Son is a large insoluble nuclear speckle protein that is required for appropriate nuclear speckle organization and cell cycle progression through mitosis. A region of unique repeat motifs in Son is the key domain necessary to mediate the proper localization of pre-mRNA processing factors and other nuclear speckle constituents.
Abstract
Subnuclear organization and spatiotemporal regulation of pre-mRNA processing factors is essential for the production of mature protein-coding mRNAs. We have discovered that a large protein called Son has a novel role in maintaining proper nuclear organization of pre-mRNA processing factors in nuclear speckles. The primary sequence of Son contains a concentrated region of multiple unique tandem repeat motifs that may support a role for Son as a scaffolding protein for RNA processing factors in nuclear speckles. We used RNA interference (RNAi) approaches and high-resolution microscopy techniques to study the functions of Son in the context of intact cells. Although Son precisely colocalizes with pre-mRNA splicing factors in nuclear speckles, its depletion by RNAi leads to cell cycle arrest in metaphase and causes dramatic disorganization of small nuclear ribonuclear protein and serine-arginine rich protein splicing factors during interphase. Here, we propose that Son is essential for appropriate subnuclear organization of pre-mRNA splicing factors and for promoting normal cell cycle progression.
INTRODUCTION
Cotranscriptional processing of pre-mRNAs in higher eukaryotic cells involves multiple biochemical processes, including capping, splicing, polyadenylation, and packaging into export-ready messenger ribonucleoprotein particles. Pre-mRNA processing factors themselves are compartmentalized in nuclear domains, including Cajal bodies and nuclear speckles in mammalian cell nuclei (reviewed in Gall, 2003; Lamond and Spector, 2003; Cioce and Lamond, 2005; Hall et al., 2006). Pre-mRNA processing machinery such as 5′ capping complexes and 3′ cleavage and polyadenylation factors and splicing factors, including small nuclear ribonuclear proteins (snRNPs) and serine-arginine rich (SR) splicing factors accumulate at transcription sites and perform their respective functions cotranscriptionally. This process is mediated by association of these factors with the carboxy-terminal domain of the largest subunit of RNA polymerase II that promotes their loading onto nascent pre-mRNAs (Misteli et al., 1999; Rosonina and Blencowe, 2004; reviewed in Hirose and Manley, 2000; Lewis and Tollervey, 2000; Phatnani and Greenleaf, 2004; Bentley, 2005; Corden, 2007; Egloff and Murphy, 2008). Many of these factors are either recruited to transcription sites from nuclear speckles, or they process messages near or in nuclear speckles (reviewed in Lamond and Spector, 2003; Hall et al., 2006).
Understanding the regulatory processes that control how and when pre-mRNA processing factors move into and out of nuclear speckles is a current challenge that will rely on learning more details about the molecular organization of nuclear speckles and the regulatory mechanisms that control individual speckle components. One model suggests that nuclear speckle assembly relies at least in part on “self-association” among components (Misteli, 2000). The dynamic exchange of one major family of splicing factors, the SR proteins, is modulated by phosphorylation of serines in arginine-serine-rich (RS) domains. Exogenous expression of SR protein kinases such as Clk/STY causes complete nuclear speckle disassembly and alters splicing efficiency (Duncan et al., 1995; Colwill et al., 1996; Yeakley et al., 1999; Prasad et al., 1999; Sacco-Bubulya and Spector, 2002). Proper nuclear speckle organization is important for coordinating transcription and pre-mRNA processing, because nuclear speckle integrity promotes cotranscriptional processing of pre- mRNAs (Sacco-Bubulya and Spector, 2002). We hypothesize that in addition to RS-RS domain interactions among splicing factors, specific nuclear speckle components could provide a platform for organizing pre-mRNA processing machinery. To understand more precisely the molecular aspects of nuclear speckle organization, we have begun to examine the nuclear speckle proteome for candidates that could perform structural roles in nuclear speckles.
Identification of nuclear speckle constituents by mass spectrometry was an important first step toward finding potential scaffolding candidates (Mintz et al., 1999; Saitoh et al., 2004). Nuclear speckle purification and proteomic analysis ultimately led to the identification of 178 nuclear speckle proteins, providing a more complete picture of nuclear speckle composition (Saitoh et al., 2004). These studies also categorized proteins based upon known or predicted functions of broad range, including those performing pre-mRNA processing, mRNA binding/packaging, and mRNA transport roles (Saitoh et al., 2004). Twelve percent of the identified proteins (∼33 proteins) are newly identified nuclear speckle proteins for which little biological information is available and that may have novel roles in nuclear speckles (Saitoh et al., 2004). Among the newly identified nuclear speckle proteins was a large 2530-amino acid protein called Son. Motif analysis showed that Son contains an RS domain, a double-stranded RNA binding domain, and a G-patch (Saitoh et al., 2004). Together with the association of Son with splicing factor SRrp53 (Cazalla et al., 2005), this implicates Son in some aspect of pre-mRNA processing.
Perhaps the most interesting characteristic of Son is that approximately one third of the primary sequence is comprised of at least six different types of tandem repeats that are unique to Son (Figure 1) and have been described previously (Wynn et al., 2000; Sun et al., 2001; Saitoh et al., 2004). Highly repetitive serine-rich sequence motifs have previously been shown to have scaffolding functions, as demonstrated by the repeated heptad sequence (∼YSPTSPS) in the carboxy-terminal domain of the largest subunit of RNA polymerase II at transcription sites (reviewed in Hirose and Manley, 2000; Lewis and Tollervey, 2000; Phatnani and Greenleaf, 2004; Bentley 2005; Egloff and Murphy, 2008). Because Son has multiple serine rich repeats and an RS domain, we hypothesized that Son might serve a similar scaffolding role in nuclear speckles.
Figure 1.
Sequence motifs in Son. The amino-terminal half of Son contains multiple novel sequence motif repeats that are unique to Son. Repeats composed of unique sequences are shown here as sets of different colored bars, with each bar representing an individual repeat. The carboxy-terminal region of Son contains motifs such as a RS domain, a double-stranded RNA binding domain, and a G patch. Amino acids corresponding to C-terminal deletion constructs are indicated.
We sought to determine whether Son is a critical component for the organization of splicing factors into nuclear speckles. Toward this end, we used an RNA interference approach to deplete Son in HeLa cells. Nuclear speckles in Son-depleted cells showed a striking disorganization of pre-mRNA processing factors. This is the first report to demonstrate a significant change in the overall organization of nuclear speckles after the depletion of a single nuclear speckle protein. In addition, Son depletion resulted in a significant decrease in cell growth, and cells were arrested in metaphase. Our data supports a role for Son in cell cycle progression as well as a novel role for Son as a key nuclear speckle scaffolding component that is essential for the structural maintenance of nuclear speckles.
MATERIALS AND METHODS
cDNA Constructs
Son cDNA sequence (KIAA1019) was obtained from the HUGE consortium, Japan (Saitoh et al., 2004). The original KIAA1019 (Saitoh et al., 2004) was truncated at the 5′ end but is complete in the updated KIAA1019 cDNA now available from the HUGE consortium. Because the 3′ end of the updated KIAA1019 cDNA does not encode the G-patch or double-stranded RNA-binding domain (DS RBD) motifs that are present in cellular Son transcripts, we substituted the correct 3′ end from a separate partial Son cDNA clone (clone fh21047 obtained from T. Nagase, HUGE consortium). The full-length Son cDNA open reading frame was then subcloned into pEYFP-C1 (Takara, Kyoto, Japan). To produce a yellow fluorescent protein (YFP)-tagged small interfering RNA (siRNA)-refractory Son mRNA (YFP-siR-Son), the open reading frame near the 5′ end of YFP-Son cDNA that corresponds to the nucleotides targeted by Son siRNA 4 was changed at seven nucleotide positions by site-directed mutagenesis using forward oligonucleotide (oligo) 5′-gccagttgtaacCatgAGCgtCgaAtaCcagatgaagtctg-3′ and reverse oligo 5′-cagacttcatctgGtaTtcGacGCTcatGgttacaactggc-3′ (the siRNA-targeted region is underlined and all mutagenized nucleotides are indicated in uppercase letters). Convenient restriction sites were used for subcloning Son deletion mutants into pEYFP-C1. The sequence of all polymerase chain reaction (PCR) products was confirmed by DNA sequencing. YFP-Magoh and YFP-MLN51 cDNAs were a kind gift from Catherine Tomasetto (Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France).
