Abstract
Steroidogenic acute regulatory protein (StAR)D4 is a member of the StAR related lipid transfer family. Homology comes from the ∼210 amino acid lipid binding domain implicated in intracellular transport, cell signaling, and lipid metabolism. StARD4 was identified as a gene downregulated 2-fold by dietary cholesterol (Soccio, R. E., R. M. Adams, K. N. Maxwell, and J. L. Breslow. 2005. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress. J. Biol. Chem. 280: 19410–19418). A mouse knockout was created to investigate StARD4’s functionality and role in lipid metabolism. Homozygous knockout mice exhibited normal Mendelian mating genetics, but weighed less than wild-type littermates, an effect not accounted for by energy metabolism or food intake. Body composition as analyzed by DEXA scan showed no significant difference. No significant alterations in plasma or liver lipid content were observed on a chow diet, but female knockout mice showed a decrease in gallbladder bile cholesterol and phospholipid concentration. When challenged with a 0.2% lovastatin diet, StARD4 homozygous mice exhibited no changes. However, when challenged with a 0.5% cholesterol diet, female StARD4 homozygous mice showed a moderate decrease in total cholesterol, LDL, and cholesterol ester concentrations. Microarray analysis of liver RNA found few changes. However, NPC1’s expression, a gene not on the microarray, was decreased ∼2.5-fold in knockouts. These observations suggest that StARD4’s role can largely be compensated for by other intracellular cholesterol transporters.
Keywords: StARD4, mouse knockout, intracellular cholesterol transport, StAR related lipid transfer
Cholesterol is essential to the consistency of mammalian cell membranes but the distribution of cholesterol varies greatly between intracellular membranes (1). Unfortunately, the exact mechanism(s) for creating and maintaining this differential has so far remained mostly a mystery. Some form of transport and sorting mechanism must exist to include or exclude cholesterol from various intracellular organelles. Understanding this transport mechanism is crucial to the understanding of overall sterol homeostasis and ultimately relates back to questions concerning the development of atherosclerosis (2).
Recent work has implicated a specialized group of nonvesicular transporters related to the steriodogenic acute regulatory protein as key players in the movement of lipids throughout the cell (1–5). StAR, the archetype of the family, is responsible for transfer of cholesterol to the mitochondria of steroid producing cells (6). StAR deficiences in humans causes a disease called congenital lipoid hyperplasia, a disorder characterized by cholesterol accumulation in lipid droplets which leads to impaired synthesis of gonadal and adrenal cortical steroid hormones (7, 8). This effect has been replicated in the mouse knockout model of StAR, which is characterized by a congential lipoid hyperplasia phenotype (7). Phospohatidylcholine transfer protein (PCTP) or StARD2 is a phophatidylcholine (PC) transporter responsible for intermembrane transport of PC (9). The common element among the family is the StAR related lipid transfer (START) domain, a ∼210 amino acid lipid binding domain implicated in intracellular transport, cell signaling, and lipid metabolism. So far, the human and mouse genomes each have 15 genes encoding start domains and further phylogenetic analysis divides the families into six subfamilies (10). The X-ray crystal structure of MLN64 (StARD3), StARD4, and PCTP have all been solved. All three share a helix-grip fold with α-helices at the N and C terminus separated by nine β-sheets and two α-helices (10).
One of the families delineated is the StARD4 subfamily, consisting of StARD4, StARD5, and StARD6. StARD4 was identified first as a gene downregulated 2-fold by dietary cholesterol in a microarray study (10). Based on the proximity to other START domain proteins and the microarray data, it is likely that the StARD4 subfamily members participate in cholesterol metabolism (11). StARD4 and StARD5 are both widely expressed with their highest levels reached in the liver. Initially, StARD6’s expression was thought to be isolated to the testis, but recent work has shown its expression in the central nervous system in places like the hippocampus, substantia gelatinosa of the spinal cord, and the cerebal cortex (11, 12). StARD4 and StARD5 share 30% amino acid homology (13). Both proteins bind cholesterol as a ligand, but StARD5 additionally binds 25-hydroxycholesterol (14, 15). Several studies have investigated the role of StAR proteins in mitochondrial p450scc steriodogenosis. Recent reports show that StARD1 and StARD6 have high steriodogenic activity, StARD3 has about 50% this amount, and StARD4, StARD5, and StARD7 have trace, to no activity (16, 17). It has also been postulated that StARD4 and StARD5 might play a role in the bile acid synthesis pathway (18). The bile acid theory has been supported by data showing that transfection of an overexpressing StARD4 plasmid in primary hepatocytes is capable of increasing the rate of bile acid production (15).
Based on these facts, the present study aimed to better understand the role of StARD4 physiologically. To this end, a gene targeting strategy to knock out the StARD4 gene in mice was undertaken. Based on observations stated above, it was thought that StARD4’s role in cholesterol transport would have a profound effect on lipid distribution throughout the cell and body. Unexpectedly, mice lacking a functional StARD4 START domain were viable and displayed overall normal sterol dynamics, indicating a role for compensatory mechanisms in place that can mask the effect of the StARD4 knockout.
MATERIAL AND METHODS
Targeted disruption of the StARD4 START domain
StARD4 is a 6 exon gene located on mouse chromosome 18 (10). A protocol adapted from Liu et al. (19) that is readily available on the NCI-Fredrick website was undertaken to create the knockout. Disruption of the StARD4 START domain was achieved by deletion of exon 3. This disruption led to excision of part of StARD4’s START domain and pretermination of transcription leading to a nonfunctional protein. Briefly, a targeting cassette with individual Lox-P recombination sites surrounding exon 3 and a downstream Flp recombination target (FRT)-neomycin-FRT marker (for selection after transfection and ultimate recombination at the FRT recombinase sites) was created and transfected into embryonic stem cells derived from C57Bl/6J mice. Correctly targeted embryonic stem cells were analyzed by Southern blotting. Only one correctly targeted embryonic stem cell line was found and ultimately used to create chimera mice which were crossed to C57BL/6J (Jackson Lab # 000664) mice to obtain heterozygous mutants. Mice used in this study were the offspring of crosses between the F1 and/or F2 generations. Mice genotyping was done by PCR. Three sets of primers were used. The first primer set corresponded to a sequence found in the second exon StARD4-A (ex2for) (5′-CTGGAAGGACTGTCTGATGT-3′). The next primer set is found in third or the deleted exon StARD4-C (ex3for) (5′-CTGCTGACCCACTTTGTAT-3′). The last primer set corresponds to DNA found in intron 3 StARD4-H (int3-4rev) (5′-CTATTCTTCTCTGAGTCCCT-3′).
