Abstract
Dopamine (DA) neurons in the substantia nigra (SNDA neurons) are among the most severely affected in Parkinson’s disease (PD). Mitochondrial complex I inhibition by rotenone or MPTP can induce SNDA neurodegeneration and recapitulate motor disability in rodents. We performed a transcriptional analysis of the midbrain response to complex I inhibition focused on selected metabolic transcripts using quantitative real-time RT-PCR in conjunction with laser-capture microdissection (LCM) of immunofluorescently-targeted SNDA and ventral tegmental area (VTA) DA neurons. There were DA neuron-selective alterations in metabolic transcripts in response to generalized complex I inhibition dependent on the behavioral response of the animal, and vulnerable SNDA neurons were more dynamic in their metabolic transcriptional response than less vulnerable VTADA neurons. The metabolic transcriptional response of DA neurons may contribute significantly to the ultimate toxicity associated with mitochondrial inhibition, and better understanding of this response may provide insight into potential targets for neuroprotection in PD.
Keywords: Parkinson’s disease, mitochondria, gene expression, rotenone, MPTP, dopamine, substantia nigra, ventral tegmental area
The underlying causes of neurodegeneration in Parkinson’s disease (PD) are not known, but mitochondrial dysfunction has been implicated by the repeated finding of decreased activity of complex I of the electron transport chain and the more recent description of mitochondrial DNA deletions in PD patients (Bender et al., 2006; Parker et al., 1989; Schapira et al., 1989; Schapira et al., 1990). Additionally, chemical inhibition of complex I by rotenone or MPTP can recapitulate dopamine (DA) neuron degeneration and associated neurological disability in experimental animals (Betarbet et al., 2000; Dawson et al., 2002; Fleming et al., 2004; Schneider et al., 1987; Sherer et al., 2003). Furthermore, several mutations that cause inherited parkinsonism affect gene products (e.g., PINK1, parkin, α-synuclein) that can impact mitochondrial function (Gispert et al., 2009; Loeb et al., 2010; Narendra et al., 2010).
Although many neuronal populations are affected, dopamine (DA) neurons in the substantia nigra pars compacta (SNDA neurons) are among the most severely depleted in both human PD and experimental chemical parkinsonism (Betarbet et al., 2000; Dawson et al., 2002; Fearnley and Lees, 1991; Hirsch et al., 1988; Okamura et al., 1995; Rinne, 1993; Schneider et al., 1987; Varastet et al., 1994). We have hypothesized that greater metabolic demand, as evidenced by overabundance of RNA transcripts related to energy metabolism, is in part responsible for the exquisite susceptibility of SNDA neuron neurons to metabolic stress (Greene et al., 2005). Interestingly, calcium-dependent pacemaker activity in SNDA neurons has been recently proposed to render these neurons more susceptible to mitochondrial inhibition by increasing metabolic demand (Chan et al., 2007).
Despite increasing knowledge about the mechanisms responsible for the vulnerability of SNDA neurons to mitochondrial stress, the downstream molecular responses to complex I inhibition are not well understood. It is only recently that regulated signaling events, such as K-ATP channel activation, as opposed to indiscriminate disruption of neuronal activities, have been implicated in determining the balance between neuronal degeneration and survival after metabolic inhibition (Liss et al., 2005). In particular, the metabolic transcriptional response of DA neurons may contribute significantly to shaping the ultimate toxicity associated with mitochondrial inhibition.
We performed a limited transcriptional analysis of the brain response to rotenone intoxication focused on selected metabolic transcripts and followed up with a similar examination in MPTP-treated mice using quantitative real-time RT-PCR in conjunction with laser-capture microdissection (LCM) of immunofluorescently-targeted SNDA and ventral tegmental area (VTA) DA neurons. Our results clearly demonstrate the necessity of using enriched cellular samples to allow meaningful interpretation of transcriptional profiling experiments and indicate that SNDA neurons have a greater dynamic response to metabolic inhibition than more resistant ventral tegmental area (VTA) DA neurons.