Cell Culture
HeLa cells and HeLa S3 cells were maintained in DMEM supplemented with penicillin/streptomycin and 10% fetal bovine serum. HeLa cells stably expressing YFP-SF2/ASF and SC35-YFP were described previously (Prasanth et al., 2003; Bubulya et al., 2004). For transcription inhibition, α-amanitin (50 μg/ml) was added for 6 h before harvesting cells.
Preparation of the Nuclear Insoluble Fraction
S3 cells (5.0 × 106) were incubated at 4°C for 3 min in cytoskeleton (CSK) buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-NaOH, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. For extracts of Son-depleted cells, cells were first transfected with 100 nM Son siGENOME1 or siGENOME4 48 h before preparation of the nuclear insoluble fraction. After centrifugation at 600 × g for 3 min, the pellets were treated with DNase I at 200 U/ml at 25°C for 30 min in digestion buffer (10 mM PIPES-NaOH, pH 6.8, 50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, and 1.2 mM PMSF). Ammonium sulfate was then added to the digested mixtures from a 1 M stock solution to a final concentration of 0.25 M, and the mixtures were incubated at 25°C for 5 min, followed by centrifugation at 600 × g for 3 min. For further extraction, the pellets were extracted in the digestion buffer containing 2 M NaCl by incubation at 4°C for 20 min, followed by centrifugation at 600 × g for 3 min. To remove RNA related components, the pellet was incubated with 0.2 mg/ml RNase A in the digestion buffer at 25°C for 30 min, and the mixtures were pulled down by centrifugation at 600 × g for 3 min to obtain a final fraction (high salt nuclear insoluble fraction). The fraction was directly dissolved in SDS sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 1% β-mercaptoethanol) for one-dimensional-SDS-polyacrylamide gel electrophoresis (PAGE).
Antibodies, Immunofluorescence, and Immunoblots
We have generated affinity-purified rabbit polyclonal anti-Son antibodies at Covance (Denver, PA). Two polyclonal antisera (WU 14 and WU15) were directed against the peptide CEESESKTKSH located near the amino terminus of Son. For immunofluorescence, HeLa cells were fixed with 2% formaldehyde, washed three times for 5 min each in phosphate-buffered saline (PBS), and then permeabilized with 0.2% Triton X-100 in PBS. After 3 × 5 min washes in PBS 0.5% normal goat serum (NGS), cells were incubated with primary antibodies WU14 (1:1000), WU15 (1:15,000), monoclonal anti-SF2/ASF AK103 (1:2500; provided by A. Krainer, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), monoclonal anti-U1-70K 2.73 (1:100; provided by S. Hoch, Agouron Institute, La Jolla, CA) and polyclonal anti-pinin A301-022A (1:1000; Bethyl Laboratories, Montgomery, TX), human anti-nucleolar antibody (anti-fibrillarin, 1:20; Sigma-Aldrich, St. Louis, MO), and monoclonal anti-PML 5E10 (1:20; provided by R. van Driel, University of Amsterdam, Amsterdam, The Netherlands) for 1 h at room temperature. Cells were washed three times for 5 min with PBS, 0.5% NGS and then incubated in fluorescently conjugated secondary antibodies (1:500; Jackson ImmunoResearch Laboratories, West Grove, PA). Peptide blocking experiments were performed by preincubation of anti-Son antibodies with 10 μM N-terminal Son peptide on a rotating platform for 1 h at room temperature before addition of the antibody to the cells. 5-Bromouridine 5′-triphosphate (bromo-UTP) labeling and oligo(dT) RNA-fluorescence in situ hybridization (FISH) were performed as described previously (Sacco-Bubulya and Spector, 2002). For imaging of fixed material, z-stacks spanning the entire nuclei were acquired on a DeltaVision RT using a 60× objective (1.4 numerical aperture; Olympus, Tokyo, Japan), and raw images were displayed as volume projections.
For immunoblotting of nuclear insoluble fractions, 10 μg of protein from each fraction were separated on a 4–20% gradient polyacrylamide gel and transferred into polyvinylidene difluoride membrane at 70 V for 3 h. Signals were detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Waltham, MA). For all other immunoblots, cells were extracted in Laemmli buffer, and equal amounts of protein were applied to 7% SDS-PAGE. Proteins were transferred to nitrocellulose membrane, blocked in PBS containing 0.1% Tween 20 and 5% nonfat dry milk and probed with primary antibodies (anti-Son WU15, anti-Son WU14, and anti-Lamin B1 were used at 1:1000; anti U1-70K and anti-SF2/ASF were used at 1:100; anti-β-actin [Sigma-Aldrich] was used at 1:10,000; and anti-histone H3 phospho S10 (Abcam, Cambridge, United Kingdom) was used at 1:500). Anti-rabbit immunoglobulin Ig (IgG) horseradish peroxidase (HRP)-conjugated antibody was used at 1:40,000 for blotting of the nuclear insoluble fraction, and anti-mouse and anti-rabbit IgG HRP-conjugated antibodies (Jackson ImmunoResearch Laboratories) were used at 1:50,000 for all other immunoblots.
RNA Interference (RNAi)
Predesigned siGENOME siRNAs directed against Son were purchased from Dharmacon RNA Technologies (Lafayette, CO) (catalog no. D-012983-01, target sequence GCUGAGCGCUCUAUGAUGU and catalog no. D-012983-04, target sequence CAAUGUCAGUGGAGUAUCA). HeLa cells (3 × 104) were plated in antibiotic free medium and transfected by using Oligofectamine (Invitrogen, Carlsbad, CA) alone (mock) or mixed with siRNAs against luciferase (60 pmol; control; catalog no. D-001210-02; Dharmacon RNA Technologies) or Son siRNAs (60 pmol). For siRNA rescue experiments, 10 μg each of YFP-siR-Son or YFP-Son plasmid was transfected using the calcium phosphate method. After a 48-h recovery period, the cells were transfected with siRNAs and harvested for analysis 48 h later. To assess Son mRNA knockdown by semiquantitative reverse transcription (RT)-PCR, reverse transcriptase reactions were performed on 100 ng of total RNA (SuperScript One-Step RT-PCR System; Invitrogen). The linear range for amplification was verified, after which 32 cycles were used to amplify both Son and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs.
Live Cell Microscopy
HeLa cells (3 × 105) stably expressing YFP-SF2/ASF were seeded onto 60-mm coverslips 24 h before RNAi. siRNA-mediated depletion of Son was done by transfection with siGENOME4. Thirteen hours after transfection, the cells were transferred to a live imaging chamber (Bioptechs, Butler, PA), perfused with L-15 medium (without phenol red) containing 20% fetal bovine serum, and placed into a 37°C environmental chamber on the stage of a DeltaVision RT imaging system. Nuclear speckle dynamics were observed by acquiring z-stacks through the entire nucleus every 5 min for at least 6 h.
Cell Growth and Flow Cytometry Analysis
For cell growth/cell cycle analysis, HeLa cells were plated in quadruplicate for a time course of both ViCell counts and flow cytometry analysis at the time of siRNA treatment and every 24 h thereafter. Cells treated in parallel were harvested 48 h after siRNA transfection for Western blots or to confirm Son knockdown by RT-PCR. A small coverslip was included in each set of cell growth and flow cytometry experiments and processed for immunofluorescence to confirm change in nuclear speckle morphology.