Western blot analysis
Proteins were isolated from cells or mouse tissues by homogenization in RIPA buffer (50 mM Tris, pH 7.4/150 mM NaCl/1 mM EDTA/0.1% SDS/1% Triton X-100/1% deoxycholate) with complete mini protease inhibitor cocktail (Roche). Crude extracts were homogenized and then centrifuged at 16,000 g for 10 min at 4°C to pellet cellular debris and nuclei. Protein concentration was measured by the bichinchoninic acid assay (BCA) assay (Pierce), and 10–50 μg of protein was used to run a Western blot gel. The following AB dilutions were used: anti-StARD4, 1:200 (Santa Cruz: sc-66663); anti-β actin (cell signaling), 1:10,000. For the detection of antibody protein complexes, the SuperSignal West Pico or Femto Kit (Pierce) was used according to the manufacturer's instructions. Band intensities were measured by using IMAGE Pro Plus.
Assessment of fertility and fecundity
All animal protocols were approved by the Rockefeller University Animal Care and Use Committee in accordance with institutional animal care and use committee policy. All mice were bred and housed at the Rockefeller University Laboratory Animal Research Center in a single humidity and temperature-controlled room with a 12 h dark-light cycle. To assess fertility and fecundity, littermate males (> 6 weeks old) were placed in cages with mature wild-type females for 1 month or longer. Littermate females were caged with wild-type females for a similar period of time. The same was done for knockout littermates. The number of mice achieving a pregnancy and the number of offspring from each mating set or pregnancy was recorded.
Mice
Wild-type C57BL/6 male mice were obtained from the Jackson Laboratory (stock no. 00664). ACTFLPe transgenic mice on the C57BL/6 background were obtained from the Jackson Laboratory (B6.Cg-Tg (ACTFLPe) 9205Dym/J, stock no. 005703, henceforth called Flp). These mice express flippase in all mouse tissues. CMV-Cre transgenic on the C57BL/6 background were obtained from the European Mutant Mouse Archive (EMMA) (B6.129P2-Tg(CMV-cre)1Cgn/CgnIbcm, stock no. 01149, henceforth called CMVCre). These mice express Cre recombinase in all mouse tissues.
DEXA scan
To assess the body fat content and bone mineral density of male mice at various ages, a dual energy X-ray absorptiometry (DEXA) scan was implemented. Mice were anesthetized with isoflurane and put on the scanning plate of the Piximus 2 (Lunar) DEXA scan. Data was collected from the output of the machine and mice were then allowed to wake up as normal following the procedure.
Mouse characterization and dietary studies
For studies involving the characterization of the StARD4 knockout phenotype, all mice were fed a normal chow diet ad libitum (PicoLab Rodent Diet 20). These mice were then euthanized at 12 weeks of age for experimentation. Unless otherwise indicated, this was the general procedure used throughout.
Briefly, mouse euthanasia followed a strict protocol. On the morning of euthanasia, food was removed from the cage early in the light cycle (9am) and mice were fasted from food but not water for the next 6 h. Prior to administration of anesthesia, blood glucose levels were measured from tail blood using a glucometer (Bayer). All mice were then sedated with ketamine/xylazine, weighed, length measured (measurements done from rump to crown), and euthanized. Next, the blood was drawn by puncturing the right and then left heart ventricles, the gallbladder bile was aspirated, and the animals were exsanguinated with heparinized PBS. Finally, tissues were harvested, weighed, and then frozen in liquid nitrogen and stored at −80°C.
For dietary studies involving the StARD4 knockout mice, 6-week-old mice were fed the semi-synthetic modified AIN76a diet containing 12% kcal as fat, added with 0.00% cholesterol (wt/wt) (Research Diets D10001) for the duration of one week's time before the start of experimentation. At week 7, mice were split into three groups and fed either the control 0.00% cholesterol diet, the same diet supplemented with 0.2% lovastatin (wt/wt) (Research Diets D09020602), or the same diet supplemented with 0.5% cholesterol (wt/wt) for the 1 week before sacrifice.
For alternative dietary studies involving the StARD4 knockout mice, 6-week-old mice's food intake and weight were weighed daily at 4 PM for 2 weeks. Mice were housed individually and careful inspection of cages was done to make sure that no food had fallen into the cage and that daily intake was accounted for. For this reason, it was deemed unnecessary to use metabolic cages to measure the rate of food intake. At 8 weeks, mice were fed ad libitum a high-fat diet, 60% kcal as fat (Research Diets D09020601-02) from week 8 to 20 before euthanasia for experimentation.
Measurements of serum, bile, and hepatic lipids
Plasma isolated during mouse euthanasia was immediately separated by centrifugation. Lipoproteins were isolated by sequential ultracentrifugation from 60 μl of plasma at d <1.006 g/ml (VLDL), 1.006 ≤d ≥1.063 g/ml (intermediate-density lipoprotein and LDL), and d >1.063 g/ml (HDL). The cholesterol in each of these fractions and in the total plasma was assayed enzymatically (Roche/Hitachi) and free cholesterol was measured enzymatically (Wako). The level of cholesterol ester was calculated by subtracting the free cholesterol content from the total cholesterol content. The triglycerides in the total plasma were assayed by enzymatic assay (Roche).
Gallbladder bile was isolated and analyzed for cholesterol, phospholipids, and bile acids by enzymatic assay (Roche, Wako).
Total liver lipids were extracted with Folch extractions. Briefly, snap-frozen liver tissues (∼100 mg) were homogenized and extracted twice with chloroform/methanol (v/v = 2:1) solution. The organic layer was dried under nitrogen gas and resolubilized in chloroform containing 2% Triton X-100. This extract was dried again and resuspended in water and then assayed for total cholesterol and triglyceride concentration using commercial kits as described above.
IPGTT
For the intraperitoneal glucose tolerance test (IPGTT), mice were fasted for 6 h following the dark (feeding) cycle. After intraperitoneal injection of glucose (in physiologic saline) (2g/kg of body weight), blood was drawn from the tail vein at 0, 15, 30, 60, and 120 min. Blood glucose levels were measured from tail blood using a handheld blood glucometer (Bayer). IPGTTs were performed on mice 3–4 days prior to euthanasia.