MATERIALS AND METHODS
Rats and rotenone treatment
All experimental procedures were in accordance with the NIH Guide for the Care and Use of Experimental Animals and approved by the Emory University Institutional Animal Care and Use Committee. Male Lewis rats (250g) were subcutaneously implanted with osmotic minipumps resulting in administration of rotenone (3 mg/kg/day) or vehicle (50:50 DMSO:polyethylene glycol). Infusion was continuous for 28 days (Greene et al., 2009; Sherer et al., 2003). During the first week of infusion, rats were given access to hydrated chow on the floor of the cage. At the end of infusion, rats were anesthetized with isofluorane and killed by decapitation. Brains were rapidly removed and bisected. Half was immediately frozen on dry ice, and the midbrain was dissected out of the other half and frozen on dry ice.
Mice and MPTP treatment
Male C57Bl/6 mice (3 months old) were administered MPTP in saline vehicle at a total dose of 60 mg/kg of MPTP given in four intraperitoneal injections 2 hours apart on a single day (Anderson et al., 2007; Teismann et al., 2003). Controls received saline using the same protocol. After 3 days, mice were anesthetized with isofluorane and killed by decapitation. Brains were rapidly removed and immediately frozen on dry ice.
General health assessment and gross motor function
Rats were observed twice daily during the first week of infusion and daily thereafter. General health and gross motor function were assessed by observing in-cage behavior and during brief gentle handling to check for rigidity (hunched posture and increased tail tone), bradykinesia (slowed movement and/or absence of rearing), dystonia (clenched paws), autonomic signs (piloerection), abnormal thermoregulation, and signs of dehydration (dry mucus membranes or tenting skin). Sick animals were killed if they were unable to care for themselves or lost more than 25% of their body weight.
Mice were observed daily after MPTP treatment. The day after treatment, all mice were moving normally around the cage.
Rapid tyrosine hydroxylase immunofluorescence (THIF)
Ten micron frozen sections through the midbrain were collected onto uncoated microscope slides, refrozen on dry ice, and placed in a −80 °C freezer. For staining, sections were fixed in ice-cold acetone for 2 min, rinsed in phosphate-buffered saline (PBS) with 0.1% Triton X-100 (PBTx), and incubated for 2 min in mouse anti-TH (1:40 dilution; Zymed, CA). After rinsing twice in PBTx, sections were incubated with Alexafluor 488-conjugated goat anti-mouse secondary antibody (1:28 dilution; Molecular Probes, OR) for 1 min. Both antibody solutions contained 400 U/ml of RNasin RNase inhibitor (Promega, WI). Slides were rinsed in PBS, dipped in cresyl violet for Nissl stain, and dehydrated to xylene. Sections were dried in a fume hood and LCM performed within 4 hours (Greene et al., 2005).
LCM and RNA isolation
Laser capture microdissection was performed using an Arcturus Pixcell IIe system with fluorescent illumination (Arcturus, CA) and the following parameters: spot size = 7 µm; power = 45–85 mW; and duration = 750–1100 µs. Approximately 500 healthy-appearing TH-positive neurons per animal from both SN (5.2–5.8 mm caudal to bregma, limited to the lateral and ventral portions of the SN pars compacta) and VTA (5.2–5.8 mm caudal to bregma, ventral to the red nucleus, dorsal to the interpeduncular nuclei, and medial to the medial terminal nucleus of the accessory optic tract, fascicles of the oculomotor nerve, or the medial lemniscus) were isolated onto separate LCM HS Caps (Arcturus). Samples were anatomically matched by identifying the above anatomical landmarks after immunostaining and prior to LCM. For each animal, TH-positive neurons from the SN were pooled into one sample, and neurons from the VTA were pooled into another, permitting a paired analysis of the two neuron types. Cells near the boundary between the regions were not dissected to ensure anatomical distinction between the regions. Total RNA was immediately extracted using the Extractsure adapter and PicoPure Isolation Kit (Arcturus) with DNase digestion (Qiagen RNase-free DNase Set). RNA was stored at −80 °C until use (Greene et al., 2005).
