. 1C). Furthermore, AP5 abolished the HFS-induced changes in eIF4E and eIF2α phosphorylation state (Fig. 1, D–F). Thus, the time course of initiation factor modulation is LTP-specific and corresponds to the window of Arc synthesis. Regulation of total eIF4E and eIF2α expression was also observed, as detailed in a later section on translation factor expression (see Fig. 6).
FIGURE 6.
mTORC1 and ERK-dependent regulation of select TOP and non-TOP encoded translation factors. A, shown is quantification of Western blot assays performed on dentate gyrus homogenates collected at 15 min, 2 h, and 4 h post-HFS in rats receiving vehicle, AP5, U0126, anisomycin (ANI), or rapamycin (Rapa) infusion. Bar graphs show the mean percent (±S.E.) changes in translation factor expression in the treated dentate gyrus relative to the contralateral control dentate gyrus. Total protein levels were normalized to GAPDH loading controls. n = 5 for treatment groups and time points. B, representative immunoblots from treated (+) and contralateral control (−) dentate gyrus in the 15 min post-HFS, AP5, and rapamycin groups.
ERK-dependent Regulation of Initiation Factor Activity and eIF4F Formation
Pharmacological and genetic approaches have demonstrated the role of MEK-ERK signaling in LTP maintenance and memory. MEK inhibitors such as U0126 block ERK phosphorylation and attenuate early and late phase LTP in the CA1 region and dentate gyrus (7, 25, 28, 33, 34). Here, we investigated the effects of MEK-ERK signaling on dynamic changes in translation factor activity during in vivo LTP. Rats were infused with U0126 (0.6 μl, 9.8 min, 100 μm, 0.06 nmol) or vehicle (DMSO-PBS) 45 min before HFS (Fig. 2A). Base-line synaptic transmission was not significantly affected by U0126 or vehicle infusion. HFS in the vehicle-infused group induced non-decremental enhancement of the fEPSP. As expected, HFS in the presence of U0126 elicited a decremental potentiation of the fEPSP slope that decayed within ∼15 min to a stable level that was not significantly different from base line. Strikingly, U0126 treatment blocked the HFS-induced increase in eIF4E phosphorylation and the decrease in eIF2α phosphorylation at both 15 min and 2 h post-HFS (Fig. 2B). The combination of U0126 and low frequency test stimulation (LFS) had no effect on initiation factor phosphorylation state (Fig. 2C).
FIGURE 2.
ERK-dependent regulation of translation initiation factor activity and eIF4F formation. A, left, shown is a time course plot fEPSP changes in rats receiving U0126 (0.6 μl, 9.8 min, 100 μm) or vehicle (PBS-DMSO) infusion (black bar) before HFS (arrows) or U0126 and LFS alone. Right, Sample field potential traces (mean of 5 sweeps) recorded at the times indicated (1 = pre-infusion, 2 = pre-HFS, 3 = post-HFS) are shown. Calibration: 5 mV, 2 ms. n = 5 for all treatment groups and time points. B, quantification of Western blot assays performed on dentate gyrus homogenates collected at 15 min and 2 h post-HFS in vehicle and U0126-infused rats. Bar graphs show percentage change (±S.E.) in the expression of phosphorylated (p) eIF4E- Ser209 and eIF2α-Ser51 in the treated dentate gyrus relative to the contralateral control dentate gyrus. Phosphoprotein levels were normalized to total protein levels for the respective translation factors. n = 5 in all groups. *, p < 0.05 (significantly different from control). C, representative immunoblots of phosphoprotein expression in treated (+) and contralateral control (−) dentate gyrus in tissue collected 15 min post-HFS (left panel) or 15 min after U0126 infusion in the LFS group (right panel). D and E, cap-pulldown assays are shown. Equal amounts (250 μg) of total extracts from homogenized DG were incubated with 7-methyl-GTP-Sepharose 4B beads. Precipitated proteins were separated on a 7–15% gradient gel and analyzed by immunoblotting. D, immunoblots for p-eIF4E- Ser209 and eIF4G1 were normalized to total levels of cap-bound eIF4E. Bar graphs show percentage change (±S.E.) in p-eIF4E and eIF4G1 expression in the treated dentate gyrus (+) relative to the contralateral control dentate gyrus (−). n = 4 in all groups. *, p < 0.05 (significantly different from control). E, shown are representative immunoblots. No GAPDH was detected in precipitated samples.
In addition to regulation of eIF4E phosphorylation, ERK signaling might function in initiation complex (eIF4F) formation, as detected by binding of eIF4E to the scaffolding protein eIF4G. To assess this possibility an m7GDP cap analogue pulldown assay was performed in homogenate samples from HFS-treated and contralateral control dentate gyrus and the resulting precipitate was immunoblotted with antibodies recognizing total eIF4E, Ser209 phosphorylated eIF4E and eIF4G1. HFS resulted in significantly enhanced eIF4G association with cap-bound eIF4E as well as enhanced phosphorylation of eIF4E (Fig. 2, D and E). These effects were blocked by UO126, demonstrating ERK-dependent enhancement of initiation complex formation.
Given the complete block of eIF4E phosphorylation and eIF4F protein complex formation, we anticipated that U0126 would inhibit LTP-associated changes in Arc expression. As shown in Fig. 3, immunoblot analysis revealed robust enhancement of Arc protein expression at 15 min and 2 h post-HFS in vehicle treated-control that was abolished by local infusion of U0126 or AP5 before HFS. Treatment with the general protein synthesis inhibitor, anisomycin, similarly blocked LTP consolidation and Arc synthesis (Fig. 3C).
FIGURE 3.
LTP-associated Arc synthesis requires NMDAR and ERK signaling. A, quantification of Western blot assays performed on dentate gyrus homogenates collected at 15 min and 2 h post-HFS in rats receiving local infusion of vehicle, AP5, U0126, or anisomycin (ANI). Bar graphs show percentage change (±S.E.) in Arc expression in the treated dentate gyrus relative to the contralateral control dentate gyrus. Arc levels were normalized to GAPDH. *, p < 0.05 (significantly different from control). n = 5 for all treatment groups and time points. B, representative immunoblots from treated (+) and contralateral control (−) dentate gyrus are shown. C, shown is a time course plot showing block of late phase LTP in the anisomycin treatment group.