RESULTS
Son Is a Bona Fide Nuclear Speckle Protein
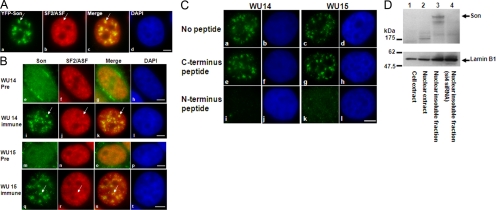
When Son was first identified in the nuclear speckle proteome, a 5′-truncated cDNA (KIAA1019) was subcloned into pEYFP, and the YFP-KIAA1019 fusion protein was expressed in HeLa cells (Sacco-Bubulya and Spector, 2002; Saitoh et al., 2004). YFP-KIAA1019 showed a precise overlay with splicing factor SC35, confirming its localization in nuclear speckles (Saitoh et al., 2004). The full-length sequence of Son has not previously been assembled. Here, we have generated a full-length Son that is fused with YFP, and expression of this fusion in HeLa cells shows precise localization with the SR splicing factor SF2/ASF in nuclear speckles (Figure 2A), consistent with previous reports of Son localization (Wynn et al., 2000; Saitoh et al., 2004). Furthermore, to ensure that endogenous Son is localized in nuclear speckles, we generated two rabbit polyclonal antibodies, designated as WU14 and WU15, against peptides corresponding to sequence near the amino terminus of Son. Immunolabeling of HeLa cells with affinity purified anti-Son antibody WU14 and WU15 shows colocalization with SF2/ASF in nuclear speckles (Figure 2B). Antibody specificity was shown by peptide competition assay in an immunofluorescence experiment (Figure 2C). In addition, the labeling of nuclear speckles with anti-Son WU14 antibody was consistently diminished in cells treated with siRNAs that target Son mRNA (Figures 3 and 4; data for WU15 were identical and are not shown). Together, the results of these experiments demonstrate specificity of our WU14 and WU15 anti-Son antibodies. Interestingly, proteomic analysis of insoluble proteins from HeLa-S3 cell nuclei revealed Son as one of the largest proteins in this fraction (Takata et al., 2009). An immunoblot with anti-Son antibody WU15 shows specific reactivity with a large protein that is enriched in a nuclear insoluble fraction of HeLa cells (Figure 2D, lane 3) compared with whole cell extract (Figure 2D, lane 1) or nuclear extract (Figure 2D, lane 2). As expected, there was enrichment of the nuclear lamina protein lamin B1 in the insoluble nuclear fraction as shown by immunoblot. Treatment of HeLa cells with Son siRNAs causes disappearance of the band that corresponds to Son (Figure 2D, lane 4), which is further evidence that the WU15 anti-Son antibody reactivity is specific to Son (similar results were obtained using WU14 anti-Son antibody; data not shown). Preincubation of the anti-Son antibodies with the peptide used to generate WU14 and WU15 blocks reactivity with Son by immunoblot, demonstrating antibody specificity (data not shown).
Figure 2.
Son is a component of nuclear speckles that is enriched in insoluble nuclear fractions. (A) Localization of YFP-Son in HeLa cells stably transfected with YFP-tagged full-length Son cDNA and processed for immunolocalization of splicing factor SF2/ASF. YFP-Son (a) colocalizes with SF2/ASF (b) at nuclear speckles (arrow). (B) Immunofluorescence using rabbit polyclonal anti-Son WU14 antibody shows that WU14 labeling (i, arrow) and WU15 labeling (q, arrow) precisely colocalize with labeling of splicing factor SF2/ASF (arrows in j and r) at nuclear speckles, whereas WU14 preimmune serum (e) and WU15 preimmune serum (m) do not label any specific nuclear domain. (C) Peptides used to generate Son antisera block immunoreactivity. The peptide corresponding to amino acid sequence in the amino terminus of Son that was used to generate the WU14 and WU15 rabbit polyclonal anti-Son antibodies blocks immunoreactivity of WU14 and WU15 by immunofluorescence. Blocking with 10 μM amino-terminal Son peptide before incubation of HeLa cells with WU14 antibody blocks WU14 immunolabeling (Ci) and WU15 immunolabeling (Ck) at nuclear speckles. Reactions without peptide (C, a and c) and with unspecific C-terminal Son peptide (C, e and g) do not block WU14 immunolabeling. (D) HeLa-S3 cells fractionated to yield nuclear insoluble proteins contain Son. An immunoblot with anti-Son antibody WU15 shows reactivity with a large protein that is enriched in a nuclear insoluble fraction of HeLa cells (lane 3) compared with whole cell extract (lane 1) or nuclear extract (lane 2). The bottom band could be an alternate splice form or breakdown product of Son. Treatment of HeLa cells with siRNA duplexes that deplete Son specifically reduced labeling of these bands (lane 4). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Bar, 5 μm.
Figure 3.
siRNA-mediated depletion of Son alters the localization of U1-70K snRNP protein. HeLa cells were transfected with empty vehicle (mock), luciferase siRNA (control), Son siRNA 1, or Son siRNA 4. 24 h posttransfection, the cells were fixed and processed for dual immunolocalization of Son and U1-70K. Son signal is nearly absent after Son depletion (i and m). U1-70K localization in nuclear speckles was altered after depletion of Son with siRNA 1 (j, arrow) but not with mock (b, arrow) or control (f, arrow) siRNAs. siRNA-mediated depletion of Son by using Son siRNA 4 showed the same alteration in U1-70K nuclear speckle localization (n, arrow) compared with mock and control siRNAs, but did not alter the U1-70K distribution in Cajal bodies (n, arrowhead). DNA was stained with DAPI. Bar, 5 μm.
Figure 4.
siRNA-mediated depletion of Son alters the localization of SF2/ASF. (A) SF2/ASF localization is altered after siRNA-mediated Son depletion. HeLa cells were treated with empty vehicle (mock), luciferase siRNA (control), or Son siRNA 1. Twenty-four hours posttransfection, HeLa cells were fixed and processed for dual immunolocalization of Son and SF2/ASF. Son signal is nearly absent after Son depletion (i and m). SF2/ASF localization in nuclear speckles was altered after depletion of Son with siRNA 1 (j, arrow) but not with mock (b, arrow) or control (f, arrow) siRNAs. siRNA-mediated depletion of Son by using Son siRNA 4 showed the same alteration in SF2/ASF nuclear speckle localization (n, arrow) compared with mock and control siRNAs. DNA was stained with DAPI. Bar, 5 μm. (B) Son expression is reduced after RNAi. Semiquantitative RT-PCR showed a significant reduction in Son mRNA expression after siRNA-mediated Son depletion (lanes 4 and 8), whereas it was not reduced in cells treated with mock transfection (lanes 2 and 6) or control siRNAs (lanes 3 and 7). Note that GAPDH mRNA was not reduced after any of the treatments (lanes 2–4 and 6–8). Neither mRNA was detected when reverse transcriptase was omitted from the reaction (lanes 1 and 5).
Nuclear Speckle Organization of pre-mRNA Splicing Factors Requires Son
Immunofluorescence experiments and YFP-Son expression show the predominant localization of Son in nuclear speckles, with little detectable diffuse nuclear signal (Figure 2, A and B). We did not detect Son in other nuclear domains such as PML bodies, nucleoli, or Cajal bodies (Supplemental Figure 1). Based on motif analysis and localization studies, we hypothesized that Son may be a structural component of the nuclear speckles. If Son is essential for nuclear speckle organization, we would expect knockdown of Son to significantly alter the distribution of pre-mRNA processing factors. We used RNAi to knock down the expression of Son in HeLa cells, and we evaluated changes in the distribution of pre-mRNA processing factors. U1-70K small nuclear ribonuclear protein (snRNP) protein localization was dramatically altered in the nuclear speckles in cells that showed significant reduction of Son immunostaining after Son depletion compared with controls (Figure 3). Son depletion by two separate siRNAs caused a significant reduction of Son immunostaining (Figure 3, i and m) as well as a complete reorganization of U1-70K to round, doughnut-shaped nuclear structures (Figure 3, j and n, arrows). Importantly, Son depletion did not alter the U1-70K distribution in Cajal bodies that are morphologically distinct from the nuclear speckles (Figure 3n, arrowhead). Similar data for nuclear speckle reorganization was obtained using human anti-Sm autoimmune serum and Y12 anti-snRNP antibody (data not shown). These data demonstrate that although snRNPs do not require Son to maintain their localization in Cajal bodies, their organization in nuclear speckles requires that Son maintain nuclear speckle structure.
