Abstract
Epigenetic modifications of chromatin play an important role in the regulation of gene expression. KMT4/Dot1 is a conserved histone methyltransferase capable of methylating chromatin on Lys79 of histone H3 (H3K79). Here we report the identification of a multisubunit Dot1 complex (DotCom), which includes several of the mixed lineage leukemia (MLL) partners in leukemia such as ENL, AF9/MLLT3, AF17/MLLT6, and AF10/MLLT10, as well as the known Wnt pathway modifiers TRRAP, Skp1, and β-catenin. We demonstrated that the human DotCom is indeed capable of trimethylating H3K79 and, given the association of β-catenin, Skp1, and TRRAP, we investigated, and found, a role for Dot1 in Wnt/Wingless signaling in an in vivo model system. Knockdown of Dot1 in Drosophila results in decreased expression of a subset of Wingless target genes. Furthermore, the loss of expression for the Drosophila homologs of the Dot1-associated proteins involved in the regulation of H3K79 shows a similar reduction in expression of these Wingless targets. From yeast to human, specific trimethylation of H3K79 by Dot1 requires the monoubiquitination of histone H2B by the Rad6/Bre1 complex. Here, we demonstrate that depletion of Bre1, the E3 ligase required for H2B monoubiquitination, leads specifically to reduced bulk H3K79 trimethylation levels and a reduction in expression of many Wingless targets. Overall, our study describes for the first time the components of DotCom and links the specific regulation of H3K79 trimethylation by Dot1 and its associated factors to the Wnt/Wingless signaling pathway.
Keywords: Dot1, Wnt signaling, Wingless, H3K79 methylation, H2B monoubiquitination
In eukaryotic organisms, gene expression patterns are spatiotemporally regulated in a manner that allows for specification of diverse cell types and their differentiation. This spatiotemporal expression is coordinated in part by transcription factors and chromatin modifiers, and by the activity of several signaling pathways, which contribute to gene expression by regulating the transcription factors. Understanding the relationship between chromatin events and signaling pathways is crucial to understanding gene regulation, development of the organism, and disease pathogenesis.
The nucleosome, the basic unit of chromatin, consists of histones H2A, H2B, H3, and H4, and 146 base pairs (bp) of DNA (Kornberg 1974; Luger et al. 1997; Kornberg and Lorch 1999). Crystal structure studies have demonstrated that the N-terminal tails of each histone protrude outward from the core of the nucleosome (Luger et al. 1997). These histone tails are subject to various post-translational modifications, including methylation, ubiquitination, ADP ribosylation, acetylation, phosphorylation, and sumoylation (Rice et al. 2003; Ehrenhofer-Murray 2004; Cosgrove and Wolberger 2005; Shilatifard 2006; Groth et al. 2007; Kouzarides 2007; Kusch and Workman 2007; Li et al. 2007), and such modifications are involved in many biological processes involving chromatin such as transcription, genome stability, replication, and repair.
Histones are methylated on either the lysine and/or arginine residues by different histone methyltransferases (HMTases) (Shilatifard 2006; Kouzarides 2007). Histone lysine methylation can occur as mono-, di-, or trimethylated forms, and several lysine residues of histones have been shown to be multiply methylated. This includes methylation on Lys4, Lys9, Lys27, Lys36, and Lys79 of histone H3, and Lys20 of histone H4 (Bhaumik et al. 2007). Almost all of the lysine HMTases characterized to date contain a SET domain, named after Drosophila Su(var)3-9, Enhancer of zeste [E(z)], and trithorax (trx). SET domain-containing enzymes can catalyze the methylation of specific lysines on histones H3 and H4, and many SET domain-containing enzymes, such as Trithorax and Enhancer of zeste, are central players in epigenetic regulation and development (Shilatifard 2008).
Histone H3 at Lys79 (H3K79) can be mono-, di-, and trimethylated by Dot1, which to date is the only characterized non-SET domain-containing lysine HMTase. Dot1 was discovered many years ago (Feng et al. 2002; Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002; Shilatifard 2006), and is highly conserved from yeast to humans. In yeast, telomeric silencing is lost when Dot1 is overexpressed or inactivated, as well as when H3K79 is mutated (Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002; Shilatifard 2006). Unlike other histone methylation patterns, the pattern of di- and trimethylation of H3K79 in yeast appears to be nonoverlapping (Schulze et al. 2009). It was also first discovered in yeast that monoubiquitination of histone H2B on Lys123 (H2BK123) by the Rad6/Bre1 complex is required for proper H3K79 trimethylation by Dot1 (Ng et al. 2002b; Wood et al. 2003; Shilatifard 2006; Schulze et al. 2009). In vivo analysis of the pattern of H2B monoubiquitination in yeast demonstrated that the H3K79 trimethylation pattern overlaps with that of H2B monoubiquitination, and that the H3K79 dimethylation pattern and H2B monoubiquitination appear to be nonoverlapping (Schulze et al. 2009). This observation resulted in the proposal that the recruitment of the Rad6/Bre1 complex and the subsequent H2B monoubiquitination could dictate diversity between H3K79 di- and trimethylation on chromatin on certain loci within the genome. In addition to a role in the regulation of telomeric silencing in yeast, Dot1 has also been shown to be involved in meiotic checkpoint control (San-Segundo and Roeder 2000) and in double-strand break repair via sister chromatid recombination (Conde et al. 2009). We recently found a relationship between cell cycle progression and H3K79 dimethylation, but not trimethylation (Schulze et al. 2009), by Dot1. Consequently, to date, very little is known about a specific biological role of histone H3K79 trimethylation.
In Drosophila, H3K79 methylation levels correlate with gene activity (Schubeler et al. 2004). Mutations in grappa, the Dot1 ortholog in Drosophila, show not only the loss of silencing, but also Polycomb and Trithorax-group phenotypes, indicating a key role for H3K79 methylation in the regulation of gene activity during development (Shanower et al. 2005). Similarly, Dot1 in mammals has been implicated in the embryonic development of mice, including a role in the structural integrity of heterochromatin (Jones et al. 2008). Genome-wide profiling studies in various mammalian cell lines have suggested that Dot1 as well as H3K79me2 and H3K79me3 localize to the promoter-proximal regions of actively transcribed genes, and correlate well with high levels of gene transcription (Steger et al. 2008). It has also been proposed that Dot1 HMTase activity is required for leukemia pathogenesis (Okada et al. 2005; Bitoun et al. 2007; Mueller et al. 2007; Krivtsov et al. 2008).
The highly conserved Wnt/Wingless (Wnt/Wg) signaling pathway is essential for regulating developmental processes, including cell proliferation, organogenesis, and body axis formation (Cadigan 2002; Gregorieff and Clevers 2005). Deregulation or ectopic expression of members of the Wnt pathway has been associated with the development of various types of cancers, including acute myeloid and B-cell leukemias (Lu et al. 2004; Muller-Tidow et al. 2004). In the canonical Wnt/Wg pathway, a cytoplasmic multiprotein scaffold consisting of Glycogen synthase kinase 3-β (GSK3-β), Adenomatous polyposis coli (APC), Casein kinase 1 (CK1), Protein phosphatase 2A, and Axin constitutively marks newly synthesized β-catenin/Armadillo for degradation by phosphorylation at the key N-terminal Ser and Thr residues (Mosimann et al. 2009). Binding of the Wnt ligands to the seven-transmembrane domain receptor Frizzled (Fz) leads to recruitment of an adaptor protein, Disheveled (Dvl), from the cytoplasm to the plasma membrane. Axin is then sequestered away from the multiprotein Axin complex, resulting in inhibition of GSK3-β and subsequent stabilization of hypophosphorylated β-catenin levels in the cytoplasm (Huang and He 2008). Stabilized β-catenin translocates into the nucleus and binds to members of the DNA-binding T-cell factor/lymphoid enhancer factor (TCF/LEF) family (Henderson and Fagotto 2002), resulting in the recruitment of several chromatin-modifying complexes, including transformation/transcription domain-associated protein (TRRAP)/HIV Tat-interacting 60-kDa protein complex (TIP60) histone acetyltransferase (HAT), ISWI-containing complexes, and the SET1-type HMTase mixed lineage leukemia 1/2 (MLL1/MLL2) complexes (Sierra et al. 2006), thereby activating the expression of Wnt/Wg target genes.