LTP Maintenance in the Dentate Gyrus Does Not Require Rapamycin-sensitive mTORC1 Signaling
Rapamycin-sensitive mTORC1 signaling is necessary for several forms of protein-synthesis-dependent synaptic plasticity, as studied extensively in the hippocampal CA1 region (17, 35–37). We, therefore, expected that rapamycin would impair LTP in the dentate gyrus. Previous work showed that brief intrahippocampal infusion of 60 nm rapamycin effectively blocks acquisition of inhibitory avoidance learning and mTORC1 activation in hippocampus (38). Using a similar protocol, we infused rapamycin 45 min before HFS. In contrast to expectation, HFS in rapamycin-treated rats evoked non-decremental LTP that was indistinguishable to that obtained in vehicle-infused controls over 4 h of post-HFS recording (Fig. 4A). A range of concentrations were tested, and the results shown are based on the maximal rapamycin concentration of 100 μm (2 μl, 32.5 min, 0.2 nmol). Systemic injection of rapamycin at high dosage (50 mg/kg intraperitoneally) 30 min before HFS similarly failed to inhibit LTP over 4 h of recording (n = 6; not shown).
FIGURE 4.
LTP maintenance and Arc synthesis does not require rapamycin-sensitive mTORC1 signaling. A, left, time course plot fEPSP changes are shown in rats receiving rapamycin (2 μl, 32.4 min, 100 μm) or vehicle (PBS-DMSO; n = 4) infusion (black bar) before HFS (arrows) or U0126 and LFS. Right, shown are sample field potential traces (mean of 5 sweeps) recorded at the times indicated (1 = pre-infusion, 2 = pre-HFS, 3 = post-HFS). Calibration: 5 mV, 2 ms. n = 5 for all treatment groups and time points. B, shown is quantification of Western blot assays of dentate gyrus homogenate samples collected at 15 min and 2 h post-HFS in vehicle and U0126-infused rats. Bar graphs show percentage change (±S.E.) in the expression of phosphorylated (p) mTOR- Ser2448, eIF4E-Ser209, eIF2α-Ser51 in the treated dentate gyrus relative to the contralateral control dentate gyrus. Phosphoprotein levels were normalized to total protein levels for the respective translation factors. C, representative immunoblots of phosphoprotein and total protein expression in treated (+) and contralateral control (−) dentate gyrus are shown. D, bar graphs show changes in Arc expression normalized to GAPDH. n = 5 for all treatment groups and time points. *, p < 0.05 (significantly different from control). E, representative Arc immunoblots from treated (+) and contralateral control (−) dentate gyrus. F and G, cap pulldown assays are shown. F, immunoblots for p-eIF4E-Ser209 and eIF4G1 were normalized to total levels of cap-bound eIF4E. Bar graphs show the percentage change (±S.E.) in p-eIF4E and eIF4G1 expression in the treated dentate gyrus (+) relative to the contralateral control dentate gyrus (−). n = 4 in all groups. *, p < 0.05 (significantly different from control). G, representative immunoblots are shown. No GAPDH was detected in precipitated samples.
Surprised by this finding, we examined mTORC1 activation using antibodies recognizing Ser2448-phosphorylated mTOR. Immunoblots showed rapid and sustained activation of mTORC1 at 15 min and 2 h post-HFS (Fig. 4, B and C). Rapamycin blocked this effect at both time points, thus verifying the efficacy of the drug in blocking mTORC1. Notably, however, rapamycin failed to inhibit the rapid modulation of eIF4E and eIF2α at 15 min post-HFS, although significant inhibition was detected at the 2-h time point. Immunoblotting for Arc further showed that rapamycin does not block the LTP-associated increase in Arc synthesis (Fig. 4, D and E), although this synthesis was abolished by anisomycin (Fig. 3C). Taken together this suggests that ERK, but not mTORC1 signaling, is required for rapid regulation of translational initiation Arc synthesis, and LTP maintenance.
These results suggest that the branch of mTORC1 signaling involved in the release of eIF4E from 4E-BP2, the main 4E-BP isoform in brain, is not necessary for LTP in the dentate gyrus. To address this issue an m7GDP cap analogue pulldown assay was performed in homogenate samples from HFS-treated and contralateral control dentate gyrus, and precipitates were immunoblotted with antibodies recognizing eIF4E and 4E-BP2. LTP was not associated with detectable reductions in the 4E-BP signal relative to eIF4E, indicating that 4E-BP2 is not being released from the complex. A quantitative analysis of the 4E-BP2 subunit/eIF4E ratio and representative blots based on similar results obtained at 15 min and 2 h post-HFS (6 rats at each time point) are shown in supplemental Fig. S1. Consistent with these observations, no signal could be detected using an antibody recognizing phosphorylated 4E-BP2 (results not shown). Analysis of eIF4G-eIF4E interaction further showed significant enhancement of initiation complex formation in rapamycin-infused rats equivalent to vehicle-infused controls (Fig. 4, F and G).
LTP Is Associated with mTORC1 Signaling to Ribosomal Protein S6 and Enhanced Translation Factor Expression
Next we examined regulation in the branch of mTORC1 signaling to p70 S6 kinase (p70S6K) and its substrate, rpS6. p70S6K is activated by mTORC1 through direct phosphorylation of Thr389 (39, 40). HFS resulted in enhanced phosphorylation of p70S6K-Thr389 at 15 min and 2 h post-HFS (Fig. 5, A and C). Rapamycin inhibited significant changes in p70S6K-Thr389 phosphorylation, with the strongest inhibition detected at 15 min post-HFS. p70S6K activation is thought to additionally require prior phosphorylation of an ERK-dependent site (Thr421/Ser424) located in the module IV region of the enzyme (41, 42). We found that U0126, but not rapamycin, blocked phosphorylation of the ERK-site on p70S6K (Fig. 5, A and C). We then assayed rpS6 regulation using antibodies recognizing the Ser240/244 site that is directly phosphorylated by p70S6K. A more than 3-fold increase in rpS6 phosphorylation was observed at 15 min, 2 h, and 4 h post-HFS (Fig. 5), and these effects were abolished by treatment with rapamycin and U0126. Taken together, we conclude that rapamycin effectively blocks HFS-evoked mTORC1 activation and downstream signaling to p70S6K and rpS6 while having no effect on LTP maintenance during 4 h of recording.