RNA isolation and qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen), followed by RNeasy cleanup (Qiagen), according to the manufacturer's instructions. For RT-PCR analysis, RNA was treated with Dnase I (Ambion), and 1–5ug was reverse transcribed using Superscript III (Invitrogen) with random haxamer primers. A 7900HT Sequence Detection System (Applied Biosystems) was used with the thermal cycling profile, 95°C for 10 min; 40 cycles of 95°C 30 s, 60°C 30 s, 72°C 1 min; 95°C 15 s, 60°C 20 s, 95°C 15 s (dissociation curve for SYBR Green reactions). The threshold was set in the linear range of normalized fluorescence, and a threshold cycle (Ct) was measured in each well. Each sample was amplified in duplicate for the genes of interest and the housekeeping gene GapDH. Each cDNA value for genes of interest was then normalized to the corresponding value for the housekeeping gene GapDH and expressed as a ratio, allowing for variability in the initial quantities of mRNA. All primers are listed in supplementary Table I.
DNA microarray analysis
Target preparation and hybridization.
All protocols were conducted as described in the Illumina GeneChip Expression Analysis technical manual. Total RNA was purified from 10 to 20 mg of wet tissue using Qiagen RNeasy Mini kit according to the manufacturer's recommendations (Qiagen, Valencia, CA). RNA quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent Technologies Inc., Palo Alto, CA). Approximately 500 ng of total RNA was used to prepare biotin-labeled RNA using Ambion Illumina TotalPrep RNA Amplification Kit (Cat# AMIL1791, Applied Biosystems, Foster City, CA).
Briefly, 500 ng of total RNA was used to synthesize the first strand of cDNA using ArrayScript reverse transcriptase and an oligo(dT) primer bearing a T7 promoter. The single-stranded cDNA was then converted into a double-stranded DNA by DNA polymerase I in the presence of E. coli RNase H and DNA ligase. After column purification, the double-stranded DNA was served as a template for in vitro transcription in a reaction containing biotin-labeled UTP, unlabeled NTPs, and T7 RNA Polymerase. The amplified, biotin-labeled antisense RNA (aRNA) was purified and quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit. A total of 750 ng of RNA in 5 ml was mixed with 10 ml of hybridization reagents and heated at 65C for 10min. After cooled to room temperature, total 15 ml of the hyb solution was applied to Illumina MouseRef-8 v1.1 chip. The chip was incubated for about 18 h at 58C. After washing and staining with streptavidin-Cy3, the chip was scanned using Illumina BeadArray Reader. The scanning was done using standard DirectHyb Gene Expression protocol with the following settings: Factor = 1, PMT = 587, Filter = 100%. The raw data was extracted using Illumina BeadStudio software without normalization.
Initial data analysis.
Genespring GX10 software was used to quantitate expression levels for targeted genes; default values provided by Illumina were applied to all analysis parameters. Border pixels were removed and the average intensity of pixels within the 75th percentile was computed for each probe. The average of the lowest 2% of probe intensities occurring in each of 16 microarray sectors was set as background and subtracted from all features in that sector. Probe pairs were scored positive or negative for detection of the targeted sequence by comparing signals from the perfect match and mismatch probe features. The number of probe pairs meeting the default discrimination threshold (0.015) was used to assign a call of absent, present, or marginal for each assayed gene, and a p-value was calculated to reflect confidence in the detection call. A weighted mean of probe fluorescence (corrected for nonspecific signal by subtracting the mismatch probe value) was calculated using the one-step Tukey's biweight estimate. This signal value, a relative measure of the expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays; after exclusion of the highest and lowest 2%, the average total chip signal was calculated and used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from one microarray were then multiplied by the appropriate scaling factor.
Data and statistical analysis
All data are expressed as mean ± SD. Statistical analysis was performed using Student's t-test or, for small sample sizes with nonnormal distribution, the nonparametric Mann-Whitney test was used to test significance. P < 0.05 was taken as the level of significance.
RESULTS
Targeted disruption of the StARD4 gene
A protocol adapted from Liu et al. (19) that is readily available on the NCI-Fredrick website was undertaken to create the StARD4 knockout mouse. We disrupted the START activity of the StARD4 gene by inserting LoxP recombination sites around exon 3 of the 6 exon StARD4 gene (Fig. 1). Embryonic stem cells carrying the StARD4 targeted construct were then injected into C57BL/6J to produce chimera mice. Through a series of mating steps, mice were bred to homozygosity to produce knockout mice. Disruption of the StARD4 gene was confirmed by PCR analysis. Western blot analysis of livers from two individual wild-type, heterozygote, and knockout mice were probed for the 23.5 kDa StARD4 protein (Fig. 2). Heterozygotes have a decrease in StARD4 protein levels corresponding to only one allele of StARD4 expression, indicating a gene dosing effect. This makes sense when the data is normalized to β-actin, as the heterozygote lanes are loaded with more protein but still have a lower level of StARD4 protein.
Fig. 1.
Gene targeting and generation of a conditional knockout (floxed) allele of the StARD4 gene. Targeting strategy employed to insert loxP sites on either side of exon 3 of StARD4. First, a targeting vector was constructed by placing a Neo selectable marker flanked by Frt sites with a loxP site in intron 3. Second, the targeted allele was created by introducing a second loxP in intron 2. Third, crossing the targeted allele mouse to the C57BL/6 ACT-FLPe mouse generated the floxed allele which was used for all subsequent crosses. Finally, to generate the null allele, floxed mice were crossed to the C57BL/6 CMV-Cre.
Fig. 2.
Confirmation of StARD4 knockout mouse creation. A: PCR analysis in order to determine the presence of the floxed allele only found in heterozygotes or homozygous mice (primer set AH. A is a 5′ primer located in exon 2 and H is a 3′ primer located ∼975bp downstream of exon 3). B: PCR analysis in order to determine the presence of the wild-type band present both in heterozygotes and in wild-type mice (primer set CH. Primer C is located in exon 3. Primer H is the same as above). C: Western blot analysis was performed on livers of 12-week-old StARD4 mice. Each phenotype was probed for StARD4 in two separate mice. StARD4 is represented by a 23.5 KDa band. D: β-Actin loading control for Western blot (Size = 45 KDa).