Quantitative real-time RT-PCR (Q-RT-PCR)
Transcripts of interest were selected to obtain information about both glycolytic and mitochondrial energy metabolism pathways (Bough et al., 2006; Greene et al., 2005). Primers were identified using Primer Quest (Integrated DNA Technologies, IA) and synthesized by IDT. All primer sets resulted in a single product as assessed by 2% agarose gel electrophoresis and melt-curve analysis. HPRT was used as the reference gene for each target gene. All primer sets had PCR efficiencies similar to the HPRT primer set (85–100%), as determined by analysis of DCt (Ct of target gene minus Ct for HPRT) across serial dilutions of template. Control reactions without added template consistently did not produce product. COX1 and PDHC were not evaluated in mouse samples because specific and efficient primer sets for mouse could not be designed after four attempts.
Total RNA from LCM samples was reverse transcribed using the Superscript II cDNA Synthesis Kit (Invitrogen, CA) with random hexamers (50 ng/µl) for priming. First strand cDNA from the equivalent of ten TH-positive neurons was used as template in a 25 µl PCR reaction with 400 nM primers, and SYBR Green PCR Master Mix (Applied Biosystems, UK). Forty-five cycles of PCR were performed using an iCyler and iQ software (Biorad, CA). Samples from each animal were run in duplicate. Threshold cycle (Ct) was determined and compared to Ct for the reference gene (HPRT), thereby normalizing for small differences in starting template concentrations (DCt). The mean DCt for control (vehicle-treated) samples was determined, and all results were normalized to the mean control using the equation 100*2(mean control DCt minus sample DCt). For direct comparison between SN and VTADA neurons in identical animals DDCt (VTADCt – SNDCt) was calculated and compared between treatments.
Statistics
Results are expressed as means. Two-way ANOVAs were used to compare relative transcript abundance across treatment group and transcript. Bonferroni post-tests were used for selected comparisons to vehicle-treated samples. A p-value of less than 0.05 was considered significant.
RESULTS
The response of rats to subcutaneous rotenone in this experiment was similar to that which we have previously described (Greene et al., 2009; Sherer et al., 2003). All animals exhibited piloerection and weight loss throughout the first 7–10 days of treatment. The majority of rats recovered, resumed gaining weight, and by 4 weeks of treatment was indistinguishable from their vehicle-treated counterparts with the exception of lower body weight. A subset of ‘sick’ animals was killed when precipitous weight loss was associated with recumbence and markedly slowed movement (Fig 1). Three out of seven sick animals displayed some TH loss in the striatum. There was no obvious TH loss in the midbrain.
Figure 1. Timeline of rotenone experiments.
Vehicle-treated rats and rotenone-treated rats were killed at both 1 and 4 weeks after the beginning of rotenone infusion. ‘Sick’ rats were sacrificed at the time points indicated when they became unable to care for themselves appropriately. Three sick rats had decreased tyrosine hydroxylase immunoreactivity in the striatum. One had a diffuse lesion (open arrow) and two had punctate lesions (closed arrows).