FIGURE 5.
mTORC1- and ERK-dependent regulation of p70S6K-rpS6 signaling. A, shown is quantification of Western blot assays performed on dentate gyrus homogenates collected at 15 min and 2 h post-HFS in rats receiving local infusion of vehicle, U0126, or rapamycin. Bar graphs show percentage change (±S.E.) in the expression of phosphorylated (p) p70S6K-Thr389, and p70S6K-Thr421/Ser424 in the treated dentate gyrus relative to the contralateral control dentate gyrus. Phosphoprotein levels were normalized to total protein levels for p70S6K. *, p < 0.05 (significantly different from control). n = 5 for all treatment groups and time points. B, bar graphs show percentage change (±S.E.) in the expression of p-rpS6-Ser240/244 in the treated dentate gyrus relative to the contralateral control dentate gyrus and normalized to total rpS6. C, representative immunoblots of phosphoprotein and total protein expression in treated (+) and contralateral control (−) dentate gyrus are shown. The arrowheads indicate measured band.
Immunohistochemical staining was used to assess the distribution of phospho-rpS6 in the dentate gyrus after induction of LTP (supplemental Fig. S2). Staining was observed in scattered granule somata of the non-stimulated, contralateral dentate gyrus. In the HFS-treated dentate gyrus, expression of phospho-rpS6 was dramatically enhanced in the granule cell layer and across the dendritic field of the molecular layer. Similar somatodendritic expression was observed at 15 min, 2 h, and 4 h post-HFS. Local infusion of AP5 blocked the enhanced expression of phospho-rpS6 in both the dorsal and ventral blades of the dentate gyrus.
mTORC1 and ERK-dependent de Novo Synthesis of Translation Factors
In situations demanding intensive protein synthesis, as during cell division, translational capacity may be maintained through the enhanced synthesis of ribosomal proteins and translation factors. Many of these proteins are encoded by TOP mRNAs and undergo enhanced translation through an mTORC1-dependent pathway (43, 44). LTP in the CA1 region in vitro is associated with mTORC1-dependent activation of p70S6K and synthesis of several TOP-containing mRNAs including rpS6, eEF2, and eEF1A (16, 18). Here, we examined changes in total translation factor expression (normalized to GAPDH levels) after LTP induction in the dentate gyrus in vivo (Fig. 6). At 15 min, expression of eIF4e, eIF2α, and rpS6 were significantly enhanced in the HFS-treated dentate gyrus relative to control, whereas expression of eEF2 and eEF1A were not significantly altered. At 2 h, no significant change in translation factor levels was observed. At 4 h, eIF4E expression was elevated, revealing a biphasic pattern of expression specific to this translation factor. AP5 infusion blocked all HFS-induced increases in translation factor expression at 15 min post-HFS as well as the enhanced expression of eIF4E at 4 h (Fig. 6A and 7B). Interestingly, however, HFS in the presence of the NMDAR antagonist led to enhancement of eIF4E expression at 2 h post-HFS, suggesting that NMDAR signaling counteracts an unknown mechanism for enhanced eIF4E expression at this time. U0126, rapamycin, and anisomycin all blocked the HFS-induced enhancement of eIF4E, eIF2α, and rpS6 expression as measured 15 min post-HFS (Fig. 6). The results show that mTORC1 and ERK signaling are both critically required for rapid de novo synthesis of translation factors during NMDAR-dependent LTP in vivo. Notably, however, inhibition of translation factor expression by rapamycin does not block LTP maintenance.
Dissociation of eEF2 Phosphorylation from LTP and Arc Expression
Although eEF2 phosphorylation is associated with reduction in global protein synthesis, translation of certain plasticity-associated mRNAs, including Arc and α-calmodulin kinase II, is maintained (20, 23, 25). In the present study, HFS resulted in rapid and sustained phosphorylation of eEF2-Thr56. This sustained increase outlasts the critical period of Arc synthesis and contrasts with the slowly declining pattern of eIF4E and eIF2α activation.
Blockade of NMDAR-dependent LTP failed to inhibit the enhanced expression of phospho-eEF2 as observed 15 min and 2 h post-HFS (Fig. 7). At 15 min, phospho-eEF2 levels were equally enhanced in the AP5-treated and PBS control groups (p > 0.05). At 2 h, phospho-eEF2 levels in the AP5 group (75.18 ± 23.57% increase) were significantly elevated ∼2-fold above rats receiving PBS infusion and HFS (35.1 + 5.3% increase, p < 0.05). Remarkably, eEF2 phosphorylation at 4 h post-HFS was completely blocked by AP5 treatment. Thus, eEF2 phosphorylation during LTP consists of an early NMDAR-independent component and a late NMDAR-dependent component. Blockade of mTORC1 signaling with rapamycin blocks the hyperphosphorylation of eEF2 at 15 min and 2 h post-HFS but has no effect on Arc synthesis. Taken together, these results dissociate eEF2 phosphorylation from enhanced Arc synthesis during LTP.
FIGURE 7.
Dissociation of eEF2 phosphorylation from LTP and Arc expression. Shown is quantification of Western blot assays of dentate gyrus homogenate samples collected at 15 min, 2 h, and 4 h post-HFS in vehicle-, AP5-, and rapamycin-infused rats. Bar graphs show percentage change (±S.E.) in the expression of phosphorylated (p) eEF2-Thr56 in AP5 infused (A) and rapamycin infused (B) in the treated dentate gyrus relative to the contralateral control dentate gyrus. Phosphoprotein levels were normalized to total protein levels for the respective translation factors. Total protein levels were normalized to GAPDH loading controls (shown in Fig. 7). n = 5 in all groups. *, p < 0.05 (significantly different from control). Representative immunoblots are shown of phosphoprotein expression in treated (+) and contralateral control (−) dentate gyrus.