Fertility and fecundity in StARD4 knockout mice
The ratio of wild-type (28 of 102; 27.5%), heterozygous (45 of 102; 44.1%), and homozygous knockouts (29 of 102; 28.4%) offspring from the mating of heterozygous females and males was not significantly different from the expected Mendelian pattern of inheritance (χ2 = 1.922 with 2 degrees of freedom, p-value = 0.38). The mice developed normally with no evidence of overall dysfunction. Furthermore, there were no gender abnormalities as both female and male mice developed normally. Additionally, both female and male heterozygous and homozygous mutant mice were fertile and had similar litter sizes compared with wild-type mice (Table 1).
TABLE 1.
Mendelian ratio of mating to homozygosity of the Neo Floxed StARD4 gene an fertility and fecundity of StARD4 mutant mice
| Female | Male | ||
|---|---|---|---|
| Wt | 14 | 14 | |
| Het | 25 | 20 | |
| K.O. | 16 | 13 | |
| No. of Breedings | No. of Pregnancies | Litter Size (mean ± SD) | |
| Female +/+× male +/− | 16 | 16 | 6.56 ± 1.93 |
| Female +/−× male +/+ | 11 | 11 | 6.36 ± 3.17 |
| Female −/−× male −/− | 5 | 5 | 7.02 ± 1.35 |
The ratio of WT/Het/KO of 102 mice born from the mating of two StARD4 mice heterozygous for the Floxed allele recombined by the cre recombinase. The progeny fit a normal Mendelian distribution for mice born under the expected 1:2:1 ratio of Wt:Het:KO (observed ratio = 28:45:29) . When scrutinized with a chi square test, χ2 = 1.922 with 2 degrees of freedom. The two-tailed p-value for this was 0.3826, meaning that by conventional criteria, this difference is considered to be not statistically significant. Both female and male heterozygous mutant mice were fertile and had similar litter sizes. All numbers are compared with the Jackson Laboratory reported findings for normal C57BL/6J.
StARD4 expression pattern by tissue
To determine where to begin analysis of the StaRD4 knockout mice, it was prudent to check expression levels of the protein in various tissues. Three individual mice were sacrificed, various organs collected, RNA isolated, and quatitiative-PCR run for StARD4 expression. StARD4 is most highly expressed in the liver and macrophages, followed by the kidney and lung (data not shown). Expression for StARD4 is found at background for muscle and adipose tissue.
StARD4 growth curve
Weeks 4–12.
Weights of StARD4 wild-type mice and knockout littermates were measured weekly from week 4 to week 12. Females were significantly lighter at week 12. Male homozygous knockouts showed a significant decrease starting at week 5 and were ∼2 g lighter at week 12. Females took longer for the weight difference to become evident, likely due to the fact that females are lighter to start and therefore it is more difficult to see the changes (Fig. 3). Heterozygote females and males were also measured and the results parallel the numbers for a normal wild-type female or male (data not shown).
Fig. 3.
Growth curve of StARD4 knockout mice. StARD4 mice (Wt/Het/KO) from 4 weeks of age after weaning until 12 weeks of age fed a chow diet. Weights of mice were recorded weekly. Weights are represented as means with the SD indicated by the vertical lines. * indicates a p-value < 0.05.
StARD4 length measurements, BMI, and organ weights
Week 12.
Lengths of male StARD4 wild-type mice and knockout littermates were measured at week 12 before euthanasia after the mice were anesthetized. Homozygous StARD4 knockout mice showed significant decrease in length. When a body mass index (BMI) was calculated as g/(mm2), mice show no significant change (Table 2). Additionally, at 12 weeks of age, basic measurements of body weight and length and organs in females and males were analyzed for changes in size. This included the liver, kidney, heart, spleen, lung, and the epididymal fat pad. Although many of the organs in knockout mice were lower in weight than comparable wild-type mice, none of the organs, when compared with overall body weight, stood out as a candidate for excessive change that could be analyzed as a morphological defect on its own (data not shown).
TABLE 2.
Weight, length, and BMI of StARD4 knockout mouse and wild-type littermate, week 12
| Weight (g) | Length (cm) | BMI (g/mm2) | |
|---|---|---|---|
| Male Wt n = 14 | 28.0 ± 2.0 | 9.3 ± 0.2 | (3.1 ± 0.1) × 10−3 |
| K.O. n = 9 | 26.0 ± 1.7 | 8.9 ± 0.3 | (3.3 ± 0.1) × 10−3 |
| p-value | 0.034 | 0.001 | 0.101 |
All values are means ± SD. n = number of mice.
Food intake of StARD4 mice weeks 6–8
To try to understand the weight difference between wild-type and StARD4 knockouts mice, five male wild-type and five male knockout mice were single caged at week 6 and their food intake and corresponding body weight was recorded day by day for the duration of the two weeks (Fig. 4). Although body weight seemed to be trending in the same manner as previously reported and eventually became significant toward week 8, food intake did not seem to play a role in explaining this apparent change in weight phenotype.
Fig. 4.
Food intake experiment on StARD4 mice: weeks 6–8. Five 6-week-old StARD4 mice's and five 6-week-old wild-type mice's food intake and body weight were recorded on a daily basis around 4PM for two weeks straight to monitor if their weight change was effected at all by a change in eating habits. Although weights seemed to diverge as previously recorded reaching and then maintaining significance from day 11 onwards, the corresponding food intake did not seem to be responsible for this change. A: Food intake. B: Body weight. C: Food intake/body weight. * indicates a p-value < 0.05.
High-fat diet study and DEXA scan
To better understand the mechanism underlying the StARD4 weight phenotype, the 10 mice used above for the food intake study were placed on a 60% fat diet (high-fat diet) from week 8 until week 20. Weights were recorded weekly (data not shown). At week 8, mice had a significant weight difference that was erased after one week of feeding the mice a high-fat diet. Both wild-type and knockout mice continued to have nonsignificant differences in weight up until week 20 when the experiment was terminated.