Although much is known about Dot1 as an H3K79 HMTase, biochemical studies isolating to homogeneity a Dot1-containing complex have not been successful during the past decade. Here, we report the first biochemical isolation of a multisubunit complex associated with Dot1, which we call DotCom. DotCom is comprised of Dot1, AF10, AF17, AF9, ENL, Skp1, TRRAP, and β-catenin. This complex is enzymatically active and can catalyze H3K79 dimethylation and trimethylation. Indeed, we demonstrate that nucleosomes containing monoubiquitinated H2B are a better substrate for DotCom in the generation of trimethylated H3K79. Given the association of Skp1, TRRAP, and β-catenin with DotCom, and the fact that these factors have been linked to the Wnt signaling pathway in previous studies, we investigated the role of the Drosophila homolog of Dot1, dDot1 (Grappa), for the regulation of Wg target genes. RNAi of dDot1 leads to a reduced expression of a subset of Wg target genes, including senseless, a high-threshold Wingless target gene. Furthermore, reduction by RNAi in the levels of the Drosophila homologs of other components of DotCom that regulate the pattern of H3K79 methylation in humans also showed a similar reduction in senseless expression and other Wg target genes. Importantly, DotCom requires monoubiquitination of H2B for H3K79 trimethylation, and, in Drosophila, the loss of Bre1, the E3 ubiquitin ligase, leads to reduction of H3K79 trimethylation and decreased expression of the senseless gene. Taken together, our data support a model in which monoubiquitinated H2B provides a regulatory platform for a novel Dot1 complex to mediate H3K79 trimethylation, which is required for the proper transcriptional control of Wnt/Wg target genes.
Results
Identification of Dot1 in a 2-MDa-containing complex
Previous studies have identified several MLL fusion partners—AF10, AF9, and ENL—as interaction partners of Dot1 (Okada et al. 2005; Zhang et al. 2006; Bitoun et al. 2007; Mueller et al. 2007). However, many of these interactions were never confirmed in an in vivo system, and very little is known about the core complexes containing Dot1. To gain a better understanding of the factors that interact directly with Dot1, we generated a HEK293T cell line harboring a Tet-inducible Flag epitope-tagged Dot1 construct. We performed a Flag affinity purification and confirmed the expression of Flag-Dot1 by Western blotting (Fig. 1A–C) and demonstrated DotCom to be active as an H3K79 HMTase. As expected, Flag-Dot1 runs at an estimated 170 kDa. In order to investigate the composition of the purified Flag-DotCom, we subjected the immunoprecipitated complex to Multidimensional Protein Identification Technology (MudPIT), and reproducibly found a statistically significant amount of peptides each for AF10, AF9, ENL, AF17, Skp1, and TRRAP (Fig. 1D). We tested the H3K79 methylase activity associated with the peak fraction containing the DotCom, and demonstrated that this complex is active as an H3K79 di- and trimethylase (Fig. 1E). This is contrary to the previously published studies suggesting that, unlike the yeast enzyme, human Dot1 is not capable of trimethylating H3K79 (McGinty et al. 2008).
Figure 1.
Dot1 forms a 2-MDa complex with AF10, AF9, ENL, Skp1, and TRRAP. (A) Schematic showing the steps involved in the purification of the DotCom. Flag-tagged Dot1, expressed under an inducible promoter in HEK293T cells, was isolated by Flag affinity chromatography. Resulting protein complexes were then separated by size exclusion on a Superose 6 column. (B) Expression and affinity purification of Dot1. Flag affinity-purified protein complexes were analyzed by silver staining and SDS-PAGE. The arrowhead indicates the position of Flag-Dot1. Flag affinity-purified preparations from nuclear extracts from non-Flag-expressing HEK293T cells were used as a control (293 Control). (C) A HMTase assay with Dot1 and 293 Flag eluates on recombinant yeast nucleosomes was performed overnight in the presence or absence of S-adenosyl methionine (SAM) and analyzed by Western blotting using antibodies specific toward H3K79me2 and H3K79me3. Signals for H3K79me2 and H3K79me3 demonstrate the expected specificity of DotCom in carrying out H3K79 methylation. Total H3 levels were assayed as a loading control. (D) Human Dot1 is in a 2-MDa complex. Silver staining of size exclusion chromatography for Flag-Dot1 preparations from A is shown. From fractions 8 to 16, a peak of Dot1 is seen at 2 MDa (fraction 10), and MudPIT analysis revealed the presence of Dot1, TRRAP, AF10, AF17, AF9, ENL, and Skp1 as indicated. Western blotting with anti-Flag and anti-Dot1 antibodies shows a similar peak at fraction 10. ENL comigrates with the Dot1 peak in these fractions. Control Flag eluates are prepared as in B. (E) Human Dot1 has HMT activity. HMT assay was performed as in C with Dot1-Flag eluate as input, Control 293 Flag eluate, and fractions 8 and 10 from the size exclusion chromatography from D. The HMT activity of fraction 10 demonstrates that the ∼2-MDa DotCom is enzymatically active. Coomassie staining shows the integrity of the four core histones in these nucleosomes.
Monoubiquitinated H2B is a better substrate for the human DotCom in H3K79 trimethylation
Previous studies in yeast demonstrated that monoubiquitination of histone H2B is required for proper H3K4 and H3K79 methylation (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002b; Sun and Allis 2002). Recent studies implemented with recombinant human Dot1 polypeptide demonstrated that human Dot1 was capable of producing mono- and dimethylated H3K79; however, no trimethylation was detected in these enzymatic studies (McGinty et al. 2008). Furthermore, McGinty et al. (2008) demonstrated that nucleosomes containing monoubiquitinated H2B were a better substrate for human Dot1 in H3K79 dimethylation; however, trimethylated products were not detected. These observations are in stark contradiction with observations made with yeast Dot1. To determine whether the purified human DotCom is capable of trimethylating H3K79 when nucleosomes are monoubiquitinated on H2B, we tested H3K79 HMTase activity of the purified DotCom toward monoubiquitinated nucleosomal substrates. Our studies demonstrate that purified human DotCom trimethylates H3K79 much more effectively when nucleosomal substrates are monoubiquitinated on H2B (Fig. 2A). We also observed an increase in dimethylation of H3K79 in the presence of monoubiquitinated H2B, which is very similar to the data reported by McGinty et al. (2008). From our studies, we do not detect any difference between the levels of H3K79 monomethylation catalyzed by the purified human DotCom when using either monoubiquitinated or nonubiquitinated nucleosomal substrates (Fig. 2A).
Figure 2.
Human Dot1 trimethylates H3K79 in the presence of monoubiquitination of H2B. (A) HMTase assays with Dot1 Flag immunocomplex and nucleosomal substrates extracted as crude yeast nuclear extracts from wild-type (DOT1), ΔDot1, and H2BK123A DOT1 strains. ΔDot1 nucleosomes serve as H2B monoubiquitinated substrates (Ubq Nuc), as they have wild-type levels of H2B monoubiquitination and are devoid of any H3K79 methylation, and H2BK123A DOT1 nucleosomes serve as nonmonoubiquitinated substrates, as they lack monoubiquitination on H2B with Lys123 mutated to alanine. Comparison of the HMTase activity of hDotCom toward H3K79me, H3K79me2, and H3K79me3 on monoubiquitinated H2B and nonmonoubiquitinated H2B nucleosomal substrates shows a preferential activity of hDotCom toward trimethylation of H3K79 and, to a lesser extent, toward dimethylation in the presence of monoubiquitinated H2B, but has no difference in the ability to mono- or dimethylate H3K79. An identical titration of hDotCom was used for both monoubiquitinated H2B and nonmonoubiquitinated H2B nucleosomal substrates. H3 was used as a loading control. (B) Fifty HMT reactions (25 μL each, with 0.88 pmol of nucleosome) were performed in the presence or absence of SAM overnight using the catalytic domain of recombinant human Dot1 (1–416), pooled, and then analyzed by Western blotting. (C–E) Detection, identification, and quantitation of histone H3 Arg-C-digested “R.LVREIAQDFKTDLR.F” peptides bearing no modification, or mono-, di-, or trimethylation on Lys79 from the reaction mixture in B. Endoproteinase Arg-C-digested peptides were analyzed with a 12-step MudPIT on an Eksigent NanoLC two-dimensional system coupled to a LTQ-Orbitrap hydrib mass spectrometer. (C) Full MS scans were acquired in FT mode at 60,000 resolution. The doubly charged ions at 852.48, 859.48, 866.49, and 873.50 m/z were fragmented in the ion trap and mapped by SEQUEST to histone H3 “R.LVREIAQDFKTDLR.F” peptides with unmodified (me0), monomethylated (me1), dimethylated (me2), and trimethylated (me3) Lys79, respectively. (D) All four differentially modified peptides coeluted within a 1- to 2-min retention time window. (E) A representative tandem mass spectrum matched to “R.LVREIAQDFK(me3)TDLR.F” was annotated. The doubly charged precursor ion with a protonated mass of 1745.99 Da was marked by the green tick along the m/z-axis. Fragment ions bearing trimethylated Lys79 (circled) were matched in both singly charged b and y ion series, which were labeled in red and blue, respectively. Doubly charged b and y ions were labeled in magenta and dark cyan, respectively. If the fragment ions lost ammonia, the resulting peaks were labeled with the same color as the source series with an asterisk (*) to indicate that they are 17 Da away from the fragment ion. Trimethylated histone H3 peptides matched by multiple spectra were observed only in the presence of SAM.