ERK Couples Arc Transcription and Translation
The foregoing data demonstrates ERK-dependent eIF4E phosphorylation, initiation complex formation, and enhanced expression of Arc during LTP. However, the effects of the U0126 on Arc protein expression could also be due to inhibition of Arc transcription (45, 46). We considered that Arc translation might be selectively blocked by applying U0126 well after the induction of Arc mRNA (26). U0126 or vehicle solution were, therefore, infused 10 min after HFS, and homogenates samples were collected for immunoblot analysis of translation initiation factor phosphorylation and Arc protein expression at 30 min post-HFS. Post-HFS administration of U0126 inhibited LTP maintenance (Fig. 8A), changes in eIF4E and eIF2α phosphorylation state (Fig. 8, B and C), and enhancement of Arc protein expression in immunoblots (Fig. 8D). A series of perfusion-fixed brains was collected for in situ hybridization and immunohistochemical analysis of Arc mRNA and protein expression, respectively. As illustrated in Fig. 8E (top panel), vehicle-infused rats exhibited robust increases in Arc mRNA and protein expression in the granule cell layer and across the dendritic field of the molecular layer. Remarkably, U0126 abolished increases in Arc mRNA expression in parallel with Arc protein (Fig. 8E, middle panel). In an attempt to elicit Arc protein expression from pre-existing RNA, HFS was applied in the presence of the transcriptional inhibitor actinomycin D, but we were unable to detect Arc protein increases in the absence of de novo Arc mRNA (Fig. 8E, bottom panel). Taken together, this suggested that Arc protein expression derives from ERK-dependent transcription well into the maintenance phase of LTP.
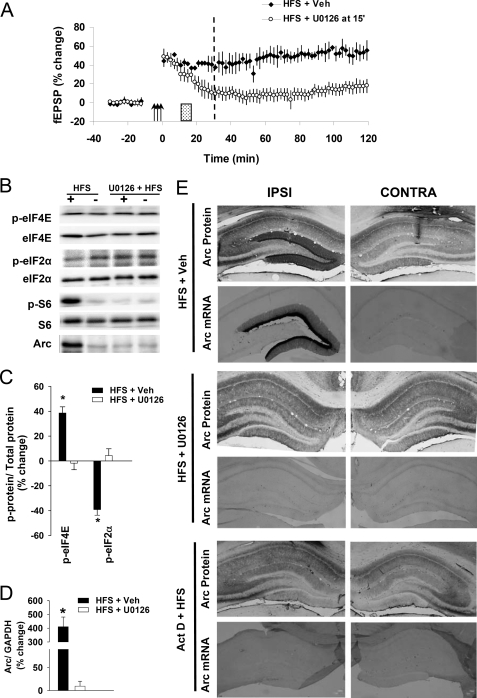
FIGURE 8.
Arc protein synthesis requires sustained, ERK-dependent Arc transcription. A, left, time course plot fEPSP changes in rats receiving U0126 (0.6 μl, 9.8 min, 100 μm) or vehicle (PBS-DMSO) infusion (black bar) post-HFS (arrows) is shown. Values are the means (±S.E.) expressed in percentage of base line. U0126 was infused 10 min post-HFS (hatched bar), and tissue was collected 30 min post-HFS (stippled line). B, representative immunoblots are shown. Bar graphs show percentage change (±S.E.) in the expression of phosphorylated (p) eIF4E-Ser209 and eIF2α-Ser51 (panel C) and Arc (panel D). n = 5–6 in all groups. E, coronal sections were obtained 30 min post-HFS and processed for Arc immunostaining and in situ hybridization. Upper panel, Arc protein and mRNA are strongly up-regulated in granule cell somata and dendrites in the ipsilateral dentate gyrus in DMSO-PBS-infused controls. Middle panel, U0126 infusion 10 min post-HFS eliminated Arc mRNA and protein expression at 30 min post-HFS is shown. Similar results were obtained in seven rats in each treatment group. Lower panel, HFS was applied in the presence of the transcriptional inhibitor actinomycin D (Act D). Inhibition of Arc transcription prevented HFS-evoked increases in Arc protein expression.
To isolate ERK effects on translation, we blocked activation of mitogen-activated protein kinase-interacting kinase (MNK), the kinase that couples ERK to phosphorylation of eIF4E (5, 13, 47, 48). Local infusion of the MNK inhibitor CGP57380 strongly attenuated LTP maintenance in a manner similar to U0126 (Fig. 9A). Western blot analysis of tissue collected 15 min post-HFS showed that CGP57380 blocks phosphorylation of MNK1-Thr197/202 and eIF4E and abolishes enhancement of Arc synthesis, whereas HFS-evoked phosphorylation of rpS6 was not affected by MNK inhibition (Fig. 9B). CGP57380 infusion in a low frequency test stimulation group had no significant effects on base-line fEPSPs or MNK1 phosphorylation state (Fig. 9B). Given these effects we further considered that ERK signaling to MNK might contribute to initiation complex formation itself. As shown in Fig. 9C, CPG57380 completely blocked enhanced loading of eIF4G onto eIF4E in the cap pulldown assay. Imunohistochemistry confirmed the block of Arc protein expression in dentate granule cells 15 min post-HFS. In contrast, in situ hybridization revealed intact HFS-evoked up-regulation of Arc mRNA in granule somata of CGP57380-infused rats (Fig. 9D). We conclude that MNK inhibition blocks eIF4F formation, eIF4E phosphorylation, and Arc synthesis without disrupting Arc RNA expression.
FIGURE 9.
Inhibition of MNK signaling blocks eIF4F formation, eIF4E phosphorylation, and Arc translation without affecting ARC transcription. A, shown is a time course plot fEPSP changes in rats receiving CGP57380 (1 μl, 16 min, 2 mm) or vehicle (PBS-DMSO) infusion (black bar) before HFS (arrows). Values are the means (±S.E.) expressed in percentage of base line. Dentate gyrus tissue for Western blotting was collected at 15 min post-HFS (stippled line) in separate experiments. B, left, bar graphs show percentage change (±S.E.) in the expression of Arc and phosphorylated (p) MNK1-Thr197/202 and eIF4E-Ser209. Phospho-proteins are normalized to respective total protein levels and GAPDH. Right, representative immunoblots are shown. n = 5–6 in all groups. C, cap pulldown assays are shown. Immunoblots for p-eIF4E-Ser209 and eIF4G1 were normalized to total levels of cap-bound eIF4E. Bar graphs show percentage change (±S.E.) in p-eIF4E and eIF4G1 expression in the treated dentate gyrus (+) relative to the contralateral control dentate gyrus (−). n = 4 in all groups. *, p < 0.05 (significantly different from control). D, coronal sections were obtained 15 min post-HFS in rats receiving CGP57380 and processed for Arc protein immunohistochemistry and mRNA in situ hybridization. In situ hybridization showed up-regulation of Arc mRNA in the dentate granule cell layer (white arrow) of CGP-treated rats. At this early time point Arc mRNA is restricted to the granule cell layer. Immunohistochemistry confirmed the block of Arc protein expression observed in the Western blot analysis.