Nuclear distribution of SR proteins SF2/ASF and SC35 was also altered by Son depletion. Figure 4A shows that SF2/ASF localization in nuclear speckles was altered after depletion of Son with siRNAs (Figure 4, j and n, arrows) but not with mock transfection (Figure 4b, arrow) or control siRNA (Figure 4f, arrow). We also observed similar results in other cell lines including IMR90 and NIH 3T3 (data not shown). Identical results were seen with HeLa cells expressing YFP-SF2/ASF (see Figure 9). The Son depletion phenotype was identical for two additional siRNAs (data not shown). The similar outcome for Son depletion using four separate siRNAs, each of which target a different region of Son mRNA, suggests that the alteration in nuclear speckle phenotype is not an off-target effect of RNA interference. HeLa cells stably expressing SC35-YFP were also treated with Son siRNAs and processed for immunolocalization of Son. SC35-YFP localization in nuclear speckles was altered to a doughnut-shaped pattern after depletion of Son (Supplemental Figure 2). Identical results were observed for the splicing factor pinin (see Supplemental Figure 2). A representative example of semiquantitative RT-PCR used to verify Son knockdown is shown in Figure 4B. Son mRNA is significantly reduced only after treatment with Son siRNAs (Figure 4B, top, lanes 4 and 8) but not after mock transfection (Figure 4B, top, lanes 2 and 6) or treatment with control siRNA (Figure 4B, top, lanes 3 and 7). Although partially reduced expression of Son at 24 h after RNAi was always sufficient to observe changes in nuclear speckle organization, continued decrease in Son expression was typically observed over 48 h. Notably, the reorganization of nuclear speckle components is not limited to splicing factors. We expressed YFP-tagged exon junction complex (EJC) central core proteins Magoh and MLN51 (Degot et al., 2004; Baguet et al., 2007) in HeLa cells to observe their response to Son depletion. Both proteins localized in nuclear speckles (Figure 5, a–h and m–t), and became redistributed into doughnut-shaped nuclear speckles after Son depletion (Figure 5, i–l and u–x).
Figure 9.
Nuclear speckles reorganize directly to a doughnut-like morphology after Son depletion. Time-lapse microscopy was performed on HeLa cells that stably express YFP-SF2/ASF and were transfected with Son siRNA 4. The inset in a indicates the region of interest enlarged in the bottom panels. Live imaging was initiated 13 h after siRNA transfection. The morphology of nuclear speckles changed directly from an irregular compacted organization (a, arrows), to an open doughnut-shaped organization (f, arrows). Time after siRNA transfection is indicated in hours. Video of this data set shows the transition from speckles to doughnuts (see Supplemental Movie). Bar, 5 μm.
Figure 5.
siRNA-mediated depletion of Son alters the localization of core EJC proteins. (A) YFP-MLN51 localization is altered after siRNA-mediated Son depletion. HeLa cells were treated with empty vehicle (mock), luciferase siRNA (control), or Son siRNA 4. Twenty-four hours posttransfection, HeLa cells were fixed and processed for immunolocalization of Son. Son signal is nearly absent after Son depletion (i). YFP-MLN51 localization in nuclear speckles was altered after depletion of Son with siRNA 4 (j, arrow) but not with mock (b, arrow) or control (f, arrow) siRNAs. (B) YFP-Magoh localization is altered after siRNA-mediated Son depletion. HeLa cells were treated with empty vehicle (mock), luciferase siRNA (control) or Son siRNA 4. Twenty-four hours posttransfection, HeLa cells were fixed and processed for immunolocalization of Son. Son signal is nearly absent after Son depletion (u). YFP-Magoh localization in nuclear speckles was altered after depletion of Son with siRNA 4 (v, arrow) but not with mock (n, arrow) or control (r, arrow) siRNAs. DNA was stained with DAPI. Bar, 5 μm.
Nuclear speckles and associated nuclear domains such as paraspeckles contain polyadenylated RNA species whose identities and functions are only beginning to be characterized. Oligonucleotide [oligo(dT)] probe has been used to detect stable polyA+ RNA in nuclear speckles (Huang et al., 1994) and was used here to monitor the response of nuclear speckle polyA+ RNA after Son knockdown. Although there was approximately equal labeling of polyA+ RNA in all conditions, Son knockdown caused dramatic redistribution of nuclear speckle polyA+ RNA (Figure 6, e–h). The similar effects of Son depletion on polyA+ RNA, snRNPs, SR proteins, pinin, and EJC core proteins suggest that Son may have general organizational functions for pre-mRNA processing factors as well as other constituents of the nuclear speckles.
Figure 6.
siRNA-mediated depletion of Son alters the localization of nuclear speckle polyA+ RNA. HeLa cells were treated with luciferase siRNA (control), or Son siRNA 4. Twenty-four hours posttransfection, HeLa cells were fixed and processed for immunolocalization of SF2/ASF to monitor nuclear speckle phenotype and RNA FISH with oligo(dT) probe to detect polyA+ RNA. Oligo(dT) probe localization in nuclear speckles was altered after depletion of Son with siRNA 4 (e, arrow) in the same way as SF2/ASF (f, arrow), whereas both showed normal nuclear speckle localization after treatment with control siRNA (a–d). DNA was stained with DAPI. Bar, 5 μm.
Because indirect effects on nuclear speckle organization might result if Son depletion alters transcription, it is important to determine whether the onset of pre-mRNA processing factor redistribution is a result of decreased global gene activity in Son-depleted cells. To address this, we performed bromo-UTP incorporation in living HeLa cells treated with Son siRNAs for 24 h. Labeling of transcripts was equivalent in Son depleted and control cells, indicating that global transcription proceeds robustly despite nuclear speckle reorganization (Figure 7A). Furthermore, if transcription were inhibited under conditions of Son depletion, we would expect to see pre-mRNA machinery in large rounded speckles and not as doughnut shapes. We confirmed that transcription inhibition of RNA polymerase II by α-amanitin treatment does not cause Son or SF2/ASF to exhibit a doughnut-like morphology. Rather, it leads to the expected larger round nuclear speckle organization of these factors (Figure 7B). We conclude that transcription activity is not reduced at the time that nuclear speckles reorganize after Son depletion, ruling out transcription-related secondary effects.
Figure 7.
Nuclear speckle reorganization after Son depletion is not a result of reduced global transcription, and transcription inhibition causes a different phenotype. (A) Global labeling of transcription sites in situ. HeLa cells were treated with control siRNA (a–d) or Son siRNAs (e–l) for 24 h and then permeabilized for uptake of Br-UTP to label nascent transcripts. Bromo-UTP incorporation was equally detectable in nuclei of Son-depleted cells (f and j) and cells treated with contol siRNA (b). Bromo-UTP labeling was absent in cells treated with α-amanitin (n). (B) Son reorganization after RNA pol II transcription inhibition. HeLa cells were treated with alpha-amanitin for 6 h and processed for immunolocalization of Son and U1-70K. U1-70K and Son redistributed to enlarged, rounded speckles upon inhibiting RNA polymerase II with α-amanitin (u, arrow) compared with untreated cells (q, arrow). DNA was stained with DAPI. Bar, 5 μm.