These enzymatic observations with the purified human DotCom are very similar to the yeast studies, indicating that the pattern of H2B monoubiquitination implemented by the Rad6/Bre1 complex could dictate the pattern of H3K79 trimethylation on chromatin (Schulze et al. 2009). To make sure that our observation regarding the capability of the human DotCom to trimethylate H3K79 is not an artifact of cross-reactivity of antibodies generated toward di- and/or trimethylated H3K79, we generated recombinant nucleosomal substrates, methylated them using human Dot1 in vitro, and tested the products for the presence of H3K79 trimethylation via mass spectrometric (MS) analyses (Fig. 2B–E). Briefly, nucleosomes were incubated with purified human Dot1 with or without S-adenosyl methionine (SAM), and a small portion of the products was analyzed by SDS-PAGE followed by Western analysis using H3K79-specific antibodies (Fig. 2B). The post-translational modifications of histone peptides resulting from this enzymatic study were then analyzed by MudPIT. We observed MS data corresponding to mostly trimethylated histone H3 peptides in multiple spectra only in the presence of SAM, confirming our Western studies that human Dot1 is capable of trimethylating its nucleosomal substrate (Fig. 2C–E). Small but significant amounts of the monomethylated form of H3K79 were seen in the (−) SAM control, likely due to a small amount of SAM that was purified along with Dot1 (Fig. 2C, bottom panel).
Biochemical analysis of the association of several of the DotCom components with Dot1
Since we demonstrated that the human DotCom is capable of trimethylating its nucleosomal substrate in an H2B monoubiquitination-dependent manner, we wanted to better understand the nature of DotCom. We therefore subjected the purified human DotCom to analytical size exclusion chromatography (Fig. 1D). Silver staining and Western blotting with Flag and Dot1 antibodies revealed a 2-MDa peak (fraction 10). Cofractionation of ENL at this peak supported the finding that ENL exists in DotCom. To identify all of the constituent proteins of the 2-MDa complex, we analyzed fraction 10 by MudPIT and found similar amounts of AF10, AF9, ENL, AF17, and Skp1. TRRAP peptides were also detected at low but significant amounts in this fraction (Fig. 1D).
To better characterize DotCom and the nature of its components, we generated stable HEK293T cell lines bearing Tet-inducible Flag epitope-tagged AF10, AF9, and ENL constructs. Flag affinity purification followed by Western blot analysis with Dot1 antibody confirmed the presence of Dot1 in all three purifications (Fig. 3A,B). MudPIT on the Flag eluates revealed the presence of most of the components of DotCom (Fig. 3E). Using yeast two-hybrid studies, it was proposed previously that Dot1 interacts with AF10 (Okada et al. 2005); however, such interactions were never confirmed with the endogenously expressed proteins in vivo. Our studies confirm this observation and also provide the identification of the other components of DotCom. In our ENL preparation, AF9 was not found, while in the AF9 preparation, we found very low amounts of ENL, supporting the idea that one or the other, but not both of these highly related proteins, may interact with one Dot1 molecule at the same time (Fig. 3). The AF10 purification does not yield any peptides for ENL, although we find significant amounts of AF9, suggesting that the Dot1–AF10 complex contains AF9 as well. We also could confirm AF17 in all of our Flag eluates of AF10, ENL, and AF9 (Fig. 3E), as well as in fraction 10 of our Dot1 size fractionations (Fig. 3D), indicating that AF17 is present in the 2-MDa DotCom, together with its paralog, AF10. Notably absent from Dot1 and AF10 complexes is AF4, the most frequent translocation partner of MLL in infant leukemias and proposed previously to participate in complexes containing Dot1 (Bitoun et al. 2007; Mueller et al. 2007, 2009). However, AF4 was found in our Flag-ENL preparations (data not shown), suggesting that earlier reports of associations between AF4 and Dot1 were due to tagging a common subunit of two distinct complexes. To demonstrate that the purified DotComs obtained by tagging its other subunits are active in H3K79 methylation, we performed HMTase assays with 3H-labeled SAM and found enzymatic activity with the Flag-AF9 complex on recombinant nucleosomes, indicating that Dot1 in the Dot1–AF9 complex is active (Fig. 3C).
Figure 3.
Purification of AF10, ENL, and AF9 complexes identify Dot1 as a common component. (A) Clonal cell lines expressing Flag-tagged AF10, AF9, and ENL under a tetracycline-inducible promoter were generated in HEK293T cells, from which Dignam nuclear extracts were prepared for Flag purification and silver staining. Arrowheads show the positions of Flag-tagged proteins. A 293 Control Flag purification from untransfected 293 cells is shown as control. (B) Flag affinity purifications of Flag-AF10, Flag-AF9, Flag-ENL, and 293 Control nuclear extracts were probed with Dot1 antibody, revealing the presence of Dot1 in all purifications except the control 293 extracts. (C) Radioactive HMTase assay was performed using immunocomplexes purified from Flag-Dot1, Flag-AF9, and 293 Control extracts on yeast recombinant nucleosomes. The fluorograph shows the activity of Flag-Dot1 and Flag-AF9, but not 293 Control preparations, toward H3. Purified recombinant yeast Dot1 was used as a positive control in the reaction. (D) MudPIT analysis of two independent Flag-Dot1 purifications (Dot1-dNSAF-A and Dot1-dNSAF-B) and of the Superose 6 size exclusion peak fraction 10 from a Flag-Dot1 purification (Dot1-SE Fract. 10). dNSAF values indicate an estimate of the relative abundance of AF9, AF10, ENL, AF17, and Skp1. The presence of similar amounts of these proteins in the 2-MDa fraction 10 indicates that these proteins exist in DotCom. (E,F) MudPIT analysis of Flag eluates of Flag-AF10, Flag-ENL, and Flag-AF9. (E) dNSAF values of proteins are plotted on the Y-axis, and are given in the table shown in F. Numbers in bold indicate the values for the tagged protein.
The role of the DotCom components in regulating its enzymatic activity toward H3K79
In order to determine the role of the DotCom components in regulating its enzymatic activity, we performed siRNA-mediated knockdown of the core components in HeLa cells (Fig. 4A,B). We observed a dramatic reduction of H3K79me2 and H3K79me3 in Dot1 and AF10 knockdowns (Fig. 4A). These observations support the findings of a previous study in which AF10 was found to help target Dot1 to chromatin (Lin et al. 2009). Mouse AF9 has been reported to interact directly with mouse Dot1a (Zhang et al. 2006) and to form a nuclear repressor complex, which in turn regulates histone H3K79 methylation at the ENaCα promoter. We also observed a modest reduction of H3K79me2 and H3K79me3 in AF9 knockdown cell extracts (Fig. 4A). However, ENL knockdown resulted in a very subtle effect on bulk H3K79 trimethylation and no substantial change in H3K79 dimethylation (Fig. 4A), although it is possible that AF9 and ENL are partially redundant with each other in forming Dot1 complexes. These observations suggest that AF10 plays an important role in Dot1’s HMTase activity through either DotCom stability, catalytic activity, or the recruitment of the complex to chromatin.
Figure 4.