DISCUSSION
The major conclusions from this work are as follows. 1) LTP consolidation in the dentate gyrus of anesthetized rats is associated with unique, time-dependent regulation of translation initiation and elongation factor activity and synthesis. 2) ERK-MNK, but not mTORC1 signaling, is required for enhanced translational initiation, Arc synthesis, and LTP maintenance. 3) Sustained Arc synthesis depends on the continued supply of new Arc mRNA during LTP maintenance. 4) mTORC1 signaling to p70S6K and rpS6 is activated and associated with enhanced synthesis of select TOP and non-TOP-encoded translation factors, yet this is dispensable for LTP maintenance. 5) eEF2 phosphorylation is neither sufficient nor necessary for Arc expression underlying LTP consolidation.
Arc has been identified as a critical regulator of multiple forms of long term synaptic plasticity and long term memory (49). Messaoudi et al. (26) reported that LTP consolidation in the dentate gyrus depends on sustained translation of Arc mRNA. The critical period of Arc translation starts within 15 min of LTP induction and persists between 2 and 4 h. Arc synthesis during this period is necessary for stable expansion of the F-actin cytoskeleton, which in turn underlies stable structural changes of the synapse. Here, we show phosphorylation of eIF4E and dephosphorylation of eIF2α on a slowly declining time course that matches the critical period of Arc translation. Local infusion of the MEK inhibitor U0126 selectively inhibited the maintenance phase of LTP and blocked modulation of eIF4E and eIF2α activity at both 15 min and 2 h post-HFS. Cap pulldown assays further demonstrated ERK-dependent enhancement of initiation complex eIF4F formation, indicated by enhanced eIF4G association with cap-bound eIF4E.
Previous studies demonstrated a critical role for ERK in Arc transcription (28, 45, 46). Huang et al. (50) further showed that U0126 blocks the localization of Arc mRNA to activated medial perforant path synapses on granule cells, probably by preventing docking of mRNA to the cytoskeleton. Here, we found that application of U0126 at 10 min post-HFS (at which time Arc mRNA is already elevated) effectively eliminates the increase in Arc mRNA and protein expression in tissue obtained 30 min post-HFS. This suggests that 1) Arc transcription is sustained for at least 10 min into the maintenance phase of LTP, 2) ERK is necessary for this transcription, and 3) Arc protein expression is intimately coupled to the provision of new mRNA. Regulation of Arc RNA stability could be involved, for instance, through ERK-dependent docking of mRNA near synapses (50), but this specific mechanism would not explain the observed loss of Arc mRNA throughout granule cell somata and dendrites in U0126-treated rats.
Interestingly, Arc mRNA is more persistently elevated in the dentate gyrus than CA1 region of the hippocampus after behavioral spatial exploration and learning, electroconvulsive seizures, or LTP induction (26, 51–55). Examination of Arc transcriptional foci by fluorescence in situ hybridization indicated prolonged (>1 h) transcriptional activation in dentate granule cells (56). Evidence from cultured hippocampal neurons further shows that Arc mRNA is rapidly degraded by non-sense mediated RNA decay by virtue of an exon junction complex in its 3′-untranslated region (57). In the extreme, Arc mRNA may be degraded after a single round of translation (58). Arc protein is likewise subject to rapid turnover (26, 46). All of this suggests a tight coupling between Arc transcription, mRNA localization, and translation, with ERK playing a central coordinating role.
To separate ERK effects on translation from transcription, we blocked activation of MNK, the kinase that couples ERK to phosphorylation of eIF4E. Local infusion of the MNK inhibitor CGP58370 blocked LTP maintenance in parallel with MNK-eIF4E signaling and Arc synthesis while leaving Arc mRNA expression in dentate granule cells intact. Remarkably, MNK activation was also required for loading of eIF4G onto eIF4E. Taken together, the data strongly support a role for ERK-MNK signaling in translational initiation, eIF4E phosphorylation, and enhanced Arc synthesis in LTP (see the model in Fig. 10). To our knowledge, this is the first evidence for ERK-MNK regulation of initiation complex formation.
FIGURE 10.
Model of translational control during Arc-dependent LTP consolidation in the dentate gyrus. The diagram depicts ERK and mTORC1 signaling pathways regulating eIF4E and p70S6K-rpS6. After LTP induction in the dentate gyrus, ERK-MNK signaling (but not rapamycin-sensitive mTORC1 signaling) is necessary for eIF4F formation, eIF4E phosphorylation, and Arc synthesis. HFS activates mTORC1 but does not promote dissociation of 4E-BP2 from eIF4E (dashed line). mTORC1 activation is necessary for p70S6K-rpS6 phosphorylation and enhanced synthesis of select translation factors, but blockade of these effects with rapamycin does not affect LTP. The present data, therefore, indicate a dominant role for ERK-MNK signaling in translational initiation and Arc synthesis underlying LTP consolidation in the dentate gyrus. Cross-talk between ERK and mTORC1 in regulation of translation factor synthesis is not depicted. The role of p70S6K-rpS6 signaling is unknown and may not be involved in translation factor synthesis.
Translational initiation in many settings, including LTP and long term depression in the hippocampal CA1 region, requires cooperation between the mTORC1 and ERK signaling pathways (6). One of the most unexpected findings was the insensitivity of LTP maintenance in the dentate gyrus to inhibition of mTORC1 by rapamycin. Our data indicate that the branch of mTORC1 signaling to 4EBP2 and eIF4E is simply not engaged (Fig. 10). m7GDP cap binding assays show that 4E-BP2 does not dissociate from eIF4E, whereas rapamycin fails to block initiation complex formation, eIF4E phosphorylation, and Arc synthesis. The results further demonstrate a major role for ERK-MNK, rather than mTORC1, in regulation of initiation complex formation. The mechanism by which MNK promotes eIF4G-eIF4E interactions in LTP remains to be studied. Phosphorylation of eIF4E is not implicated in initiation complex formation. Work in non-neuronal cells has identified a number of MNK substrates in addition to eIF4E, including eIF4G and PSF (polypyrimidine tract-binding protein-associated splicing factor), a protein which binds to AU-rich elements of target mRNAs (48, 59, 60). ERK-MNK regulation of novel eIF4E-binding proteins in brain such as neuroguidin and CYFIP1 (61, 62) is another possibility. Finally, although the mTORC1-BP2 pathway was not activated, we do not rule out a role for rapamycin-insensitive mTORC1 signaling in LTP.