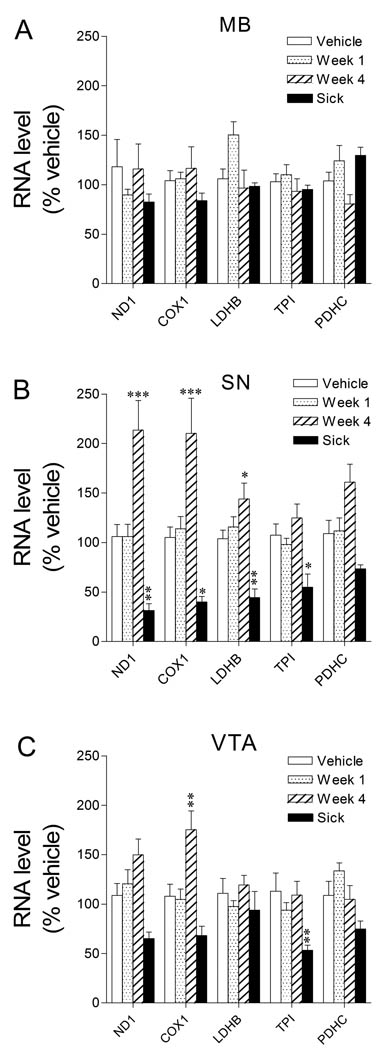
Examination of metabolic transcripts in microdissected midbrain samples revealed no alterations during rotenone intoxication (Fig 2A); however, there were dramatic alterations in the same transcripts when they were assessed in IFLCM-isolated dopamine neurons (Fig 2B, C). This is particularly informative since the micro- and IFLCM-dissected samples were obtained from opposite sides of the midbrain from the same animals.
Figure 2. Dynamic, neuron-selective metabolic transcriptional responses during rotenone infusion.
A. No changes in metabolic transcripts were evident when microdissected midbrain tissue was analyzed by Q-RT-PCR. B. There was a large dynamic range for metabolic transcript level in SNDA neurons. Metabolic transcripts were increased up to 200% in SNDA neurons after 4 weeks of exposure to rotenone. SNDA neurons from sick animals had dramatically lower levels of metabolic transcripts. C. While the trend was similar in VTADA neurons, the magnitude of differences was lower. ND1, complex I ND1 subunit; COX1, cytochrome oxidase subunit 1; LDHB, lactate dehydrogenase B; TPI, triose phosphate isomerase; PDHC, pyruvate dehydrogenase complex E1 subunit. *p < 0.05, **p < 0.01, ***p < 0.001 by post-hoc Bonferroni after two-way ANOVA. N = 12 (Vehicle), 8 (Week 1), 8 (Week 4), and 7 (Sick).
Relative levels of metabolic transcripts in SNDA and VTADA neurons from rats treated with rotenone for 1 week were similar to samples from vehicle-treated rats (Fig 2B, C).
SNDA neurons had marked variation in levels of metabolism related transcripts between experimental groups with an overall increased abundance of metabolic transcripts after 4 weeks of treatment, but decreased abundance in sick animals (Fig 2B). ANOVA revealed a highly significant main effect of treatment group F(3,150) = 48.6, p < 0.0001, no main effect of transcript F(4,150) = 1.4, p > 0.05, and a slightly significant group times transcript interaction F(12,150) = 2.2, p < 0.02. Post hoc analysis revealed significant increases between 4 week rotenone- and vehicle-treated rats for ND1, COX1, LDH, and PDHC and also significant decreases between rotenone-treated sick and vehicle-treated rats for ND1, COX1, LDH, and TPI.
VTADA neurons had less dramatic but still significant variation in levels of metabolism related transcripts between experimental groups with an overall increased abundance of metabolic transcripts after 4 weeks of rotenone treatment, but decreased abundance in sick animals (Fig 2C). ANOVA revealed a highly significant main effect of treatment group F(3,155) = 14.2, p < 0.0001, no main effect of transcript F(4,155) = 1.4, p > 0.05, and a slightly significant group times transcript interaction F(12,155) = 1.9, p < 0.05. Post hoc analysis revealed a significant increase in COX1 between 4 week rotenone- and vehicle-treated rats and a significant decrease in TPI between rotenone-treated sick and vehicle-treated rats.
Given the unexpected decrease in metabolic transcripts in sick rotenone-treated rats, we repeated the experiment in mice treated with MPTP. We have previously reported that mice treated using this protocol have a 60% decrease in TH immunoreactivity in the striatum (Anderson et al., 2007). There was a qualitative decrease of TH immunofluorescence in the midbrain from MPTP-treated mice, but only healthy-appearing neurons were captured for analysis.