SiRNA-mediated knockdown of Dot1, AF10, AF9, and ENL affects global H3K79 methylation levels. (A) HeLa cells were treated with scrambled, ENL, AF9, AF10, and Dot1 siRNAs as indicated. Total cell lysates were subjected to immunoblot analyses with the indicated antibodies. Knockdown demonstrates that AF10 is the major regulator of H3K79 methylation by Dot1, whereas AF9 and ENL effects on H3K79 methylation are affected to a lesser extent, with AF9 having a stronger effect than ENL. Tubulin and H3 were used as loading controls. (B) qRT–PCR of the siRNA-treated HeLa cells shows the extent of knockdown after 72 h of incubation. Relative expression to Gapdh is plotted. (C) Flag affinity purification from Flag-Dot1 and Flag-AF10 293 cell extracts coimmunoprecipitate β-catenin, a key mediator of the Wnt signaling pathway. The presence of β-catenin in both the Flag-Dot1 and Flag-AF10, but not in Flag purification from HEK 293T extracts, indicates that β-catenin interacts with DotCom.
Skp1 is a component of the Skp1–Cullin–F-box (SCF) E3 ubiquitin ligase (Cardozo and Pagano 2004) complex and promotes ubiquitination of its target substrates.
TRRAP has been identified as a subunit common to the p300/CBP-associated factor complex (PCAF), Tip60, TATA-binding protein-free TAFII-containing complex, novel SAGA-related complex, and SAGA-like complexes (Carrozza et al. 2003; Murr et al. 2007). TRRAP has been implicated in transcription and DNA repair through its interaction between several transcriptional activators and the SAGA/GCN5/PCAF and NuA4/Tip60 complexes (Carrozza et al. 2003; Peterson and Cote 2004). It was also reported that the TRRAP–TIP60 complex interacts directly with β-catenin (Sierra et al. 2006), a key mediator of the Wnt signaling pathway. Recently, TRRAP was shown to interact with Skp1, and it was proposed that, by interacting with the SCF complex, TRRAP mediates ubiquitination of β-catenin in the nucleus and leads to its degradation (Finkbeiner et al. 2008). Based on these results, we asked whether DotCom interacts with β-catenin. Flag affinity immunoprecipitation of Flag-Dot1 and Flag-AF10 from HEK293T cells leads to significantly high enrichment of β-catenin (Fig. 4C). This interaction did not depend on DNA, as digestion of nucleic acids with Benzonase did not affect the enrichment. Taken together, our data suggest that DotCom interacts with β-catenin and might have a role in the regulation of Wnt target genes.
The Drosophila DotCom regulates expression of the Wingless target senseless
To investigate the role of DotCom in the regulation of Wnt signaling, we used the more genetically amenable system of Drosophila in which we could address the role of the components of DotCom in a developmental context in a living organism. In Drosophila, Wnt signaling, known as Wingless signaling, has been studied extensively. Wingless signaling is required for growth and morphogenesis in wing imaginal discs, which constitute the primordia in larval stages that later give rise to the adult wings. Wingless (Wg), the eponymous ligand for Wingless signaling, is expressed along the dorsal/ventral (D/V) boundary. The ability of Wg to act as a morphogen allows the formation of a Wg gradient with high levels of Wg at the D/V boundary and decreasing levels toward the periphery of the wing pouch. The establishment of this Wg gradient is crucial for the activation of various Wg target genes in adjacent cells (Zecca et al. 1996; Strigini and Cohen 1997).
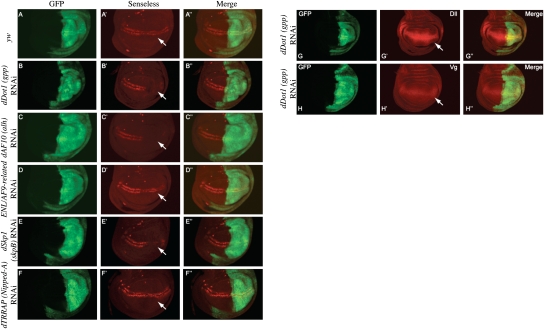
In order to test the role of dDot1 in wingless signaling, we down-regulated dDot1 expression by RNAi. dDot1 shares significant homology at the catalytic domain with yeast Dot1 and hDot1 (Min et al. 2003), and is functionally capable of methylating H3K79 (Shanower et al. 2005). Ubiquitous knockdown of dDot1 with two independent RNAi lines at 25°C resulted in either early larval lethality before the third instar stage (line v16001) or lethality in the pupal stages (line v47199). This confirms previous findings that dDot1 is an essential gene required for viability of the organism. We then proceeded to use the engrailed-GAL4 driver to knock down dDot1 (en-Gal4:dDot1-RNAi-v16001) in the posterior compartment of wing imaginal discs. In these imaginal discs, the anterior compartment serves as the wild-type control. We monitored the expression of the high-threshold Wg target senseless, which is expressed in two stripes flanking the Wg expression domain at the D/V boundary (Fig. 5A′; Nolo et al. 2000; Johnston and Sanders 2003), and two low-threshold Wg target genes, Distalless (Dll) (Fig. 5G′) and Vestigial (Vg) (Fig. 5H′), which are expressed throughout the wing pouch anlage in a graded manner with peak levels at the D/V boundary (Zecca et al. 1996; Strigini and Cohen 1997). We observed a robust reduction of senseless expression in the posterior compartment of the wing discs where dDot1 RNAi was expressed (Fig. 5B–B″, white arrow). However, we could not find any detectable change in the expression level of dll and vg (Fig. 5G–H″) These observations show that dDot1’s function is essential for the expression of senseless, and suggest that dDot1 is needed for the expression of genes that require high levels of Wg signaling.
Figure 5.
dDot1 (gpp), dAF10 (Alhambra), ENL/AF9-related, and dSkp1 (skpB) specifically regulate the expression of a wingless target gene, senseless. (A′–F′) Staining of wing imaginal discs from Drosophila third instar larvae with antibodies to the Wingless target gene senseless. Using the UAS-GAL4 system, en-GAL4 was used to drive the expression of UAS-hairpin constructs targeting Drosophila Dot1 and its interactors. (A–H) The domain of knockdown is marked by GFP, which is also expressed from a UAS. (A′–F′) Arrows point to the location where senseless is normally expressed in two stripes. (A–A″) yw crossed to the en-GAL4 driver was used as a control and shows the wild-type expression of senseless in the wing pouch. (A″–G″) Merge of GFP and Senseless signals. Knockdowns of dDot1 (B–B″), dAF10 (C–C″), and dSkp1 (E–E″) lead to major reductions in Senseless, with lesser reductions in Senseless levels observed for knockdown of dENL/AF9 (D–D″). (F–F″) No change in Senseless levels after TRRAP RNAi were observed in this genetic background, but reductions could be seen in an enhanced RNAi background overexpressing Dicer-2 (see Supplemental Fig. 2). (G″–H″′) Immunofluorescence analysis of Distalless (Dll) and Vestigial (Vg), the products of two low-threshold wingless target genes; distalless (G–G″) and vestigial (H–H″) are not affected by the knockdown of dDot1 (gpp-v16001), as seen by similar broad stripes outside and inside the domain of RNAi knockdown as indicated by GFP staining in the domain of en-Gal4-driven expression.
To establish the possible function of a putative dDotCom in flies, we proceeded to test several Drosophila homologs for similar effects on Wg target genes in vivo. Alhambra was identified as the Drosophila homolog of AF10 and AF17, and was shown to inhibit Polycomb response element (PRE)-mediated repression (Perrin et al. 2003). We used two different UAS-RNAi constructs targeting Alhambra (dAF10) and observed a strong reduction in senseless expression in the wing imaginal discs (Fig. 5C–C″, arrow), suggesting that dDot1 and dAF10 are in a complex and regulate the expression of Wg target genes.
ENL and AF9 are very similar proteins and are represented by a single protein in Drosophila, aptly named EAR (ENL/AF9-related). Knockdown of AF9 and ENL in HeLa cells leads to only a moderate loss of H3K79 methylation (Fig. 4A). RNAi against EAR in wing discs resulted in a very weak but reproducible loss of senseless expression (Fig. 5D–D″, white arrow).