Immunofluorescence labeling of splicing factors in the nucleoplasm sometimes seemed reduced when the doughnut-shaped nuclear speckles were observed after Son depletion. To determine whether splicing factors are destabilized by disruption of nuclear speckle organization, HeLa cells treated with siRNAs against Son for 48 h were extracted for immunoblot analysis of splicing factor abundance. Expression of splicing factors U1-70K and SF2/ASF was not significantly different in extracts from Son-depleted cells (Figure 8A, lanes 3 and 4) compared with cell treated with mock transfection (Figure 8, lane 1) or control siRNA (Figure 8, lane 2). Son mRNA was nearly absent after treatment of cells with Son siRNAs (Figure 8, lanes 8 and 9), whereas it was not reduced in cells treated with mock transfection (Figure 8, lane 6) or control siRNA (Figure 8, lane 7). These data suggest that the decrease in immunofluorescence signal of splicing factors observed after morphological alteration of Son-depleted nuclear speckles is due to general redistribution of these proteins and not to decreased protein levels.
Figure 8.
Pre-mRNA splicing factor protein expression level is not altered by Son depletion. (Top) HeLa cells treated with siRNAs against Son were extracted for immunoblot analysis of splicing factors. Expression of splicing factors U1-70K and SF2/ASF were not significantly reduced in extracts from Son depleted cells (lanes 3 and 4) compared with cell treated with mock (lane 1) and control (lane 2) siRNAs. The actin immunoblot is shown to demonstrate equal loading of total protein in each lane. (Bottom) Son depletion was confirmed by semiquantitative RT-PCR. A duplicate siRNA depletion experiment was performed in parallel to confirm complete reduction in Son mRNA expression after 48 h of siRNA-mediated Son depletion. Son mRNA was nearly abolished after treatment of cells with Son siRNA 1 (lane 8) and Son siRNA 4 (lane 9), whereas it was not reduced in cells treated with mock (lane 6) or control (lane 7) siRNAs. GAPDH mRNA was not reduced after any of the treatments (lanes 6–9). Neither mRNA was detected when reverse transcriptase is omitted from the reaction (lane 5).
Nuclear Speckle Disorganization Occurs Quickly and Directly upon Son Depletion
Immunofluorescence experiments performed on Son-depleted cells indicated that the redistribution of pre-mRNA splicing factors into a doughnut-shaped structure began reproducibly across the entire coverslip of cells at ∼14–16 h after siRNA treatment when Son expression begins to decline (data not shown). We performed time-lapse microscopy to determine whether the nuclear speckles completely disassembled and then reorganized into these structures, or whether each nuclear speckle remained intact but simply reorganized directly to a different shape. Live imaging of HeLa cells stably expressing YFP-SF2/ASF showed that morphologically normal nuclear speckles change their organization directly into a “doughnut” over time (Figure 9). At 13 h post-siRNA treatment, the YFP-SF2/ASF is distributed in nuclear speckles (Figure 9a) and then begin to reorganize into “doughnuts” at ∼15–16 h (Figure 9, c and d), which are completely formed by 18 h after Son depletion is initiated (Figure 9f). A time-lapse video of YFP-SF2/ASF localization showed direct transition from nuclear speckles to doughnut-shaped structures (see Supplemental Movie S1). Importantly, this morphological change over time was not seen when YFP-SF2/ASF HeLa cells were treated with control siRNA (data not shown). Time-lapse imaging has therefore ruled out a disassembly/reassembly model for the structural alteration observed after Son depletion. Furthermore, it indicates that the onset of morphological change in nuclear speckle structure occurs within a few hours of the onset of Son depletion.
The Unique Son Repeat Domain Rescues Nuclear Speckle Organization in Cells Depleted of Endogenous Son
Seven silent mutations were introduced into the nucleotide sequence of YFP-tagged Son cDNA (see Materials and Methods) in the 5′ coding region that corresponds to the position in Son mRNA targeted by Son siRNA 4 to produce a siRNA-refractory YFP-tagged Son (referred to here as YFP-siR-Son). Expression of YFP-Son-siR in HeLa cells showed that it colocalizes with SF2/ASF in nuclear speckles (Figure 10A), which is expected because it has the same amino acid sequence as YFP-Son. Experiments were performed to analyze SF2/ASF distribution in Son-depleted cells that are protected by expression of YFP-siR-Son (Figure 10A). As expected, treatment of cells with control siRNA (Figure 10, a–d) did not affect distribution of SF2/ASF. Treatment of cells with Son siRNA 4 yielded doughnut-like redistribution of SF2/ASF in cells that were not protected by siRNA-resistant Son (Figure 10, e–h, arrow), while yielding normal nuclear speckle localization for SF2/ASF in cells that were protected by expression of YFP-siR-Son (Figure 10, e–h, the top cell is transfected with YFP-siR-Son). In addition, YFP-siR-Son expression did not rescue SF2/ASF distribution in cells treated with siRNA 1, because the region of Son targeted by this oligo has not been modified to resist its effects (data not shown). Likewise, YFP-Son could not rescue SF2/ASF distribution because it was targeted by siRNAs (Figure 10B, lanes 4 and 5, top; immunofluorescence data not shown). Semiquantitative RT-PCR was used to determine the levels of exogenous and endogenous Son expression in this experiment. RT-PCR to detect the YFP portion of exogenously expressed constructs verified siRNA-resistant expression of YFP-siR-Son (Figure 10, lane 10, top), whereas RT-PCR against endogenous Son shows depletion of endogenous Son (Figure 10B, lane 10, middle). YFP-siR-Son resisted depletion by siRNA 4 but not siRNA 1 (Figure 10B, compare lanes 9 and 10 in upper panel). Both endogenous Son and nonresistant YFP-Son were reduced by both siRNAs (Figure 10B, lanes 4 and 5). This set of experiments demonstrates that siRNA-resistant Son expression prevents alterations in nuclear speckle phenotype in Son-depleted cells.
Figure 10.
Expression of siRNA-resistant Son rescues nuclear speckle organization in cells depleted of endogenous Son. HeLa cells transfected with plasmid encoding YFP-tagged siRNA-resistant Son (YFP-siR-Son) were plated for siRNA treatment. The YFP tag allowed visualization of exogenous Son in transiently transfected cells, as well as detection of exogenous Son mRNA by RT-PCR oligos that target the YFP-encoding region. Endogenous Son was detected by RT-PCR oligos that target the 3′-untranslated region that is absent in YFP-Son and YFP-siR-Son. Duplicate experiments were performed to simultaneously analyze nuclear speckle phenotype (A) and endogenous and siRNA-resistant Son expression (B). (A) YFP-siR-Son (a–h) and YFP-siR-Son (1-1493; i–p) colocalized with SF2/ASF in nuclear speckles. Treatment of cells with control siRNAs (a–d and i–l) did not affect speckle morphology. Treatment with Son siRNA 4 showed the expected nuclear speckle reorganization (i–l, arrow), but neighboring cells protected by YFP-siR-Son FL or YFP-siR-Son (1-1493) expression showed a normal nuclear speckle phenotype (YFP-positive cells in e–f and m–n, respectively). (B) YFP-siR-Son resists depletion by Son siRNA 4. Semiquantitative RT-PCR was performed to target the YFP-encoding region that amplifies both YFP-Son and YFP-siR-Son (YFP, top row), the 3′-untranslated region of endogenous Son mRNA (Son, middle row), or GAPDH (bottom row). HeLa cells were transfected with either YFP-Son (YFP-Son; left set of panels), siRNA resistant YFP-Son (YFP-siR-Son FL, middle set of panels), or siRNA-resistant Son repeats (YFP-siR-Son 1-1493, right set of panels) before siRNA treatment. YFP-Son was used as a control for evaluating siRNA depletion of a non-siRNA resistant YFP-tagged Son mRNA by RT-PCR only. Individual RT-PCR reactions were performed on total RNA from each sample. Endogenous Son mRNA is significantly reduced after siRNA treatment with Son-specific siRNA 4 (middle row, lanes 4–5, 9–10, and 14–15) but not with mock transfection or control siRNAs (middle row, lanes 2–3, 7–8, and 12–13). While the nonresistant YFP-Son was reduced by treatment with both Son siRNAs (top row, left panel, lanes 4–5), the siRNA resistant YFP-siR-Son constructs were not reduced by treatment with siRNA 4 (top row, lanes 10 and 15) but were depleted by siRNA 1 (top row, lanes 9 and 14). GAPDH mRNA was not reduced after any of the treatments (bottom row, lanes 2–5, 7–10, and 12–15). No mRNA was detected when reverse transcriptase was omitted from the reaction (lanes 1, 6, and 11).