Results from MPTP-treated mice were similar to sick rotenone-treated rats in that SNDA neurons displayed a marked overall decrease in abundance of metabolic transcripts (Fig 3). ANOVA revealed a highly significant main effect of treatment group F(3,40) = 10.6, p < 0.0001, but no main effect of transcript F(3,40) = 1.7, p > 0.05 or treatment times transcript interaction F(9,40) = 0.9, p > 0.05. Post hoc analysis revealed a significant decrease in ND1 and LDH transcripts between MPTP-treated and vehicle-treated mice in SNDA neurons.
Figure 3. Neuron-selective metabolic transcriptional responses after MPTP treatment.
SNDA neurons from MPTP-treated mice had lower levels of metabolic transcripts than those from vehicle-treated mice. There were no significant changes in VTADA neurons after MPTP treatment. Dashed line represents vehicle-treated transcript abundance. ND1, complex I ND1 subunit; LDHB, lactate dehydrogenase B; TPI, triose phosphate isomerase; ANT2, adenine nucleotide translocator 2. *p < 0.05, **p < 0.01 by post-hoc Bonferroni after two-way ANOVA. N = 4 (Vehicle) and 3 (MPTP).
Direct comparison between neuron populations from the same animals revealed a decrease in the ratio of metabolic transcript abundance between SNDA and VTADA neurons in MPTP-treated mice and sick rotenone rats (Fig 4). For MPTP-treated mice, ANOVA revealed a highly significant main effect of treatment group in F(1,20) = 35.9, p < 0.0001, but no main effect of transcript F(3,20) = 1.3, p > 0.05 or treatment times transcript interaction F(3,20) = 0.4, p > 0.05. Post hoc analysis revealed a significant decrease in the SN:VTA ratio of ND1 and LDHB transcripts in MPTP-treated mice. In other words, in vehicle-treated mice metabolic transcripts were more abundant in SNDA neurons, but after MPTP treatment, they were more abundant in VTADA neurons (Fig 4A).
Figure 4. Dopamine neuron subtype-selective metabolic transcriptional responses after complex 1 inhibition.
A. When compared in the same mice, the ratio of expression level for metabolic transcripts shifts from favoring SNDA neurons to favoring VTADA neurons in MPTP-treated animals. N = 4 (Vehicle) and 3 (MPTP). B. When compared in the same rats, the ratio of expression level for metabolic transcripts shifts from favoring SNDA neurons to favoring VTADA neurons in sick animals. N = 12 (Vehicle) and 7 (Rotenone). DDCt, delta-delta cycle threshold, ND1, complex I ND1 subunit; LDHB, lactate dehydrogenase B; TPI, triose phosphate isomerase; ANT2, adenine nucleotide translocator 2. *p < 0.05, **p < 0.01 by post-hoc Bonferroni after two-way ANOVA.
The situation was similar in sick rotenone-treated rats (Fig 4B). ANOVA revealed a significant main effect of treatment group F(1,57) = 5.5, p < 0.05, but no main effect of transcript F(3,57) = 1.8, p > 0.05 or treatment times transcript interaction F(3,57) = 0.7, p > 0.05. In other words, in vehicle-treated rats metabolic transcripts overall were more abundant in SNDA neurons, but after rotenone treatment leading to illness, that overabundance was less pronounced. Although there was a general shift of metabolic transcript abundance toward VTADA neurons, post hoc analysis revealed no significant differences in individual transcripts.