The Drosophila genome contains six Skp1-related genes (Murphy 2003). We tested the involvement of one of these, SkpB, for involvement in Wg signaling. Knockdown of SkpB resulted in a decreased expression of senseless, suggesting that SkpB regulates the expression of wingless target genes similar to Dot1. Nipped-A is the Drosophila homolog of the yeast Tra1 and human TRRAP proteins and is a key component of both the SAGA and Tip60 (NuA4) chromatin-modifying complexes (Allard et al. 1999; McMahon et al. 2000). Nipped-A (dTRRAP) is an essential gene and has also been implicated in Notch signaling. Reducing the gene dosage of dTRRAP by half reduces both the mastermind and Notch activities during wing development (Gause et al. 2006). Ubiquitous expression of different dTRRAP RNAi constructs leads to pupal lethality in all cases (Supplemental Table 1). Quantitative RT–PCR (qRT–PCR) of the dTRRAP RNAi larvae (line v40789) showed only a small reduction in the transcript level, suggesting that even a small reduction of dTRRAP is detrimental to the fly viability. Two of the stronger RNAi lines (v40789 and 2905R-3), when crossed to en-GAL4, did not result in any significant reduction in senseless (Fig. 5F, white arrow). As this might be due to low efficiency of the RNAi, we enhanced the RNAi by coexpressing Dicer-2 (Dietzl et al. 2007) and found a robust reduction of senseless expression (Supplemental Fig. 2). Taken together, this indicates a role for dTRRAP in dDotCom to mediate the regulation of the Wg target gene senseless. Reduction of the members of dDotCom does not affect the expression of Wingless protein in the wing imaginal discs (Supplemental Fig. 3), although we found that knockdown of dDot1 leads to a small reduction in Wg (Supplemental Fig. 3B–B″). This may be due to a possible role for dDot1 in the regulation of expression of Wg directly or indirectly.
Regulation of senseless expression by dDotCom suggests that this complex might have a broader role in the regulation of Wg signaling. Thus, we investigated the role of individual members of dDotCom on other Wg target genes, frizzled 3 (dfz3) (Sivasankaran et al. 2000), CG5895 (Mosimann et al. 2006), CG6234 (Fang et al. 2006), Notum (Liu et al. 2008), and Homothorax (hth). To analyze the role of dDot1, we chose a hypomorphic allele of dDot1 (gpp1A) that has been shown to enhance the Abdominal-B phenotype (Shanower et al. 2005). We extracted total RNA from third instar wandering larvae and analyzed the expression levels by qRT–PCR. All of the tested genes were down-regulated (Supplemental Fig. 4A). To substantiate our results, we drove the strong dDot1 RNAi line (v16001) in wing discs with A9-GAL4 and analyzed the expression levels of Wg target genes. We observed similar knockdowns of the Wg target genes as in dDot1 (gpp1A) mutant larvae (Supplemental Fig. 4B). We also asked if other putative components of dDotCom would have similar effects on Wg target gene expression. Indeed, it appears that components of DotCom could regulate the expression of some of the Wg target genes, but not as strongly as either dDot1 or dAF10, both of which are required for proper H3K79 methylation (Supplemental Fig. 4C–F). Taken together, a consistent reduction in the expression of most of the Wg target genes was observed upon reduction of dDotCom subunits, with the strongest effects seen when either dDot1 or dAF10, levels were altered. This provides evidence that dDot1 and several of its interacting proteins are novel regulators of Wg signaling. However, extensive global expression and localization analyses under wild-type and different Wnt activation conditions must be performed before a full set of gene targets for Dot1 and member of its complex can be identified.
H2B monoubiquitination by Bre1 links H3K79 trimethylation and not di- or monomethylation to Wingless signaling
Histone H2B is monoubiquitinated at the Lys123 residue (Lys123) in yeast or the corresponding Lys120 in humans (Zhang 2003; Shilatifard 2006). In yeast, Rad6, an E2 ubiquitin-conjugating enzyme, and Bre1, an E3 ubiquitin ligase, are required for monoubiquitination of H2B (Shilatifard 2006). Monoubiquitination of H2B is required for H3K79 trimethylation by Dot1 in yeast (Schneider et al. 2004; Lee et al. 2007; Schulze et al. 2009) and H3K79 methylation in humans. It has also been proposed that monoubiquitination of H2B directly stimulates Dot1 to carry out H3K79 mono- and dimethylation (McGinty et al. 2008). However, our studies in this paper demonstrate that human Dot1 can trimethylate H3K79 and that nucleosomes bearing monoubiquitinated H2B are better substrates for purified human Dot1, thus validating the studies in yeast and confirming conservation of this histone modification pathway from yeast to humans.
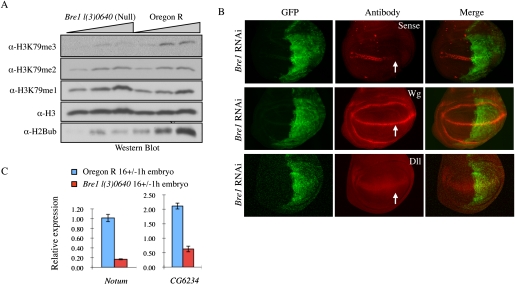
We therefore asked whether H2B monoubiquitination in Drosophila is also required for H3K79 trimethylation. Probing total protein extract from 16 ± 1-h homozygous-null Bre1 [l(3)01640] embryos (Bray et al. 2005) with H3K79 methylation and monoubiquitinated H2B-specific antibodies, we confirm that reduction in H2B monoubiquitination levels results in a specific reduction in H3K79 trimethylation, but not in mono- or dimethylation (Fig. 6A). This observation regarding the role of H2B monoubiquitination and H3K79 trimethylation in Drosophila is similar to our observation of this process in yeast (Schulze et al. 2009). The presence of small amounts of monoubiquitinated H2B in the Bre1 mutant embryos (Fig. 6A) is perhaps the result of the maternal contribution of Bre1 mRNA. Approximately 10% of Bre1 mRNA remained in these embryos, as seen by qRT–PCR (data not shown). Analysis of later stages of null Bre1 embryos was not possible, as the embryos either did not hatch or died soon after hatching.
Figure 6.
H2B monoubiquitination by Bre1 specifically links H3K79 trimethylation to Wingless signaling. (A) Bre1 mutant embryos show reduced levels of bulk monoubiquitinated H2B and H3K79me3. H3K79me and H3K79me2 are not affected. Histones were analyzed from 16 ± 1-h embryos of homozygous Bre1 mutants [l(3)01640] and wild-type controls for the presence of H3K79 methylation as indicated. (B) RNAi-mediated knockdown of Bre1 in wing imaginal discs leads to reduced expression of senseless. Expression of Wingless protein itself is not affected, and there is no change in the low-threshold Wg target distalless (dll). en-Gal4 was used to drive the expression of UAS-Bre1-RNAi, and the domain of knockdown is marked by UAS-GFP. The arrow shows the loss of senseless expression. (C) Bre1 mutant embryos show reduced expression of Notum and CG6234. RNA was extracted from 16 ± 1-h embryos of homozygous Bre1 mutants [l(3)01640] and wild-type controls, and expression levels were determined by qRT–PCR. Relative expression to Tubulin is plotted.
These data suggest that, in vivo in flies, only trimethylation requires the presence of H2B monoubiquitination, and not mono- or dimethylation. This implies that the sole known H3K79 methyltransferase enzyme in flies, dDot1, is stimulated by the presence of monoubiquitination on H2B to carry out trimethylation of H3K79, thus providing us with a tool to analyze the function of H3K79 trimethylation in gene expression. Using Bre1 RNAi in the posterior half of the wing discs, driven by en-Gal4 coexpressing Dicer 2, we find a strong reduction of the high-threshold Wg target gene senseless (Fig. 6, top panel). However, we did not observe any effect on the expression of either Wg or Dll (Fig. 6B, middle and bottom panels). We then analyzed the expression of other Wg target genes in the 16 ± 1-h Bre1-null l(3)0640 mutant embryos, and could confirm that the expression of Notum and CG6234 was significantly affected (Fig. 6C). The differential effect of Bre1 loss on expression of these Wg target genes suggests that the requirement of H2B monoubiquitination in facilitating the Wg transcriptional program might be dependent on the context. Overall, these studies demonstrate a specific role for H3K79 trimethyation, but not mono- or dimethylation, in Wnt signaling.
Discussion
The histone methylase Dot1 is a conserved enzyme with a similar function in mono-, di-, and trimethylating H3K79 from yeast to human. Molecular studies during the past several years have demonstrated that trimethylation of H3K79 on chromatin by Dot1 requires proper H2BK123 monoubiquitination by the Rad6/Bre1 complex (Briggs et al. 2002; Ng et al. 2002b; Wood et al. 2003; Schulze et al. 2009). H3K79 di- and trimethylation have a nonoverlapping pattern on chromatin, and the exclusion of monoubiquitination of H2B at the dimethylated sites has been proposed to be the regulator of this process (Schulze et al. 2009).