To address our hypothesis that the unique repeats of Son mediate proper nuclear speckle organization, similar experiments were performed to determine whether the repeat region of Son could rescue nuclear speckle organization in Son-depleted cells. Several C-terminal deletion constructs were made in YFP-siR-Son at amino acid positions indicated on Figure 1. These constructs were transiently expressed before Son depletion to determine whether they could protect nuclear speckles from reorganizing. Importantly, cells that expressed YFP-siR-Son amino acids 1-1493 containing the unique repeat region (Rpt) of Son showed unaltered nuclear speckle localization for SF2/ASF after Son depletion (Figure 10, m–p, the cell on the right is transfected with YFP-siR-Son repeats) compared with controls (Figure 10, i–l). RT-PCR confirmed that the YFP-siR Son resisted depletion by siRNA 4 (Figure 10, lane 15, top), whereas endogenous Son was depleted (Figure 10, lane 15, middle). An intermediate deletion construct encoding amino acids 1-2008 includes the RS domain but lacks the G-patch and DS RBD and was also able to rescue nuclear speckle organization (Supplemental Figure 3); however, because amino acids 1-1493 are sufficient for rescue, the RS domain, G-patch, and DS RBD of Son are clearly dispensable. Finally, a construct comprised of the extreme amino terminus of Son (amino acids 1-334) did not rescue nuclear speckle organization (Supplemental Figure 3). We therefore conclude that the unique repeat region of Son mediates proper nuclear speckle organization.
Son Is Essential for Cell Viability and Cell Cycle Progression
Treatment of HeLa cells with four separate siGENOME oligos (Dharmacon RNA Technologies) directed against Son leads to a significant reduction of Son mRNA and protein by 24 h posttransfection. To determine longer term effects of Son depletion, we performed a time course of siRNA treatment. Visual observations by phase contrast microscopy indicated that longer siRNA treatments correlated with reduced cell numbers, suggesting that Son depletion interferes with cell growth. This is consistent with a previous report that also noted Son is required for cell growth (Ahn et al., 2008). However, exactly which phase of the cell cycle is affected has not been determined. To begin to understand the effects of Son depletion on cell viability and cell cycle, we performed cell counts in untreated cells and mock-treated cells versus cells treated with control siRNA or Son siRNAs (Figure 11A). For these experiments, a small coverslip was inserted into each culture dish and processed for immunofluorescence to confirm the expected change in nuclear speckle morphology in Son-depleted cells (data not shown). We counted the total numbers of cells in mock, control siRNA oligo and Son siRNA-treated samples at different time points after siRNA treatment, and automated trypan blue staining was used to score cell viability in the same experiment. Growth curves indicate that cell growth was stalled in Son-depleted cells by 48 h, compared with cells treated with mock transfection and control siRNA that continued to grow with the normal doubling time of ∼24 h. The graph depicts the average numbers of viable cells counted at each time point from three separate groups of experiments.
Figure 11.
Son depletion causes cell growth defects and cell cycle arrest. (A) Cell growth curves. Sixteen sets of siRNA experiments (set = mock, control, si1, and si4) were performed and cell counts were obtained on a ViCell after a time course. Growth curves indicate that cell growth stalled in Son-depleted cells at 48 h, whereas cells continued to grow beyond 48 h after mock and control siRNA treatments. The graph depicts average numbers of cells from three separate groups of experiments. (B) Cell cycle is altered in Son-depleted cells. The siRNA experiments from the 48-h time point in part A were also analyzed by flow cytometry; data shown is representative of one of the four experimental groups. Untreated cells were used to set the gating on the flow cytometer and showed expected peaks at G1 and G2/M. Graphs of DNA content are shown for mock transfected, control siRNA treated cells and cells treated with Son siRNAs 1 and 4. A corresponding table of the percent of cells in each cell cycle stage is shown. (C) Immunoblot analysis of Son-depleted cells shows an increase in histone H3 modification (serine 10 phosphorylation). Cells from the 48-h time point in A were extracted in Laemmli buffer for immunoblot analysis. There was an increase in phosphorylation of histone H3Ser(10) in extracts from Son-depleted cells (lanes 3 and 4) as compared with mock (lane 1) and control extracts (lane 2). Extract from nocodazole arrested HeLa cells (N, lane 5) was used as a positive control for elevated H3 Ser(10)P in mitosis. Actin immunoblotting was used to confirm equal loading of total protein in each lane.
To determine whether the stalled growth after 48 h of Son depletion was due to changes in the cell cycle, we performed flow cytometry analysis that indicated the cell cycle distribution of control siRNA-treated cells was similar to that of untreated cells. siRNA experiments from the 48-h time point in part A were analyzed by flow cytometry; data shown are representative of one of the four experimental groups (Figure 11B). A corresponding table of the percent of cells in each cell cycle stage indicates a 9–12% decrease in G1 stage cells after Son depletion with two separate siRNAs (compared with control siRNA), with a concomitant increase (8–12%) of cells in G2/M stages. Flow cytometry analysis of cells at 72 h after Son depletion showed similar results, also with a similar decrease in G1 and increase in G2/M phase cells (data not shown).
Immunoblot analysis of Son-depleted cells was performed to distinguish between growth arrest at G2 versus M phase after Son depletion. We assessed whether there was an increase in phosphorylated histone H3 after Son depletion, as this would indicate an increase in mitotic cells. Cells harvested from 48 h after Son depletion in the same growth curve experiment shown in Figure 11, A and B, were extracted in Laemmli buffer for immunoblot analysis of histone H3Ser(10)P (Figure 11C). A significant increase in phosphorylation of histone H3Ser(10) was detected in extracts from cells treated with Son siRNAs (Figure 11C, lanes 3 and 4) compared with extracts from mock-treated cells (Figure 11C, lane 1) and cells treated with control siRNA oligos (Figure 11C, lane 2). Extract from nocodazole arrested HeLa cells (Figure 11C, lane 5) was used as a positive control for elevated H3Ser(10)P in mitosis. These results suggest that Son depletion leads to interrupted cell growth patterns presumably by interfering with progression through mitosis.
The increase in mitotic cells was also observed directly by microscopy, and the number of cells in each phase of mitosis was scored in Son-depleted cells (Figure 12). HeLa cells expressing H2B-GFP were treated with mock transfection, a mix of siRNAs against Son, or Son siRNA oligo 4. An increase of cells in mitosis was readily observed in Son-depleted cells after 36–48 h (Figure 12A, i–j and n–o) compared with mock-treated cells (Figure 12A, d and e). The mitotic index of cells scored over time after siRNA treatment (Figure 12B) also indicates a 9% increase in mitotic cells by 48 h that is consistent with the flow cytometry data shown in Figure 10B. Finally, the percentage of cells in each mitotic phase was determined (Figure 12C). After 48 h of Son depletion, the number of prometaphase/metaphase cells nearly doubles (increases from 54 to 96% of the mitotic cells), whereas the number of cells in anaphase and telophase decreases dramatically. This demonstrates that Son is necessary for cell cycle progression and that the absence of Son causes mitotic arrest at metaphase.
Figure 12.
Son depleted cells arrest in metaphase. Son siRNA mixture or Son siRNA 4 were transfected to HeLa cells stably expressing histone H2B-GFP at 10 and 100 nM, respectively. (A) After transfection, GFP fluorescence was observed every 12 h. An increase in mitotic cells was seen after 36 h of Son depletion (i–j and n–o). (B) Number of cells in mitosis scored in A plotted over time. (C) Mitotic index and number of cells in each mitotic phase were scored for >1000 cells. Son depletion caused arrest of mitosis in metaphase as determined by the dramatic doubling of cells in prometaphase/metaphase and almost complete absence of cells in anaphase and telophase.