Previous work in the CA1 region of hippocampal slices has established a critical role for rapamycin-sensitive mTORC1 signaling in stable LTP and enhanced translation of several TOP mRNAs including rpS6 (7, 16, 18). Enhanced synthesis of translation factors and ribosomal proteins may boost translational capacity during periods of intensive protein synthesis (44, 63, 64). In the present study in the dentate gyrus, the arm of mTORC1 signaling to p70S6K and rpS6 is persistently activated and associated with enhanced synthesis of select TOP (rpS6) and non-TOP-encoded translation factors (eIF4E and eIF2α), yet this regulation is dispensable for LTP maintenance. Another TOP-encoded translation factor, eEF1A, is strongly up-regulated after LTP induction in the CA1 region (11) but was not regulated in the present study. In the dentate gyrus, synthesis of eEF1A has been linked to metabotropic glutamate receptor activation and long term depression (65).
Although earlier studies implicated rpS6 regulation in translation of 5′-TOP mRNAs (66, 67), recent analysis of knock-in mice bearing alanine substitutions of all five phosphorylatable serine residues in rpS6 shows that mTORC1-dependent TOP translation occurs in the absence of rpS6 regulation (5, 68, 69). This suggests that rpS6 does not regulate TOP translation although functions of the individual phosphorylation sites have not been ruled out. The clear conclusion from the present study, however, is that mTORC1 signaling to p70S6K and rpS6 is dispensable for LTP maintenance over the 4-h period examined here (Fig. 10). This leaves open the possibility that mTOR-p70S6K-rpS6 signaling functions at a later stage of the consolidation process beyond the time-window of Arc synthesis. In the taste cortex, learning is associated with biphasic increases in mTOR activity in distinct neuronal compartments (70). In Aplysia, rapamycin-sensitive signaling is required for stabilization of long term synaptic facilitation 72 h after its onset (71).
eEF2 phosphorylation acts to slow peptide chain elongation, but synthesis of some plasticity-associated proteins is enhanced in this context. N-Methyl-d-aspartate treatment of synapto-neurosome preparations phosphorylates eEF2 and reduces global protein synthesis while permitting enhanced expression of α-calmodulin kinase II. Consolidation of taste memory is similarly associated with enhanced phosphorylation of eEF2 and synaptic expression of α-calmodulin kinase II in the taste cortex (72). Forskolin-induced LTP in region CA1 (23) and brain-derived neurotrophic factor-LTP in the dentate gyrus (25) are both associated with eEF2 phosphorylation and enhanced Arc expression. Here, we found that NMDAR block inhibits LTP induction and Arc expression without affecting eEF2 phosphorylation at 15 min and 2 h post-HFS (although late phosphorylation of eEF2 at 4 h was blocked). Conversely, rapamycin blocked eEF2 phosphorylation without affecting LTP. These results uncouple eEF2 phosphorylation from early LTP maintenance and Arc expression. However, eEF2 could function at later time points or regulate other forms of plasticity. For example, HFS of the perforant path can activate metabotropic glutamate receptor mechanisms involved in phenomena such as depotentation, metaplasticity, and homeostatic plasticity (73–75). Metabotropic glutamate receptor-induced long term depression in the CA1 region of hippocampal slices requires local synthesis of Arc from pre-existing mRNA (76), and this synthesis is mechanistically linked to eEF2 phosphorylation (24). The mechanism by which NMDAR-dependent mTORC1 signaling promotes eEF2 phosphorylation during LTP is unknown. mTORC1 typically inhibits eEF2 kinase indirectly via activation of S6K. However, eEF2 kinase is positively and negatively regulated by multiple signaling pathways, suggesting a complex, context-specific regulation (5).
The present work demonstrates that translational control mechanisms engaged in long term synaptic plasticity differ fundamentally between brain regions. Unlike the hippocampal CA1 region and several other brain regions, translational initiation during LTP consolidation in the dentate gyrus is regulated by ERK-MNK signaling without a detectable contribution from mTORC1. This rapamycin-insensitive form of regulation is coupled to sustained translation of Arc, which also appears to be unique to dentate gyrus LTP. This represents an important first step in identifying the post-transcriptional mechanisms that shape the duration and efficacy of Arc translation and LTP consolidation. The exact function of eIF4E phosphorylation remains enigmatic. Increases in eIF4E activity increase translation of subsets of mRNAs rather than affecting global translation (14). It has been suggested that decreased binding of eIF4E to the cap structure as a result of phosphorylation could serve to speed scanning of the pre-initiation complex to the translation start site or accelerate recruitment of new initiation complexes (5). Such a mechanism could be decisive for transcripts like Arc that are only weakly initiated due to the strong secondary structure of their 5′-untranslated region.
Supplementary Material
This work was funded by the Norwegian Research Council and the University of Bergen.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- LTP
- long term potentiation
- eIF
- eukaryotic initiation factor
- 4E-BP
- eIF4E-binding proteins
- mTORC1
- mammalian target of rapamycin protein complex 1
- rpS6
- ribosomal protein S6
- eEF2
- eukaryotic elongation factor 2
- CA1
- cornu ammonis 2
- AP5
- 2-amino-5-phosphonopentanoic acid
- p70S6K
- p70 S6 kinase
- ERK
- extracellular signal-regulated kinase
- NMDAR
- N-methyl-d-aspartate receptor
- HFS
- high frequency stimulation
- LFS
- low frequency stimulation
- PBS
- phosphate-buffered saline
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- fEPSP
- field excitatory postsynaptic potential
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- MNK
- mitogen-activated protein kinase-interacting kinase
- TOP
- terminal oligopyrimidine.