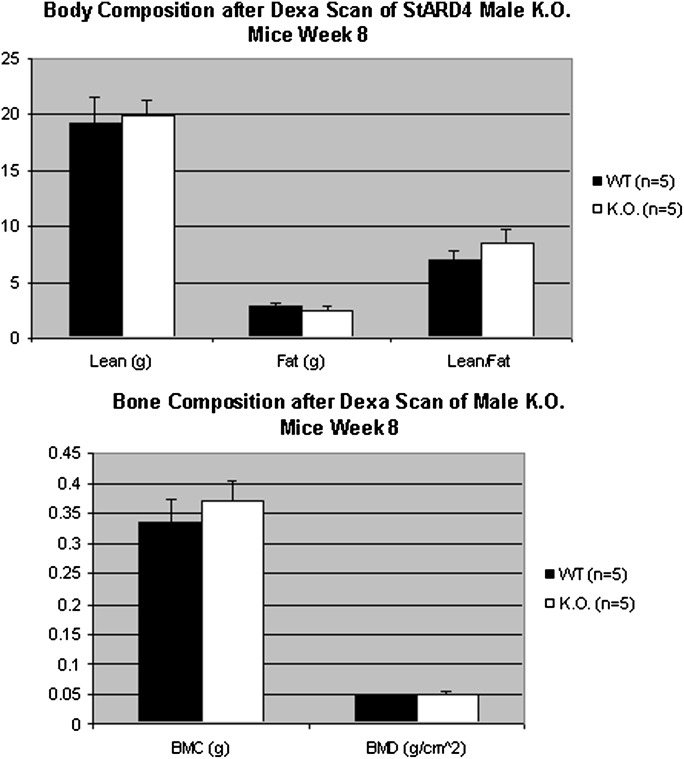
At 8 and 20 weeks, these mice were scanned using DEXA to measure bone composition and lean body mass versus total fat to try to parse out elements of the weight phenotype (Fig. 5 only shows week 8. Week 20 results are not included, but show insignificant changes). No significant findings for lean body mass, total fat, bone mineral content, or bone mass density arose from the DEXA scan.
Fig. 5.
DEXA scan of StARD4 mice at week 8 after 12 weeks of high-fat diet feeding. Five 8–20-week-old StARD4 mice and five wild-type littermates were fed a 60% high-fat diet. At week 20, a day before euthanasia, mice were subjected to a DEXA scan to elucidate their body and bone composition. There appeared to be no difference in lean versus normal fat or between bone mineral content and density of knockout and wild-type littermate controls.
Pathology of StARD4 mice
A complete pathological examination was performed on 12-week-old StARD4 female knockout mice and their wild-type littermates by the Tri-institutional Core Facility located in the Memorial Sloan Kettering Cancer Center Pathology Department. No anatomical or histological changes were found in any of the organs analyzed, which included brain, heart, lung, spleen, liver, gallbladder, stomach, duodenum, jejunum, ileum, colon, cecum, thymus, tongue, kidney, esophagus, pancreas, renal lymph nodes, salivary glands, brown fat, adrenal glands, sciatic nerve, spinal chord, thyroid glands, etc. Further, a full panel of blood chemistry was run including alkaline phosphatase, alanine amino transferase, aspartate amino transferase, gamma glutamyl transferase, albumin, globulin, creatinine, phosphorous, chloride, potassium, sodium, albumin/globulin ratio, blood urea nitrogen/creatine ratio, osmalarity, anion gap, red blood cells, white blood cells, neutrophils, lymphocytes, monocytes, eosinophils, and platelets with no significant changes found (data not shown).
Chow diet: serum, hepatic, and gallbladder lipid concentrations
There were no significant differences in serum cholesterol, triglycerides, cholesterol esters, free cholesterol, and glucose among wild-type and StARD4 knockout mice for mice, both female and male, 12 weeks of age, fed a chow diet ad libitum (Table 3). Hepatic cholesterol, free cholesterol, cholesterol ester, and triglyceride levels showed no significant differences between wild-type mice and their homozygous knockout littermate controls (Table 3). Gallbladder bile cholesterol, phospholipids, and bile acid levels were measured in StARD4 knockout mice at 12 weeks of age fed a chow diet ad libitum (Table 3). Although no change was found in the bile acids of either females or males or in cholesterol or phospholipids of males, a significant decrease was found in the knockout females in total bile cholesterol and phospholipids.
TABLE 3.
Plasma, hepatic, and gallbladder bile lipid concentrations of StARD4 mice at 12 weeks of age fed a chow diet
| Female |
Male |
||||
| Wt | K.O. | Wt | K.O. | ||
| Plasma | Total cholesterol (mg/dl) | 62 ± 12 | 63 ± 2 | 64 ± 13 | 68 ± 11 |
| HDL-C (mg/dl) | 25 ± 4 | 25 ± 5 | 37 ± 8 | 32 ± 8 | |
| Non-HDL-C (mg/dl) | 37 ± 8 | 38 ± 3 | 27 ± 5 | 36 ± 3 | |
| Free cholesterol (mg/dl) | 10 ± 3 | 11 ± 3 | 21 ±10 | 17 ± 4 | |
| Cholesterol ester (mg/dl) | 50 ± 10 | 53 ± 3 | 46 ± 15 | 56 ± 12 | |
| Triglycerides (mg/dl) | 21 ± 6 | 27 ± 8 | 30 ± 8 | 35 ± 8 | |
| Glucose (mg/dl) | 138 ± 24 | 116 ± 20 | 160 ± 36 | 163 ± 28 | |
| Hepatic | Total cholesterol (mg/g) | 2.8 ± 0.5 | 2.4 ± 0.4 | 1.6 ± 0.3 | 2.1 ± 0.7 |
| Free cholesterol (mg/g) | 2.2 ± 0.5 | 1.9 ± 0.4 | 1.2 ± 0.3 | 1.5 ± 0.6 | |
| Cholesterol ester (mg/g) | 0.6 ± 0.4 | 0.54 ± 0.3 | 0.4 ± 0.1 | 0.4 ± 0.2 | |
| Triglycerides (mg/g) | 14.2 ± 4.9 | 12.2 ± 4.6 | 7.9 ± 3.5 | 7.4 ± 2.5 | |
| Gall bladder bile | Total cholesterol (mg/dl) | 190 ± 50 | 135 ± 64* | 186 ± 59 | 161 ± 88 |
| Phospholipids (mg/dl) | 2065 ± 573 | 1576 ± 591* | 1446 ± 256 | 1532 ± 424 | |
| Bile acids (mM) | 94 ± 35 | 76 ± 6 | 43 ± 15 | 46 ± 16 | |
Values are means ± SD. n = 8–15 mice per group. * p-value < 0.05. All other p-values nonsignificant. Non-HDL is calculated from the difference between total cholesterol and HDL.