DISCUSSION
Son is the largest RS-domain-containing protein identified in the proteomic analysis of nuclear speckles (Saitoh et al., 2004). Initial studies on RS domains in the Drosophila proteins Transformer and suppressor-of-white-apricot revealed this motif to be sufficient for nuclear speckle targeting (Li and Bingham, 1991; Hedley et al., 1995). The RS domain has been further characterized in the classical SR protein family of splicing factors, such as SF2/ASF and SC35 that are key players in constitutive and alternative splicing. Although the RS domain mediates the interaction of SR proteins in nuclear speckles (Sacco-Bubulya and Spector, 2002), phosphorylation of the RS domain is required to release SR proteins from nuclear speckles and to assemble SR proteins into the spliceosome (Mermoud et al., 1994; Colwill et al., 1996; Yeakley et al., 1999; Sacco-Bubulya and Spector, 2002). Importantly, the classical SR proteins contain RNA-recognition motifs and RNA binding domains that are crucial for their splicing-related functions (Caceres et al., 1997). Thus, the presence of an RS domain and other RNA binding domains is diagnostic for nuclear speckle constituents, and the presence of these motifs in Son as well as the nuclear speckle localization of Son suggests a putative role for Son in some aspect of pre-mRNA splicing. A recent study that identified Son as a binding partner for splicing factor SRrp53 by using a yeast two-hybrid screen supports this idea (Cazalla et al., 2005). In addition, the double-stranded RNA binding domain in Son suggests RNA binding functions for Son. We do not yet know what specific RNAs might interact with Son; however, our results indicate that maintaining the correct distribution of stable polyA+ RNA that resides in nuclear speckles requires Son.
Here, we show that the novel nuclear speckle protein Son is required for nuclear speckle integrity and the proper localization of pre-mRNA processing factors. The morphological effects associated with siRNA-mediated Son depletion implicate Son in maintaining assembly and/or function of pre-mRNA processing machineries. In light of our finding that Son remains in the insoluble nuclear fraction (Takata et al., 2009), it is tempting to speculate a role for Son in scaffolding the nuclear speckle interior into a network that can tether pre-mRNA splicing factor complexes. The change to a doughnut-shaped morphology is much different from what has been found in previous studies that have addressed nuclear speckle organization. This is typically achieved by manipulating either transcription and/or splicing activity by adding drugs/inhibitors to cells, or by changing global splicing factor phosphorylation by overexpressing SR protein kinases or using phosphatase inhibitors. For example, speckles become enlarged in cells that have been treated with transcription inhibitors, or by injecting cells with oligonucleotides or antibodies to inhibit pre-mRNA splicing (Spector et al., 1983; O'Keefe et al., 1994). In other studies, exogenous expression of the SR protein kinase Clk/STY completely disrupted nuclear speckle components to a diffuse nuclear localization (Colwill et al., 1996; Sacco-Bubulya and Spector, 2002). Interestingly, YFP-KIAA1019 (Son) redistributed from nuclear speckles to a diffuse nuclear distribution upon exogenous expression of Clk/STY, possibly indicating that serine/threonine phosphorylation regulates the distribution and/or function of Son (Sacco-Bubulya and Spector, 2002). The RS domain of Son, or the serine residues in its tandem repeats, could be phosphorylated by SR protein kinases. Ultrastructural analysis of nuclei in cells treated by overexpression of Clk/STY did not reveal any apparent residual nuclear speckle scaffolding such as a network of filaments upon dispersal of splicing factors (Sacco-Bubulya and Spector, 2002). One idea is that Son might be a linker between individual interchromatin granules or splicing factor complexes in the nuclear speckles rather than forming a filamentous scaffold. In this linker model, it is possible that Son may bind to pre-mRNA processing machinery or multimerize via its multiple novel repetitive motifs, perhaps in a manner that is consistent with the self-assembly model for nuclear organelles.
Most data supporting the current model for nuclear speckle assembly point to a self-association and self-assembly mechanism for nuclear speckle components rather than assembly onto a meshwork or interconnection of filamentous proteins (and Colwill et al., 1996; Yeakley et al., 1999; Sacco-Bubulya and Spector, 2002; reviewed in Misteli, 2000; Lamond and Spector, 2003; Misteli, 2007). Cline and Nelson (2007) proposed a nuclear speckle scaffold role for Par 6. However, depletion of Par 6 did not lead to disorganized speckles but instead only reduced the intensity of a single epitope in nuclear speckles (as shown by anti-SC35 phospho-epitope immunostaining). Furthermore, the distribution of other pre-mRNA processing factors following Par 6 RNAi was not reported in that study (Cline and Nelson, 2007), and it is not known whether Par-6 expression could rescue decreased SC35 phosphorylation. Our data show Son as the first example of a nuclear speckle protein whose depletion causes a dramatic reorganization of nuclear speckles. Importantly, this reorganization is prevented by expression of a siRNA-resistant YFP-siR-Son. The unique series of repeat motifs in Son can rescue nuclear speckle organization, whereas the RS domain, G-patch, and DS RBD are dispensable in this regard. We therefore conclude that the unique repeat domain in Son is necessary for proper maintenance of nuclear speckle organization and proper nuclear speckle localization of pre-mRNA processing factors.
Repetitive sequences in gene regulatory proteins have previously been implicated in important molecular scaffolding roles. For example, the carboxy-terminal domain (CTD) of the largest subunit of human RNA polymerase II has 52 repeats of the heptad YSPTSPS that coordinate transcription and pre-mRNA processing of protein coding transcripts. The RNA polymerase II CTD is required for cotranscriptional recruitment of splicing factors (Misteli and Spector, 1999), which are recruited to transcription sites in an intron-dependent manner (Jiminez-Garcia et al., 1993; Huang and Spector, 1996; Misteli et al., 1998). The 5′ end pre-mRNA capping factors, pre-mRNA splicing factors, and 3′ end processing factors are also loaded from the RNA polymerase II CTD onto pre-mRNAs as they emerge from the polymerase (reviewed in Hirose and Manley, 2000, Lewis and Tollervey, 2000; Bentley, 2005). More recent studies suggest that different steps of pre-mRNA processing, and even the control of alternative splicing, are regulated by different groups of heptad repeats within the RNA polymerase II CTD. We show that the repeat region of Son is required for proper nuclear speckle organization. Although there is not direct sequence similarity between Son and RNA polymerase II CTD, the nature of Son's short tandem serine-rich repeats suggests that Son might serve an analogous role as a scaffolding protein. This would perhaps be more suited to the “deterministic” model for nuclear assembly and could be played out in nuclear speckles if Son seeds the assembly of splicing complexes. Likewise, Son repeats may complement or antagonize the functions of the RNA polymerase II CTD if it is corecruited with pre-mRNA processing factors to transcription sites. Although we await results of additional studies, a gene regulatory role for Son is promising. Sun et al. (2001) have shown that Son binds to regulatory elements in human hepatitis B virus promoter where it can regulate viral transcription.