Although H3K79 methylation is a ubiquitous mark associated with actively transcribed genes, and its presence is a clear indicator for the elongating form of RNA polymerase II (Krogan et al. 2003; Steger et al. 2008), Dot1 itself has a very low abundance and is very hard to detect in cells. This indicates that Dot1 is an active enzyme with a very high specific activity toward its substrate, H3K79. Due to the low abundance of Dot1 in cells, its molecular isolation and biochemical purification have been hindered for the past decade. In this study, we report the biochemical isolation of a Dot1-containing complex (DotCom) and demonstrate a specific link between H3K79 trimethylation by DotCom and the Wnt signaling pathway. Our study reports (1) the identification and biochemical isolation of a large macromolecular complex (∼2 MDa) containing human Dot1, in association with human AF10, AF17, AF9, ENL, Skp1, TRRAP, and β-catenin; (2) the biochemical demonstration that the human DotCom is capable of trimethylating H3K79, and the analysis of the role of histone H2B monoubiquitination in the enhancement of this H3K79 trimethylase activity of the human DotCom; (3) identification of the role of the components of DotCom in the regulation of its H3K79 methylase activity; (4) demonstration of a role for the Drosophila homolog of Dot1 and its associated factors in the Wnt signaling pathway; and, finally, (5) the identification of a specific requirement of H3K79 trimethylation, but not mono- or dimethylation, in the regulation of Wnt target transcription, thereby linking H3K79 trimethylation to Wnt signaling.
A role for human Dot1 in H3K79 trimethylation
Dot1 was initially isolated from yeast, and these studies demonstrated that the enzyme is capable of mono-, di-, and trimethylating H3K79 (Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002). Subsequent molecular and biochemical studies demonstrated that prior H2B monoubiquitination by the Rad6/Bre1 complex is required for proper H3K79 trimethylation by yeast Dot1 (Briggs et al. 2002; Ng et al. 2002b; Wood et al. 2003; Schulze et al. 2009). A recent analysis of the human homolog of Dot1 suggested that its HMTase domain is not capable of trimethylating H3K79, and that this enzyme can only dimethylate its substrate (McGinty et al. 2008). McGinty et al. (2008) also demonstrated that reconstitution of monoubiquitinated H2B into chemically defined nucleosomes, followed by enzymatic treatment with Dot1, resulted only in dimethylation of H3K79. Since these observations are in contrast with the published studies in yeast, we tested the enzymatic activity of our purified human DotCom toward monoubiquitinated and nonmonoubiquitinated nucleosomes. Our studies demonstrate that the human DotCom can indeed trimethylate H3K79 (Fig. 1C,E), and that monoubiquitination of histone H2B enhances this enzymatic property of the human DotCom (Fig. 2A). Since our enzymatic studies employ antibodies generated toward mono-, di-, and trimethylated H3K79 to identify the products of the enzymatic reactions containing human Dot1, we wanted to make certain that our observations are not the result of cross-reactivity between these antibodies. We therefore generated recombinant nucleosomes and treated them with human Dot1 in the presence and absence of SAM, and analyzed the products by MS (Fig. 2C–E). The chemical analysis of the products from this enzymatic reaction confirmed that human Dot1 is capable of trimethylating H3K79 (Fig. 2C–E). We purposely chose to digest our hDot1-treated nucleosome samples with Endoproteinase Arg-C because previous unpublished work on analyzing yeast histone modifications by MudPIT had shown that the trimethylated peptide containing H3K79 was not detected when digesting with trypsin. Notably, McGinty et al. (2008) performed their digestions with trypsin, which might explain their failure to detect this modification by MS.
Components of the human DotCom regulate its H3K79 HMTase activity and the Wnt signaling pathway
Our studies identified several factors—including ENL, AF9, AF17, AF10, SKP1, TRA1/TRAPP, and β-catenin—as components of the human DotCom (Figs. 1, 2, 4). To test the role of these factors in regulating Dot1’s catalytic activity, we reduced their levels via RNAi. These studies demonstrated that AF10 functions with Dot1 to regulate its catalytic properties in vivo (Fig. 4A,B). We are unable to detect significant differences in Dot1’s H3K79 HMTase activity in vivo when reducing the levels of ENL, AF9, and AF17 (Fig. 4A; data not shown). As discussed below, factors that significantly alter the H3K79 methylation pattern by Dot1 are also linked to its transcriptional regulatory functions at Wnt target genes.
Since Dot1 also appears to interact with β-catenin (Fig. 3C), and given the known role for β-catenin, Skp1, and TRRAP in the Wnt signaling pathway, we set out to test the role for Dot1 and the components of its complex in Wnt signaling. Drosophila melanogaster is an outstanding model system for the study of the Wnt signaling pathway. Given the power of genetics and biochemistry in Drosophila, we tested the role of dDot1 and the members of its complex in wingless signaling. From this study, we learned that down-regulation of Drosophila Dot1 and Drosophila AF10 had the most significant effects in the regulation in the expression of the Wg target senseless (Fig. 5). Given the fact that our molecular studies demonstrated that Dot1 and AF10 have the strongest effect in the regulation of H3K79 methylation in vivo (Fig. 4A,B), we wanted to determine whether a specific form of H3K79 methylation is required for Wnt target gene expression.
Histone H2B monoubiquitination is required for proper H3K79 trimethylation (Shilatifard 2006; Schulze et al. 2009). The E2/E3 complex Rad6/Bre1 is required for the proper implementation of H2B monoubiquitination on chromatin, and this complex is highly conserved from yeast to humans. Deletion of the Drosophila homolog of Bre1 results in the loss of H2B monoubiquitination and the specific loss of H3K79 trimethylation (Fig. 6A). Interestingly, reduction in the levels of H3K79 trimethylation results in a defect in expression of one of the Wnt target genes, senseless (Fig. 6B), although the H3K79 mono- and dimethylation in this mutant background appear to be normal (Fig. 6A). In addition to senseless, we tested the role of H3K79 methylation at other Wnt targets and observed the same effect for Notum and CG6234. Overall, these studies demonstrate a link between H3K79 trimethylation by the DotCom and the Wnt signaling pathway.
Wnt/Wg signaling serves a critical role in tissue development, proliferation of progenitor cells, and many human cancers (Reya and Clevers 2005). The key player in the Wnt pathway is β-catenin, which is shuttled into the nucleus at the onset of activation of the pathway. Various proteins that interact with β-catenin in the nucleus—such as CBP/p300, TRRAP, MLL1/MLL2, Brg1, telomerase, Hyrax, Pygopus, and CDK8—modulate the transcriptional output of Wg/Wnt target genes (Willert and Jones 2006; Carrera et al. 2008; Firestein et al. 2008; Park et al. 2009). These proteins probably provide the context specificity to Wnt response directing proliferation or differentiation effects of Wnt signaling. Our finding that dDotCom is required for expression of a subset of Wg targets suggests that dDotCom might also facilitate Wg-regulated programs of transcriptional regulation in specific contexts. As most human cancers have elevated levels of Wnt signaling and require Wnt signaling for continued proliferation, DotCom might play a role in supporting the high rate of expression of Wnt target genes in such cancers.
Several studies have found interactions between Dot1 and many translocation partners of MLL (Okada et al. 2005; Bitoun et al. 2007; Mueller et al. 2007, 2009). While these associations suggest a link between Dot1 methylation and leukemogenesis, it was not clear how Dot1 methylation would participate in this process. Recently, GSK3, a regulator of β-catenin and Wnt signaling, was found to be essential for proliferation of MLL-transformed cells and for progression of a mouse model of MLL-based leukemia (Wang et al. 2008). Our studies linking Dot1 H3K79me3 with Wnt signaling provide insight into the role of Wnt signaling and Dot1 methylation in MLL translocation-based leukemia.