IPGTT
Three to five days before euthanasia at 12 weeks of age, the StARD4 mice, both female and male, were fed a chow diet and subjected to an IPGTT. This exam measures the mouse's responsiveness and recovery to a glucose stimulus. Glucose was measured at 0, 15, 30, 60, and 120 min after injecting a glucose bolus of 2g/kg (supplementary ). The StARD4 knockout mice exhibited no difference from their wild-type littermates.
0.2% Lovastatin and 0.5% cholesterol diets’ effect on StARD4 mRNA and protein
In order to perturb the system, two approaches were taken. Both approaches followed the same regimen as follows: from week 6–7, mice were fed a 0.0% cholesterol AIN76a diet and then from week 7–8, mice were either fed the AIN76a supplemented with 0.2% lovastatin or 0.5% cholesterol and euthanized at week 8. mRNA and protein from liver was extracted and probed for StARD4 expression or protein levels (data not shown). The 0.2% lovastatin diet elicited a significant 3-fold increase in StARD4 expression found also in the protein levels whereas the high cholesterol diet did not seem to alter StARD4 levels directly (supplementary Fig. II).
0.2% Lovastatin diet: serum and hepatic lipid concentrations
Female and male mice on the 0.2% lovastatin diet showed no significant differences in serum cholesterol, triglycerides, cholesterol esters, and free cholesterol. Additionally, hepatic cholesterol, free cholesterol, cholesterol ester, and triglyceride levels showed no significant differences between wild-type mice and their homozygous knockout littermates (supplementary Table II).
0.5% Cholesterol diet: serum and hepatic lipid concentrations
Male mice on the 0.5% cholesterol diet mostly showed no change as well in serum lipid concentration, whereas the female homozygous mice had mildly decreased total cholesterol (20%), LDL (45%), and cholesterol esters (18%) as compared with wild-type controls (all P < 0.04) (supplementary Table II). Hepatic cholesterol, free cholesterol, cholesterol ester, and triglyceride levels showed no significant differences between wild-type mice and their homozygous knockout littermates (supplementary Table III).
Global transcription profiling of liver
Illumina mouse array gene chips were used to analyze transcript profile of RNA extracted from livers of four different male wild-type mice and four different male homozygous knockout mice at 12 weeks of age on a 0.0% cholesterol diet. Of the ∼23,000 transcripts detected in this analysis, only two transcripts showed a >2-fold increase, LCN2 and Hamp2. No genes survived Student's t-tests until the fold change was lowered to below 1.1-fold change, indicating a lack of significant difference in mRNAs related to sterol metabolism. In addition, qPCR on wild-type and knockout mice fed a 0.0% cholesterol diet were used to assess expression of genes related to intracellular cholesterol transport. As with the microarray experiment, no differences in expression were found for NPC2, MLN64, StARD5, and Cav-1/2. qPCR showed NPC1, a gene not included in the microarray, was ∼2.5-fold lower in expression in knockout compared with WT mice [P = 0.05(Fig. 6)]. Most of the same genes mRNA levels, excluding Cav1/2, were also checked on the 0.2% lovastatin diet and the 0.5% cholesterol (supplementary ). Moderate increases in StARD5 expression was found on both diets with little change elsewhere.
Fig. 6.
RNA expression of StARD4 WT and KO mice. At 8 weeks of age, mice were euthanized. RNA was extracted from mice fed 0.0% cholesterol diet and expression levels were checked for StARD5, MLN64, NPC1, NPC2 Cav-1, and Cav-2. NPC1 RNA was significantly lower in the knockout mouse. All other comparisons of wild-type to knockout had insignificant p-values. * indicates a p-value < 0.05.
DISCUSSION
The START domain superfamily of proteins has been shown to be involved in many pathways of intracellular cholesterol trafficking (4, 17, 20). It has been proposed that all proteins with a START domain contain a similar binding pocket that binds varying ligands or cholesterol derivatives based on modification of that binding domain (21). StARD4 belongs to the StARD4 subfamily, a START domain subfamily containing StARD4, StARD5, and StARD6. StARD5 has previously been shown to bind cholesterol and 25-hydroxycholesterol whereas StARD4, StARD1, and MLN64 have been shown to only bind cholesterol (14, 15). Unlike StARD1 and MLN64, StARD4 and StARD5 do not have N-terminal localization sequences and are therefore predicted to be cytoplasmic proteins. Start domain cytoplasmic localization is not uncommon, exemplified by PCTP/StARD2, a protein that plays a crucial role in phosphatidylcholine intracellular transport (22). Additionally, StARD4 and StARD5 share over 30% homology. The present studies used targeted mutation of the StARD4 gene in mice to probe the functional role of StARD4 in lipid metabolism.
Much to our surprise, mice homozygous for mutation disrupting StARD4’s StART domain were healthy and displayed next to no abnormalities in plasma lipid dynamics or liver cholesterol metabolism. Dietary manipulations as performed in this study had no effect on overall lipid metabolism. However, it cannot be ruled out that a different dietary manipulation might be able to exacerbate the phenotype of the StARD4 knockout. Additionally, the time of exposure to a particular diet may also be of importance. Soccio et al. (11) previously published that wild-type mice fed a 0.5% cholesterol diet for 3 weeks decreases StARD4 mRNA levels by 2–3-fold, whereas we report here that the same diet fed to wild-type for 1 week has no such effect.
In total, six START family members have been knocked out in mice: StARD1, StARD2 (PCTP), StARD3 (MLN64), StARD11 (CERT), and StARD12 (DLC1-deleted in liver cancer 1), and now StARD4. StARD1 knockout has provided a striking and clinically relevant phenotype, as the knockout is the model used to study the human disease congenital lipoid adrenal hyperplasia (8). However, it has proven difficult to uncover and characterize lipid phenotypes in the other StAR protein mouse knockout models. Initial studies of PCTP disruption showed no difference in phospholipid metabolism (23). However, when pushed with a lithogenic diet, PCTP knockouts showed an impaired excretion of lipid into bile and an improper balance in the bile of cholesterol to phospholipids (24, 25). Mice with MLN64 mutations also have no apparent abnormalities in lipid metabolism or storage when pushed with a chow, high-cholesterol, or high–fat diet (26). StARD11 and StaRD12 are both embryonic lethal. StARD11 knockouts are embryonic lethal at day E11.5, caused by degeneration of the mitochondria (27). StARD12 knockouts are embryonic lethal at day E10.5 with defects in neural tube, brain, heart, placenta, actin filaments, and focal adhesions (28). This hints toward the likely conclusion that there is a level of redundancy between various START domain proteins that make a single mouse knockout model difficult to use for investigation of the START domain related proteins.