To envision Son as a scaffolding protein in nuclear speckles does not exclude the possibility that regulatory mechanisms, such as phosphorylation of serines in the RS domain by SR proteins kinases, might release Son for dynamic exchange between nuclear speckles and the nucleoplasm. If Son has a role in pre-mRNA transcription or splicing, then similar phosphorylation events may promote assembly of Son into transcription/splicing complexes at transcription sites. Alternatively, they may lead to dismantling of the nuclear speckles as cells enter mitosis. Our flow cytometry and microscopy results showed stalled cell growth in Son-depleted cells. Immunoblots of splicing factor levels remain constant after Son depletion, so stalled growth is unlikely a result of reduced splicing factor abundance. This is important because reducing the level of other splicing factors can lead to cell cycle arrest (Kittler et al., 2004; Li et al., 2005; Pacheco et al., 2006). Rather, it is more likely that Son has a direct role in mitosis, as many nuclear proteins have moonlighting roles during mitosis. Because Son depletion leads to an increase of cells in metaphase, it is clear that Son is an essential target for normal cell cycle progression. Consistent with our results, a genome-scale RNAi screen to search for genes that altered mitotic progression or caused mitotic defects identified Son as one of 2146 putative proteins having this role (Kittler et al., 2007). Our study has now more precisely determined that the absence of Son leads to increased numbers of prometaphase/metaphase cells as well as enrichment of the mitotic cell marker histone H3(Ser10)P in Son-depleted cells. The model that Son is a central scaffolding protein for nuclear speckle components would make it a likely target for regulation of nuclear speckle disassembly that must occur as cells enter mitosis. Furthermore, it would likely be important for the continuous assembly/disassembly of nuclear speckle components during mitotic progression (Lamond and Spector, 2003; Prasanth et al., 2003; Bubulya et al., 2004). The assembly of mitotic interchromatin granule clusters (MIGs), which are the mitotic equivalent of nuclear speckles, begins at metaphase (Thiry, 1995; Turner and Franchi, 1997; Bubulya et al., 2004). Live imaging of MIGs during mitosis showed that MIGs grow larger and more numerous from metaphase until telophase and then are disassembled as splicing factors begin to enter daughter nuclei (Bubulya et al., 2004). Interestingly, because Son depletion causes mitotic arrest at metaphase, precisely when the onset of MIG assembly in mitotic cells occurs, this suggests that Son is essential for MIG assembly to proceed. In this case, the mechanism for how Son seeds assembly of MIGs could be quite similar to how it maintains nuclear speckle organization during interphase.
The presence of Son in the insoluble nuclear fraction is consistent with a role for Son as a potential scaffolding protein for nuclear speckle assembly. Unique Son domains implicate novel functions for this nuclear speckle protein and putatively could allow Son to accommodate many binding partners. This may occur either in nuclear speckles during the assembly and modification of pre-mRNA processing complexes or at transcription sites during cotranscriptional pre-mRNA processing. Pinpointing Son as a key component for nuclear speckle organization during interphase as well as the importance of Son for normal mitotic progression opens many directions for future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Takahiro Nagase for kindly providing KIAA1019 and other Son cDNAs; Hiroshi Kimura (Osaka University) for providing HeLa cells expressing H2B-GFP; Catherine Tomasetto for providing YFP-Magoh and YFP-MLN51; Eric Romer and Courtney Sulentic (Boonshoft School of Medicine, Wright State University) for technical assistance and use of ViCell; Ramakrishna Kommangani and Madhavi Kadakia (Boonshoft School of Medicine) for technical assistance with flow cytometry; and Michael Leffak, Mark Mamrack, David Spector, Prasanth Kannanganattu, Michael Markey, and Bubulya laboratory members for helpful discussions. This work was supported by National Institutes of Health grant R15GM-084407, startup funds (Wright State University [WSU] 262135), Ohio Board of Regents New Investigator (WSU 666435), Research Initiation (WSU 262143), and Research Challenge (WSU 666768, WSU 667129) awards (to P.A.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0126) on January 6, 2010.
REFERENCES
- Ahn E.-H., Yan M., Malakhova O. A., M.-Lo C., Boyapati A., Ommen H. B., Hines R., Hokland P., Zhang D. E. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A., Degot S., Cougot N., Bertrand E., Chenard M. P., Wendling C., Kessler P., Le Hir H., Rio M. C., Tomasetto C. The exon-junction-complex-component metastatic lymph node 51 function in stress-granule assembly. J. Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- Bentley D. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bubulya P. A., Prasanth K. V., Deerinck T. J., Gerlich D., Beaudouin J., Ellisman M. H., Ellenberg J., Spector D. L. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J. F., Misteli T., Screaton G. R., Spector D. L., Krainer A. R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D., Newton K., Caceres J. F. A novel SR-related protein is required for the second step of pre-mRNA splicing. Mol. Cell. Biol. 2005;25:2969–2980. doi: 10.1128/MCB.25.8.2969-2980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M., Lamond A. I. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Cline E. G., Nelson W. J. Characterization of mammalian Par 6 as a dual-localization protein. Mol. Cell. Biol. 2007;27:4431–4443. doi: 10.1128/MCB.02235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J. L., Bell J. C., Duncan P. I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Corden J. L. Patterns of phosphorylation in a region of RNA polymerase II may constitute a code that controls the recruitment of regulatory factors to control gene expression. Science. 2007;318:1735–1736. [Google Scholar]
- Degot S., Le Hir H., Alpy F., Kedinger V., Stoll I., Wendling C., Seraphin B., Rio M. C., Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J. Biol. Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Duncan P. I., Howell B. W., Marius R. M., Drmanic S., Douville E. M., Bell J. C. Alternative splicing of STY, a nuclear dual specificity kinase. J. Biol. Chem. 1995;270:21524–21531. doi: 10.1074/jbc.270.37.21524. [DOI] [PubMed] [Google Scholar]
- Egloff S., Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Gall J. G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- Hall L. L., Smith K. P., Byron M., Lawrence J. B. The molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley M. L., Amrein H., Maniatis T. An amino acid motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Manley J. L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Huang S., Deerinck T. J., Ellisman M. H., Spector D. L. In vivo analysis of the stability and transport of nuclear poly(A) + RNA. J. Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Spector D. L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 1996;131:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiminez-Garcia L. F., Spector D. L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Kittler R., Putz G., Pelletier L., Buchholtz F. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Kittler R., et al. Genome-scale genomic profiling of cell division in human tissue culture cells. Nat. Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Spector D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Li H., Bingham P. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Li X., Wang J., Manley J. L. Loss of splicing factor SF2/ASF induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud J. E., Cohen P. T., Lamond A. I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz P. J., Patterson S. D., Neuwald A. F., Spahr C. S., Spector D. L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci. 2000;113:181–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misteli T., Cáceres J. F., Clement J. Q., Krainer A. R., Wilkinson M. F., Spector D. L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, Cáceres J. F., Spector D. L. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T., Spector D. L. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- O'Keefe R. T., Mayeda A., Sadowski C. L., Krainer A. R., Spector D. L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco T. R., Moita L. F., Gomes A. Q., Hacohen N., Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of cdc25 transcripts. Mol. Biol. Cell. 2006;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H. P., Greenleaf A. L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Prasad J., Colwill K., Pawson T., Manley J. L. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. V., Sacco-Bubulya P. A., Prasanth S. G., Spector D. L. Sequential entry of components of gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E., Blencowe B. J. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA. 2004;10:581–589. doi: 10.1261/rna.5207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya P., Spector D. L. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 2002;156:425–436. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N., Spahr C. S., Patterson S. D., Bubulya P., Neuwald A. F., Spector D. L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L., Schrier W. H., Busch H. Immunoelectron microscopic localization of snRNPs. Biol. Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Sun C. T., Lo W. Y., Wang I. H., Lo Y. H., Shiou S. R., Lai C. K., Ting L. P. Transcription repression of human hepatitis B virus genes by NREBP/SON. J. Biol. Chem. 2001;276:24059–24067. doi: 10.1074/jbc.M101330200. [DOI] [PubMed] [Google Scholar]
- Takata H., Nishijima H., Ogura S. I., Sakaguchi T., Bubulya P. A., Mochizuki T., Shibahara K. I. Proteome analysis of human nuclear insoluble fractions. Genes Cells. 2009;14:975–990. doi: 10.1111/j.1365-2443.2009.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M. Behavior of interchromatin granule clusters during the cell cycle. Eur. J. Cell Biol. 1995;68:14–24. [PubMed] [Google Scholar]
- Turner B. M., Franchi L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J. Cell Sci. 1987;87:269–282. doi: 10.1242/jcs.87.2.269. [DOI] [PubMed] [Google Scholar]
- Wynn S. L., et al. Organization and conservation of the GART/SON/DONSON locus in mouse and human genomes. Genomics. 2000;68:57–62. doi: 10.1006/geno.2000.6254. [DOI] [PubMed] [Google Scholar]
- Yeakley J. M., Tronchere H., Olsen J., Dyck J. A., Wang H. Y., Fu X. D. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.