Materials and methods
Antibodies and cell lines
Antibodies recognizing the Flag tag were obtained from Sigma; H3, H3K79me1, H3K79me2, and H3K79me3 were obtained from Abcam. Antibodies against monoubiquitinated H2B were from Medimabs, and the β-catenin antibody was from BD Transduction Laboratories. α-Senseless was a kind gift from Hugo Bellen; α-Vestigial and α-Distalless were generously provided to us by Sean B. Carroll. Dot1 cDNA was a kind gift from the Roeder laboratory. Dot1 (1303–1537) and ENL (281–497) were expressed as His tag fusion proteins in PET-16b, purified on a NTA-agarose according to Qiagen's protocol, and sent to Pocono Rabbit Farm and Laboratories for immunization into rabbits. HeLa cell lines were purchased from the American Type Culture Collection. The cDNAs of the human Dot1, AF10, AF9, and ENL genes were cloned into pCDNA5/FRT vectors with an N-terminal Flag tag. The plasmids were then transfected into 293FRT cells and selected by hygromycin. Single clones were picked and cultured into roller bottles. The 293FRT cells were grown in suspension with CD293 medium (Invitrogen) as described by the manufacturer. Nuclear extracts were prepared and subjected to anti-Flag agarose immunoaffinity purification.
siRNA
HeLa cells were treated with siRNA duplex (Dharmacon) using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Total RNA was extracted using Qiagen's RNeasy kit, and DNA was digested before using the RNA in qRT–PCR.
Immunoprecipitation
Approximately 107 cells for each assay were collected, washed with phosphate-buffered saline, and lysed in a high-salt lysis buffer (20 mM HEPES at pH 7.4, 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.5% Triton X-100, 1 mM dithiothreitol) containing proteinase inhibitors (Sigma). After incubation for 30 min at 4°C, the lysate was centrifuged thoroughly at 4°C twice. The balance buffer (20 mM HEPES at pH 7.4, 1 mM MgCl2, 10 mM KCl) was added to the resulting supernatant to make the final NaCl concentration 300 mM. The lysate was then mixed with antibodies and protein A beads or with anti-Flag agarose (Sigma). After incubation for 4 h at 4°C, the beads were spun down and washed three times with wash buffer (10 mM HEPES at pH 7.4, 1 mM MgCl2, 300 mM NaCl, 10 mM KCl, 0.2% Triton X-100) before eluting by boiling in an SDS gel sample buffer.
MudPIT analysis
Trichloroacetic acid-precipitated protein mixtures from purifications were digested with endoproteinase Lys-C and trypsin (Roche) as described previously. Peptide mixtures were loaded onto triphasic 100-mm fused silica microcapillary columns as described previously. Loaded microcapillary columns were placed in-line with a Quaternary Agilent 1100 series high-pressure liquid chromatography pump and a Deca-XP ion trap mass spectrometer (Thermo Fisher) equipped with a nano-LC electrospray ionization source. Fully automated MudPIT runs were carried out on the electrosprayed peptides. Tandem mass spectra were interpreted by using SEQUEST against a database containing Homo sapiens protein sequences downloaded from the National Center for Biotechnology Information. In addition to estimating the false discovery rates, each sequence was randomized (keeping the same amino acid composition and length), and the resulting “shuffled” sequences were added to the “normal” human database and searched at the same time. Peptide/spectrum matches were sorted and selected using DTASelect, keeping false discovery rates at 2% or less, and peptide hits from multiple runs were compared using CONTRAST. To estimate protein levels, spectral counts of nonredundant proteins were normalized by using the in-house-developed script NSAF7.
MudPIT analysis for H3K79 methylation analyses
TCA-precipitated proteins were resuspended in 30 μL of 100 mM Tris-HCl (pH 8.5), 8 M urea, reduced with 5 mM TCEP [Tris(2-carboxylethyl)-phosphine hydrochloride; Pierce], and alkylated with 10 mM CAM (Chloroacetamide; Sigma). Urea was then diluted to 4 M with 100 mM Tris-HCl (pH 8.5), and Endoproteinase Arg-C (Roche) was added to 0.5 μg, overnight at 37°C. The reaction was quenched by adding formic acid to 5%. The peptide mixtures were then loaded onto 250-μm fused silica microcapillary columns packed first with 3 cm of 5-μm Strong Cation Exchange material (Partisphere SCX; Whatman), followed by 1 cm of 5-μm C18 reverse phase (Aqua; Phenomenex). Loaded 250-μm columns were connected using a filtered union (UpChurch) with 100-μm fused silica columns pulled to a 5-μm tip using a P2000 CO2 laser puller (Sutter Instruments) and packed with 9 cm of reverse-phase material (McDonald et al. 2002).
The loaded microcapillary columns were placed in-line with an Eksigent NanoLC 2D system. The four channels of the nanoLC were filled with 5% acetonitrile, 0.1% formic acid (channels 1A and 2A); 0.5 M ammonium acetate, 5% acetonitrile, 0.1% formic acid (channel 1B); and 80% acetonitrile, 0.1% formic acid (channel 2B). Fully automated 12-step chromatography runs were carried out with a flow rate of 500 nL/min, measured at 250 nL/min at the tip of the column. Peptides were sequentially eluted from the SCX resin to the reverse-phase resin by 10-min-long salt steps of increasing concentrations (0%, 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 100%, and 100% for steps 1–12, respectively), followed by the organic gradient to 75% solvent.
The application of a 2.5-kV distal voltage electrosprayed the eluting peptides directly into a LTQ-Orbitrap hybrid mass spectrometer equipped with a nano-LC electrospray ionization source (ThermoFinnigan). Full MS spectra were recorded on the peptides over a 400–1600 m/z range in the Orbitrap at 60,000 resolution, followed by fragmentation in the ion trap (at 35% collision energy) on the first to fifth most intense ions selected from the full MS spectrum. Dynamic exclusion was enabled for 120 sec (Zhang et al. 2009) with an exclusion window of 0.03–1.03. Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan).
MS/MS spectra were first searched using SEQUEST (Eng et al. 1994) with a peptide mass tolerance of 20 ppm, and without specifying differential modifications against a protein database consisting of 27,027 sequences, consisting of human dot1, Saccharomyces cerevisiae histones, and Escherichia coli proteins (downloaded from NCBI on November 14, 2007), as well as 177 usual contaminants such as human keratins, IgGs, and proteolytic enzymes. To estimate false discovery rates, each protein sequence was randomized (keeping the same amino acid composition and length), and the resulting “shuffled” sequences were added to the database used for the SEQUEST searches. To account for alkylation by CAM, +57 Da were added statically to cysteine residues for all searches.
As described in Shi et al. (2009), differential modification searches were set up to query a protein database containing only the sequences for S. cerevisiae histones for peptides containing (1) methylated lysines and arginines (+14.0157) and oxidized methionines (+15.9949); (2) dimethylated lysines and arginines (+28.0314) and oxidized methionines; (3) trimethylated lysines (+42.0470) and oxidized methionines; (4) acetylated lysines, serines, and threonines (+42.0106) and oxidized methionines; (5) phosphorylated lysines, serines, and tyrosines (+79.9663) and oxidized methionines; and (6) ubiquitinated lysines (+114.0429) and oxidized methionines. The maximum number of modified amino acids per differential modification in a peptide was limited to four. After this round of searches, an in-house-developed script, sqt-merge (Zybailov et al. 2005), was used to combine the eight sets of SEQUEST output files (sqt files) generated from the normal search (i.e., without modifications) and post-translational modification (PTM) searches described above into one set. This merging step allowed only the best matches out of n differential SEQUEST queries to be ranked first. For the second round of searches, only spectra matching modified peptides were selected (−m 0 −t 0 DTASelect parameters), and their coordinates were written out into smaller ms2 files using the “−copy” utility of DTASelect (Tabb et al. 2002). These subsetted ms2 files contained at best a few hundred MS/MS spectra and were subjected to the same seven differential searches against the complete database described above (including shuffled sequences). Again, sqt-merge was used to bring together the results generated by these different searches. This step allowed us to check that spectra matching modified peptides from histone sequences did not find a better match against the larger protein database. All spectra matching modified peptides were assessed visually and given an evaluation flag (Y/M/N, for yes/maybe/no). The “no” matches were removed from the final data (−v 2 parameter in DTASelect). Spectra/peptide matches were only retained if they had a DeltCn of at least 0.08, and minimum XCorr of 1.8 for singly, 2.0 for doubly, and 3.0 for triply charged spectra. In addition, the peptides had to comply with the Arg-C enzyme specificity (the preceding and last amino acids were arginines), and be at least seven amino acids long. DTASelect (Tabb et al. 2002) was used to select and sort peptide/spectrum matches passing this criteria set. U_SPC6 software (in-house by Tim Wen) was used to extract total and modified spectral counts for each amino acid within yeast histones and to calculate modification levels based on local spectral counts (Xiang et al. 2007).