In support of this idea, a family of proteins related to the StAR proteins is the oxysterol binding proteins (OSBPs). OSBPs are part of a large family of lipid binding proteins of which there are at least 12 members that play numerous roles in lipid distribution and metabolism (4). A large amount of the work on oxysterol-related proteins has been done in yeast Sacchromyces cerevisiae, which has seven OSBP homolog proteins (4). Yeast mutants for any one of the seven OSBP homolog genes have little to no defect in plasma membrane to endoplasmic reticulum sterol transfer, suggesting a level of redundancy (29). However, conditional deletion of all seven genes slows exogenous sterol transport, causes vacuolar fragmentation and accumulation of lipid droplets ultimately ending in cell death (30, 31). Overall, there are many families of intracellular transporters, including, Niemann Pick C proteins, the caveolins, OSBPs, and sterol carrier protein 2, and all might play some role in compensating for the missing StARD4 protein. It has even been suggested that only a limited number of the genes involved in lipid transport and associated membrane trafficking have to date been identified (12). Just recently, TMEM97 has been identified by an RNAi screen as a novel transporter binding to the Niemann Pick C 1 protein in the regulation of endosomal uptake of cholesterol from LDL particles (32).
It is important to emphasize that part of the construction of this knockout left room for the creation of tissue-specific knockouts. Although this paper used a CMV-cre promoter to create a ubiquitous knockout, it is possible that tissue specific knockouts, perhaps a LysM-cre promoter for macrophage specific knockout, could be used to test for the role of StARD4 as it relates to atherosclerosis. Thorp et al. (33) has shown successful use of gene specific knockouts to show increased apoptosis in mice with macrophage specific deletion of bcl2. Other LysM-cre specific crosses were done for the PCTP (StARD2) protein, whose initial lipid phenotype was unclear. In this case, macrophage specific deletion of PCTP resulted in decreased cholesterol efflux to ApoA-I particles (34). It is possible that StARD4 might play an important role in the development of atherosclerosis by influencing macrophage cholesterol efflux.
Interestingly, StARD4 mice seemed to have a weight- related phenotype in that they were about 2 g lighter than wild-type littermate controls. It seemed that this was not due to issues involving food intake or energy metabolism as shown in the high-fat diet feeding study. The lengths of the animals were smaller as well, indicating that the size observation might be due to a developmental defect of the StARD4 mice. It has been shown that nonsteroidogenic COS-1 cells cotransfected with P450scc, 3β-hydroxysteroid reductase, necessary proteins for progesterone production, and StARD4 have mildly increased steroidogenesis (1). So, one could speculate that the absence of StARD4 may impair steroid hormone production during mouse development and compromise growth. However, the decreased weight and length are in proportion with the mice overall being smaller, a phenotype that is very complex and could be related to any number of factors not discussed.
Finally, female knockout mice showed a significant impairment of cholesterol and phospholipid excretion into gallbladder bile. This is in agreement with previously published data that showed that overexpression of StARD4 increased bile synthesis (15). As mentioned before, PCTP knockout mice also show an imbalance in cholesterol and phospholipids in the bile, indicating a complex mechanism governing intracellular sterol dynamics. This same study showed that StARD4 enhanced cholesterol ester synthesis in hepatocytes from exogenous cholesterol sources. It is then tempting to postulate that the decrease in total cholesterol, LDL-C, and cholesterol ester seen in females on the high-cholesterol diet could somehow be mechanistically related to this earlier finding. Perhaps StARD4 plays some role in shuttling between the ER and lipid droplets.
In conclusion, mutation of the START domain of StARD4, a protein thought to play a vital role in intracellular cholesterol dynamics and transport, does not appear to impair cholesterol homeostasis intracellularly. These findings suggest that proteins with redundant functions exist that can supplement the role of StARD4 in vivo and that the whole intracellular cholesterol milieu remains a complicated and underdeveloped research topic for the study of cholesterol regulation and the development of atherosclerosis.
Supplementary Material
Acknowledgments
The authors thank Dr. Changcheng Zhou and Dr. Ralph Burkhardt for their patience, guidance and wisdom. Without both of their contributions in the form of time, mentorship, comradeship, and the occasional prank, life would not have been the same over the past 3–4 years. Microinjection was performed by the Rockefeller University Gene Targeting Resource Center. A special thanks to Dr. Chingwen Yang, Jing Gao, and Katarina Zafranskaia. Clinical pathology was performed at the Memorial Sloan Kettering Pathology Laboratory. A special thanks to Dr. Suzanne Couto.
Footnotes
Abbreviations:
- BMI
- body mass index
- CERT
- ceramide transfer protein
- DLC1
- deleted in liver cancer
- DEXA
- dual energy X-ray absorptiometry
- FRT
- Flp recombination target
- IPGTT
- intraperitoneal glucose tolerance test
- MLN64
- metastatic lymph node 64
- OSBP
- oxysterol binding protein
- PC
- phosphatidylcholine
- PCTP
- phosphatidylcholine transport protein
- StAR
- steroidogenic acute regulatory protein
- START
- StAR-related lipid transfer
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables and three figures.