REFERENCES
- 1.Nelson S. B., Turrigiano G. G. (2008) Neuron 60, 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bliss T., Collingridge G., Morris R. (2007) in The Hippocampus Book (Andersen P., Morris R., Amaral D., Bliss T., O'Keefe J. eds), pp. 343–474, Oxford University Press, New York [Google Scholar]
- 3.Bramham C. R., Wells D. G. (2007) Nat. Rev. Neurosci. 8, 776–789 [DOI] [PubMed] [Google Scholar]
- 4.Sutton M. A., Schuman E. M. (2006) Cell 127, 49–58 [DOI] [PubMed] [Google Scholar]
- 5.Proud C. G. (2007) Biochem. J. 403, 217–234 [DOI] [PubMed] [Google Scholar]
- 6.Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelleher R. J., 3rd, Govindarajan A., Jung H. Y., Kang H., Tonegawa S. (2004) Cell 116, 467–479 [DOI] [PubMed] [Google Scholar]
- 8.Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C., Sossin W., Kaufman R., Pelletier J., Rosenblum K., Krnjević K., Lacaille J. C., Nader K., Sonenberg N. (2007) Cell 129, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa-Mattioli M., Gobert D., Harding H., Herdy B., Azzi M., Bruno M., Bidinosti M., Ben Mamou C., Marcinkiewicz E., Yoshida M., Imataka H., Cuello A. C., Seidah N., Sossin W., Lacaille J. C., Ron D., Nader K., Sonenberg N. (2005) Nature 436, 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banko J. L., Poulin F., Hou L., DeMaria C. T., Sonenberg N., Klann E. (2005) J. Neurosci. 25, 9581–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knauf U., Tschopp C., Gram H. (2001) Mol. Cell. Biol. 21, 5500–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheper G. C., Proud C. G. (2002) Eur. J. Biochem. 269, 5350–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter J. D., Sonenberg N. (2005) Nature 433, 477–480 [DOI] [PubMed] [Google Scholar]
- 15.Bianchini A., Loiarro M., Bielli P., Busà R., Paronetto M. P., Loreni F., Geremia R., Sette C. (2008) Carcinogenesis 29, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 16.Tsokas P., Grace E. A., Chan P., Ma T., Sealfon S. C., Iyengar R., Landau E. M., Blitzer R. D. (2005) J. Neurosci. 25, 5833–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cammalleri M., Lütjens R., Berton F., King A. R., Simpson C., Francesconi W., Sanna P. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14368–14373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsokas P., Ma T., Iyengar R., Landau E. M., Blitzer R. D. (2007) J. Neurosci. 27, 5885–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutton M. A., Taylor A. M., Ito H. T., Pham A., Schuman E. M. (2007) Neuron. 55, 648–661 [DOI] [PubMed] [Google Scholar]
- 20.Scheetz A. J., Nairn A. C., Constantine-Paton M. (2000) Nat. Neurosci. 3, 211–216 [DOI] [PubMed] [Google Scholar]
- 21.Ryazanov A. G., Shestakova E. A., Natapov P. G. (1988) Nature 334, 170–173 [DOI] [PubMed] [Google Scholar]
- 22.Nairn A. C., Palfrey H. C. (1987) J. Biol. Chem. 262, 17299–17303 [PubMed] [Google Scholar]
- 23.Chotiner J. K., Khorasani H., Nairn A. C., O'Dell T. J., Watson J. B. (2003) Neuroscience 116, 743–752 [DOI] [PubMed] [Google Scholar]
- 24.Park S., Park J. M., Kim S., Kim J. A., Shepherd J. D., Smith-Hicks C. L., Chowdhury S., Kaufmann W., Kuhl D., Ryazanov A. G., Huganir R. L., Linden D. J., Worley P. F. (2008) Neuron 59, 70–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanhema T., Dagestad G., Panja D., Tiron A., Messaoudi E., Håvik B., Ying S. W., Nairn A. C., Sonenberg N., Bramham C. R. (2006) J. Neurochem. 99, 1328–1337 [DOI] [PubMed] [Google Scholar]
- 26.Messaoudi E., Kanhema T., Soulé J., Tiron A., Dagyte G., da Silva B., Bramham C. R. (2007) J. Neurosci. 27, 10445–10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messaoudi E., Ying S. W., Kanhema T., Croll S. D., Bramham C. R. (2002) J. Neurosci. 22, 7453–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying S. W., Futter M., Rosenblum K., Webber M. J., Hunt S. P., Bliss T. V., Bramham C. R. (2002) J. Neurosci. 22, 1532–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez J. J., Davies H. A., Silva A. T., De Souza I. E., Peddie C. J., Colyer F. M., Lancashire C. L., Fine A., Errington M. L., Bliss T. V., Stewart M. G. (2005) Eur. J. Neurosci. 21, 2384–2396 [DOI] [PubMed] [Google Scholar]
- 30.Steward O., Wallace C. S., Lyford G. L., Worley P. F. (1998) Neuron 21, 741–751 [DOI] [PubMed] [Google Scholar]
- 31.Gingras A. C., Raught B., Sonenberg N. (2004) Curr. Top. Microbiol. Immunol. 279, 169–197 [DOI] [PubMed] [Google Scholar]
- 32.Scheper G. C., van Kollenburg B., Hu J., Luo Y., Goss D. J., Proud C. G. (2002) J. Biol. Chem. 277, 3303–3309 [DOI] [PubMed] [Google Scholar]
- 33.English J. D., Sweatt J. D. (1997) J. Biol. Chem. 272, 19103–19106 [DOI] [PubMed] [Google Scholar]
- 34.Rosenblum K., Futter M., Voss K., Erent M., Skehel P. A., French P., Obosi L., Jones M. W., Bliss T. V. (2002) J. Neurosci. 22, 5432–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klann E., Dever T. E. (2004) Nat. Rev. Neurosci. 5, 931–942 [DOI] [PubMed] [Google Scholar]
- 36.Tang S. J., Reis G., Kang H., Gingras A. C., Sonenberg N., Schuman E. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelinas J. N., Banko J. L., Hou L., Sonenberg N., Weeber E. J., Klann E., Nguyen P. V. (2007) J. Biol. Chem. 282, 27527–27535 [DOI] [PubMed] [Google Scholar]
- 38.Bekinschtein P., Katche C., Slipczuk L. N., Igaz L. M., Cammarota M., Izquierdo I., Medina J. H. (2007) Neurobiol. Learn. Mem. 87, 303–307 [DOI] [PubMed] [Google Scholar]