Recombinant histone preparation
Recombinantly expressed yeast histones H2A, H2B, H3, and H4 were prepared by the method described previously (Luger et al. 1999). E. coli BL21(DE3)pLysS cells were used for histone protein expression. Transformed cells were grown to mid-log phase in 6 L of LB media at 37°C, followed by induction with 0.4 mM IPTG for 3 h at the same temperature. Cells were harvested and washed in wash buffer (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 1 mM EDTA, 1mM banzamidine). Cells were disrupted by sonication in wash buffer. Inclusion bodies were then spun down and washed twice with wash buffer containing 1% Triton X-100. Pellets were then soaked in DMSO (500 μL) and incubated for 30 min, followed by extraction in 15–20 mL of extraction buffer (7 M guanidinium chloride, 20 mM Tris-HCl at pH 7.5, 10 mM dithiothreitol [DTT]) for 1 h. After clearing by centrifugation at 23,000g for 10 min, the supernatants were purified using a Sephacryl S-200 column eluting with SAU-1000 buffer (7 M deionized urea, 20 mM sodium acetate at pH 5.2, 1 M NaCl, 5 mM 2-mercaptoethanol, 1 mM EDTA). Peak fractions containing histones were combined and dialyzed in refolding buffer (2 M NaCl, 10 mM tris-HCl at pH 7.5, 1 mM EDTA, 5 mM 2-mercaptoethanol).
Nucleosome assembly
Recombinant yeast histones were denatured in unfolding buffer (6 M guanidinium chloride, 20 mM Tris-HCl at pH 7.5, 5 mM DTT), mixed in equimolar ratio, and dialyzed in refolding buffer. Octamers formed were then purified by size exclusion chromatography. Six-hundred-one targeting sequences were then mixed with octamers in 1:1.5 ratio, followed by a stepwise dilution into low-salt buffer. Nucleosomes thus formed were analyzed by native and SDS-PAGE gels (Supplemental Fig. 1). The native gel was stained with ethidium bromide to show the retarded migration of DNA wrapped around the octamer in a nucleosome. Coomassie staining of SDS-PAGE gels show the presence of all four histones—H2A, H2B, H3, and H4—in equimolar ratio (Supplemental Fig. 1).
Histone lysine methyltransferase assays
For a single HMT assay, nucleosomes (0.88 pmol) were mixed with 1× MAB buffer (50 mM Tris at pH 8.5), 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 250 mM sucrose), and enzyme in the presence (0.08 mM) or absence of SAM and incubated overnight at 30°C. In case of radioactive HMT assay, 3H-Sam was used. Mixtures were then resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting. For analyzing the H3K79me3 by MS, 44 pmol of nucleosomes were incubated with recombinant human Dot1 (catalytic domain, 1–416 amino acids) overnight and precipitated.
Fly stocks
Dot1 (gpp-v16001), Dot1 (gpp-v47199), and dTRRAP (Nipped-A 40789) were from DGRC; dAF10 (Alh 1070R-1), EAR (4913R-1), and dSkp1 (skpB 8881R-4) were from NIG,. Act5C-Gal4, en-Gal4:GFP, Dicer-2, en-Gal4, and Bre1 (3) 01640 were from Bloomington Stock Center. Bre1-RNAi was generated by cloning Bre1 cDNA cDNA (1037–1580) into Sym-pUAST vector, and fly lines were made by BestGene. All of the RNAi crosses were done at 27°C. Bre1 (3) 01640 was balanced over CyO, twist-Gal4:GFP. Embryos were collected every 2 h and aged for 15 h at 25°C before sorting manually or by using a flow cytometry.
Acknowledgments
We thank Drs. Hugo Bellen, Sean Carroll, and Paul Schedl for reagents. We thank Dr. Edwin Smith for conversations and consultation on this study and for critical reading of this manuscript. We thank the Stowers Institute Tissue Culture Facility for assistance with growing cell lines. We are also grateful to Laura Shilatifard for editorial assistance. C.L. is a PhD student registered with the Open University This work was supported by a post-doctoral fellowship by the Jane Coffins Child Memorial Fund to H.M.H., and a grant (R01CA89455) from the National Institute of Health to A.S.H.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1898410.
Supplemental material is available at http://www.genesdev.org.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. Wnt signaling—20 years and counting. Trends Genet. 2002;18:340–342. doi: 10.1016/s0168-9525(02)02707-5. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Conde F, Refolio E, Cordon-Preciado V, Cortes-Ledesma F, Aragon L, Aguilera A, San-Segundo PA. The Dot1 histone methyltransferase and the Rad9 checkpoint adaptor contribute to cohesin-dependent double-strand break repair by sister chromatid recombination in Saccharomyces cerevisiae. Genetics. 2009;182:437–446. doi: 10.1534/genetics.109.101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83:468–476. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates Iii JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 2006;25:2735–2745. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Finkbeiner MG, Sawan C, Ouzounova M, Murr R, Herceg Z. HAT cofactor TRRAP mediates β-catenin ubiquitination on the chromatin and the regulation of the canonical Wnt pathway. Cell Cycle. 2008;7:3908–3914. doi: 10.4161/cc.7.24.7354. [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Eissenberg JC, Macrae AF, Dorsett M, Misulovin Z, Dorsett D. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol Cell Biol. 2006;26:2347–2359. doi: 10.1128/MCB.26.6.2347-2359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: From endoderm to cancer. Genes & Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Fagotto F. The ins and outs of APC and β-catenin nuclear transport. EMBO Rep. 2002;3:834–839. doi: 10.1093/embo-reports/kvf181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, He X. Wnt/β-catenin signaling: New (and old) players and new insights. Curr Opin Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Sanders AL. Wingless promotes cell survival but constrains growth during Drosophila wing development. Nat Cell Biol. 2003;5:827–833. doi: 10.1038/ncb1041. [DOI] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL–AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Kusch T, Workman JL. Histone variants and complexes involved in their exchange. Subcell Biochem. 2007;41:91–109. [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C, Zhang KL, Zhang Y, Xu GL, Bohlander SK. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood. 2009;114:651–658. doi: 10.1182/blood-2009-03-209395. [DOI] [PubMed] [Google Scholar]
- Liu YI, Chang MV, Li HE, Barolo S, Chang JL, Blauwkamp TA, Cadigan KM. The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets. Dev Biol. 2008;323:41–52. doi: 10.1016/j.ydbio.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, Kipps TJ, Corr M, Carson DA. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: Single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom. 2002;219:245–251. [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. β-Catenin hits chromatin: Regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Tidow C, Steffen B, Cauvet T, Tickenbrock L, Ji P, Diederichs S, Sargin B, Kohler G, Stelljes M, Puccetti E, et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol Cell Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD. Drosophila skpA, a component of SCF ubiquitin ligases, regulates centrosome duplication independently of cyclin E accumulation. J Cell Sci. 2003;116:2321–2332. doi: 10.1242/jcs.00463. [DOI] [PubMed] [Google Scholar]
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: Mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes & Dev. 2002a;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002b;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin L, Bloyer S, Ferraz C, Agrawal N, Sinha P, Dura JM. The leucine zipper motif of the Drosophila AF10 homologue can inhibit PRE-mediated repression: Implications for leukemogenic activity of human MLL-AF10 fusions. Mol Cell Biol. 2003;23:119–130. doi: 10.1128/MCB.23.1.119-130.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes & Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Dover J, Johnston M, Shilatifard A. Global proteomic analysis of S. cerevisiae (GPS) to identify proteins required for histone modifications. Methods Enzymol. 2004;377:227–234. doi: 10.1016/S0076-6879(03)77013-X. [DOI] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Vivian CJ, Lee KJ, Ge C, Morotomi-Yano K, Manzl C, Bock F, Sato S, Tomomori-Sato C, Zhu R, et al. DNA-PKcs-PIDDosome: A nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell. 2009;136:508–520. doi: 10.1016/j.cell.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes & Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankaran R, Calleja M, Morata G, Basler K. The Wingless target gene Dfz3 encodes a new member of the Drosophila Frizzled family. Mech Dev. 2000;91:427–431. doi: 10.1016/s0925-4773(99)00313-5. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development. 1997;124:4697–4705. doi: 10.1242/dev.124.22.4697. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K, Jones KA. Wnt signaling: Is the party in the nucleus? Genes & Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Takeo S, Florens L, Hughes SE, Huo LJ, Gilliland WD, Swanson SK, Teeter K, Schwartz JW, Washburn MP, et al. The inhibition of Polo kinase by Matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 2007;5:e323. doi: 10.1371/journal.pbio.0050323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a Wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes & Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a–AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCα in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal Chem. 2009;81:6317–6326. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]