REFERENCES
- 1.Soccio R. E., Adams R. M., Maxwell K. N., Breslow J. L. 2005. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress. J. Biol. Chem. 280: 19410–19418. [DOI] [PubMed] [Google Scholar]
- 2.Ikonen E. 2008. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 9: 125–138. [DOI] [PubMed] [Google Scholar]
- 3.Soccio R. E., Breslow J. L. 2004. Intracellular cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 24: 1150–1160. [DOI] [PubMed] [Google Scholar]
- 4.Prinz W. A. 2007. Non-vesicular sterol transport in cells. Prog. Lipid Res. 46: 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maxfield F. R., Wustner D. 2002. Intracellular cholesterol transport. J. Clin. Invest. 110: 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikonen E. 2006. Mechanisms for cellular cholesterol transport: defects and human disease. Physiol. Rev. 86: 1237–1261. [DOI] [PubMed] [Google Scholar]
- 7.Stocco D. M. 2001. StAR protein and the regulation of steroid hormone biosynthesis. Annu. Rev. Physiol. 63: 193–213. [DOI] [PubMed] [Google Scholar]
- 8.Caron K. M., Soo S. C., Parker K. L. 1998. Targeted disruption of StAR provides novel insights into congenital adrenal hyperplasia. Endocr. Res. 24: 827–834. [DOI] [PubMed] [Google Scholar]
- 9.Bose H. S., Sugawara T., Strauss J. F., 3rd, Miller W. L. 1996. The pathophysiology and genetics of congenital lipoid adrenal hyperplasia. International Congenital Lipoid Adrenal Hyperplasia Consortium. N. Engl. J. Med. 335: 1870–1878. [DOI] [PubMed] [Google Scholar]
- 10.Wirtz K. W. 1991. Phospholipid transfer proteins. Annu. Rev. Biochem. 60: 73–99. [DOI] [PubMed] [Google Scholar]
- 11.Soccio R. E., Adams R. M., Romanowski M. J., Sehayek E., Burley S. K., Breslow J. L. 2002. The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc. Natl. Acad. Sci. USA. 99: 6943–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang T. Y., Chang C. C., Ohgami N., Yamauchi Y. 2006. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22: 129–157. [DOI] [PubMed] [Google Scholar]
- 13.Soccio R. E., Breslow J. L. 2003. StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J. Biol. Chem. 278: 22183–22186. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Agudo D., Ren S., Hylemon P. B., Montanez R., Redford K., Natarajan R., Medina M. A., Gil G., Pandak W. M. 2006. Localization of StarD5 cholesterol binding protein. J. Lipid Res. 47: 1168–1175. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Agudo D., Ren S., Wong E., Marques D., Redford K., Gil G., Hylemon P., Pandak W. M. 2008. Intracellular cholesterol transporter StarD4 binds free cholesterol and increases cholesteryl ester formation. J. Lipid Res. 49: 1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose H. S., Whittal R. M., Ran Y., Bose M., Baker B. Y., Miller W. L. 2008. StAR-like activity and molten globule behavior of StARD6, a male germ-line protein. Biochemistry. 47: 2277–2288. [DOI] [PubMed] [Google Scholar]
- 17.Ponting C. P., Aravind L. 1999. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24: 130–132. [DOI] [PubMed] [Google Scholar]
- 18.Pandak W. M., Ren S., Marques D., Hall E., Redford K., Mallonee D., Bohdan P., Heuman D., Gil G., Hylemon P. 2002. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J. Biol. Chem. 277: 48158–48164. [DOI] [PubMed] [Google Scholar]
- 19.Liu P., Jenkins N. A., Copeland N. G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauss J. F., 3rd, Kishida T., Christenson L. K., Fujimoto T., Hiroi H. 2003. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol. Cell. Endocrinol. 202: 59–65. [DOI] [PubMed] [Google Scholar]
- 21.Iyer L. M., Koonin E. V., Aravind L. 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 43: 134–144. [DOI] [PubMed] [Google Scholar]
- 22.Feng L., Chan W. W., Roderick S. L., Cohen D. E. 2000. High-level expression and mutagenesis of recombinant human phosphatidylcholine transfer protein using a synthetic gene: evidence for a C-terminal membrane binding domain. Biochemistry. 39: 15399–15409. [DOI] [PubMed] [Google Scholar]
- 23.van Helvoort A., de Brouwer A., Ottenhoff R., Brouwers J. F., Wijnholds J., Beijnen J. H., Rijneveld A., van der Poll T., van der Valk M. A., Majoor D., et al. 1999. Mice without phosphatidylcholine transfer protein have no defects in the secretion of phosphatidylcholine into bile or into lung airspaces. Proc. Natl. Acad. Sci. USA. 96: 11501–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scapa E. F., Pocai A., Wu M. K., Gutierrez-Juarez R., Glenz L., Kanno K., Li H., Biddinger S., Jelicks L. A., Rossetti L., et al. 2008. Regulation of energy substrate utilization and hepatic insulin sensitivity by phosphatidylcholine transfer protein/StarD2. FASEB J. 22: 2579–2590. [DOI] [PubMed] [Google Scholar]
- 25.Kanno K., Wu M. K., Scapa E. F., Roderick S. L., Cohen D. E. 2007. Structure and function of phosphatidylcholine transfer protein (PC-TP)/StarD2. Biochim. Biophys. Acta. 1771: 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishida T., Kostetskii I., Zhang Z., Martinez F., Liu P., Walkley S. U., Dwyer N. K., Blanchette-Mackie E. J., Radice G. L., Strauss J. F., 3rd 2004. Targeted mutation of the MLN64 START domain causes only modest alterations in cellular sterol metabolism. J. Biol. Chem. 279: 19276–19285. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Rao R. P., Kosakowska-Cholody T., Masood M. A., Southon E., Zhang H., Berthet C., Nagashim K., Veenstra T. K., Tessarollo L., et al. 2009. Mitochondrial degeneration and not apoptosis is the primary cause of embryonic lethality in ceramide transfer protein mutant mice. J. Cell Biol. 184: 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durkin M. E., Yuan B. Z., Zhou X., Zimonjic D. B., Lowy D. R., Thorgeirsson S. S., Popescu N. C. 2007. DLC-1:a Rho GTPase-activating protein and tumour suppressor. J. Cell. Mol. Med. 11: 1185–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raychaudhuri S., Im Y. J., Hurley J. H., Prinz W. A. 2006. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 173: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beh C. T., Rine J. 2004. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 117: 2983–2996. [DOI] [PubMed] [Google Scholar]
- 31.Beh C. T., Cool L., Phillips J., Rine J. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 157: 1117–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartz F., Kern L., Erz D., Zhu M., Gilbert D., Meinhof T., Wirkner U., Erfle H., Muckenthaler M., Pepperkok R., et al. 2009. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10: 63–75. [DOI] [PubMed] [Google Scholar]
- 33.Thorp E., Li Y., Bao L., Yao P. M., Kuriakose G., Rong J., Fisher E. A., Tabas I. 2009. Brief report: increased apoptosis in advanced atherosclerotic lesions of Apoe-/- mice lacking macrophage Bcl-2. Arterioscler. Thromb. Vasc. Biol. 29: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baez J. M., Tabas I., Cohen D. E. 2005. Decreased lipid efflux and increased susceptibility to cholesterol-induced apoptosis in macrophages lacking phosphatidylcholine transfer protein. Biochem. J. 388: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.