- 39.Isotani S., Hara K., Tokunaga C., Inoue H., Avruch J., Yonezawa K. (1999) J. Biol. Chem. 274, 34493–34498 [DOI] [PubMed] [Google Scholar]
- 40.Navé B. T., Ouwens M., Withers D. J., Alessi D. R., Shepherd P. R. (1999) Biochem. J. 344, 427–431 [PMC free article] [PubMed] [Google Scholar]
- 41.Lehman J. A., Calvo V., Gomez-Cambronero J. (2003) J. Biol. Chem. 278, 28130–28138 [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Dong Z., Nomura M., Zhong S., Chen N., Bode A. M., Dong Z. (2001) J. Biol. Chem. 276, 20913–20923 [DOI] [PubMed] [Google Scholar]
- 43.Jefferies H. B., Fumagalli S., Dennis P. B., Reinhard C., Pearson R. B., Thomas G. (1997) EMBO J. 16, 3693–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stolovich M., Tang H., Hornstein E., Levy G., Cohen R., Bae S. S., Birnbaum M. J., Meyuhas O. (2002) Mol. Cell. Biol. 22, 8101–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waltereit R., Dammermann B., Wulff P., Scafidi J., Staubli U., Kauselmann G., Bundman M., Kuhl D. (2001) J. Neurosci. 21, 5484–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao V. R., Pintchovski S. A., Chin J., Peebles C. L., Mitra S., Finkbeiner S. (2006) Nat. Neurosci. 9, 887–895 [DOI] [PubMed] [Google Scholar]
- 47.Waskiewicz A. J., Flynn A., Proud C. G., Cooper J. A. (1997) EMBO J. 16, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyronnet S., Imataka H., Gingras A. C., Fukunaga R., Hunter T., Sonenberg N. (1999) EMBO J. 18, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bramham C. R., Alme M. N., Bittins M., Kuipers S. D., Nair R. R., Pai B., Panja D., Schubert M., Soule J., Tiron A., Wibrand K. (August19, 2009) Exp. Brain Res. 10.1007/s00221-009-1959-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang F., Chotiner J. K., Steward O. (2007) J. Neurosci. 27, 9054–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramírez-Amaya V., Vazdarjanova A., Mikhael D., Rosi S., Worley P. F., Barnes C. A. (2005) J. Neurosci. 25, 1761–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyashita T., Kubik S., Haghighi N., Steward O., Guzowski J. F. (2009) J. Neurosci. 29, 898–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larsen M. H., Olesen M., Woldbye D. P., Hay-Schmidt A., Hansen H. H., Rønn L. C., Mikkelsen J. D. (2005) Brain Res. 1064, 161–165 [DOI] [PubMed] [Google Scholar]
- 54.Vazdarjanova A., McNaughton B. L., Barnes C. A., Worley P. F., Guzowski J. F. (2002) J. Neurosci. 22, 10067–10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly M. P., Deadwyler S. A. (2003) J. Neurosci 23, 6443–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vazdarjanova A., Brennan A. M., Houston F. P., Worley P. F., Barnes C. A., Guzowski J. F. (2002) Soc. Neurosci. Abstr. 678.2 [Google Scholar]
- 57.Giorgi C., Yeo G. W., Stone M. E., Katz D. B., Burge C., Turrigiano G., Moore M. J. (2007) Cell 130, 179–191 [DOI] [PubMed] [Google Scholar]
- 58.Maquat L. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 89–99 [DOI] [PubMed] [Google Scholar]
- 59.Wang X., Flynn A., Waskiewicz A. J., Webb B. L., Vries R. G., Baines I. A., Cooper J. A., Proud C. G. (1998) J. Biol. Chem. 273, 9373–9377 [DOI] [PubMed] [Google Scholar]
- 60.Buxadé M., Morrice N., Krebs D. L., Proud C. G. (2008) J. Biol. Chem. 283, 57–65 [DOI] [PubMed] [Google Scholar]
- 61.Jung M. Y., Lorenz L., Richter J. D. (2006) Mol. Cell. Biol. 26, 4277–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Napoli I., Mercaldo V., Boyl P. P., Eleuteri B., Zalfa F., De Rubeis S., Di Marino D., Mohr E., Massimi M., Falconi M., Witke W., Costa-Mattioli M., Sonenberg N., Achsel T., Bagni C. (2008) Cell 134, 1042–1054 [DOI] [PubMed] [Google Scholar]
- 63.Tang H., Hornstein E., Stolovich M., Levy G., Livingstone M., Templeton D., Avruch J., Meyuhas O. (2001) Mol. Cell. Biol. 21, 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao L., Pilotte J., Xu T., Wong C. C., Edelman G. M., Vanderklish P., Yates J. R., 3rd (2007) J. Proteome Res. 6, 1059–1071 [DOI] [PubMed] [Google Scholar]
- 65.Huang F., Chotiner J. K., Steward O. (2005) J. Neurosci. 25, 7199–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamilton T. L., Stoneley M., Spriggs K. A., Bushell M. (2006) Biochem. Soc. Trans. 34, 12–16 [DOI] [PubMed] [Google Scholar]
- 67.Meyuhas O. (2000) Eur. J. Biochem. 267, 6321–6330 [DOI] [PubMed] [Google Scholar]
- 68.Ruvinsky I., Meyuhas O. (2006) Trends Biochem. Sci. 31, 342–348 [DOI] [PubMed] [Google Scholar]
- 69.Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. (2005) Genes Dev. 19, 2199–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belelovsky K., Kaphzan H., Elkobi A., Rosenblum K. (2009) J. Neurosci. 29, 7424–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casadio A., Martin K. C., Giustetto M., Zhu H., Chen M., Bartsch D., Bailey C. H., Kandel E. R. (1999) Cell 99, 221–237 [DOI] [PubMed] [Google Scholar]
- 72.Belelovsky K., Elkobi A., Kaphzan H., Nairn A. C., Rosenblum K. (2005) Eur. J. Neurosci. 22, 2560–2568 [DOI] [PubMed] [Google Scholar]
- 73.Naie K., Tsanov M., Manahan-Vaughan D. (2007) Eur. J. Neurosci. 25, 3264–3275 [DOI] [PubMed] [Google Scholar]
- 74.Wu J., Rowan M. J., Anwyl R. (2004) Neuroscience 123, 507–514 [DOI] [PubMed] [Google Scholar]
- 75.Kulla A., Manahan-Vaughan D. (2008) Hippocampus 18, 48–54 [DOI] [PubMed] [Google Scholar]
- 76.Waung M. W., Pfeiffer B. E., Nosyreva E. D., Ronesi J. A., Huber K. M. (2008) Neuron 59, 84–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.