DISCUSSION
There are three main findings from this study. First, we have directly confirmed in the same animals the widely-held belief that performing transcriptional profiling experiments of this type using enriched neuronal populations instead of regional microdissections greatly increases the sensitivity with which changes can be detected, and thus the likelihood of generating interpretable results. Second, there were selective alterations in metabolic transcripts in midbrain dopamine neurons in response to generalized complex I inhibition dependent on the behavioral and neuropathological response of the animal. In particular, rats that behaviorally recovered from rotenone toxicity displayed an increase in metabolic transcripts while those that did not recover displayed a marked decrease. Third, vulnerable SNDA neurons were more dynamic in their metabolic transcriptional response than less vulnerable VTADA neurons.
We and others have previously hypothesized that sample composition is critical to performing transcriptional profiling experiments from heterogeneous tissues such as the brain (Chung et al., 2005; Greene, 2006; Greene et al., 2005; Meurers et al., 2009). This concept is particularly important as these techniques become more readily available and more easily applied not only to animal samples, but postmortem human tissue (Duke et al., 2007; Hauser et al., 2005; Papapetropoulos et al., 2007; Simunovic et al., 2009). In the current experiment, analysis of microdissected midbrain tissue in the same animals masked the substantial effects of complex I inhibition on the transcriptional profile of midbrain dopamine neurons (Fig 2). While LCM does not provide an absolutely pure population of dopamine neurons, significant enrichment of the samples was critical to interpreting the experiment and demonstrating the interesting result that SNDA neurons are selectively impacted by complex I inhibition. Errors of interpretation could conceivably occur in both directions, raising caution concerning experiments that employ dissected tissue samples, even when targeting other cell types, such as astrocytes or microglia. The impact of these results on other experimental methods commonly used such as western blot or LCMS proteomics is not clear, but should be given consideration in future experiments.
Our sampling of metabolic transcripts was not exhaustive, but included both nuclear-(LDHB, TPI, PDHC, ANT2) and mitochondrially-encoded (ND1, COX1) genes involved in both glycolysis (LDHB, TPI, PDHC) and oxidative phosphorylation (ND1, COX1, ANT2). All participate in critical regulatory checkpoints in energy metabolism; for example, regulation of PDHC controls metabolic flux from glycolysis into the tricarboxylic acid cycle. Our previous results indicating more robust expression in SNDA versus VTADA neurons for metabolic transcripts were also taken into consideration (Greene et al., 2005). ND1 was specifically targeted since rotenone and MPP+ bind to that subunit to inhibit complex I activity (Greenamyre et al., 1992; Nicklas et al., 1985).
We initially hypothesized that midbrain DA neurons would respond to complex I inhibition by increasing transcripts related to energy metabolism in a compensatory fashion. This was partially correct, in that DA neurons from rats treated with rotenone for 4 weeks displayed a greater abundance of metabolic transcripts than vehicle-treated rats. The results are particularly interesting in that there was a concerted increase in the metabolic transcripts examined, not a transcript-specific response. In this regard, DA neurons in vivo have a similar response to rotenone as neuroblastoma cells in vitro (Greene et al., 2008). Of course, our sampling of metabolic transcripts was not exhaustive, but it included both nuclear- (LDH, TPI, PDHC) and mitochondrially-encoded (ND1, COX1) transcripts involved in both oxidative phosphorylation (ND1, COX1) and glycolysis (LDH, TPI, PDHC). Interestingly, we have described a similar concerted upregulation of energy metabolism transcripts associated with mitochondrial biogenesis in hippocampal neurons from rats exposed to a calorie-restricted ketogenic diet (Bough et al., 2006). Furthermore, increasing metabolic demand (as opposed to inhibiting supply) by increasing synaptic activity in cultured neurons has been shown to induce metabolism transcripts encoded in the mitochondria and the nucleus via the nuclear respiratory transcription factors, NRF-1 and NRF-2 (Dhar et al., 2008; Dhar and Wong-Riley, 2009). Exploration of the neuronal signaling mechanism responsible for this concerted transcriptional response is beyond the scope of these experiments, but it appears that the ratio of metabolic supply to metabolic demand may be a critical factor in regulating metabolic capacity.
Unexpectedly, rats that became irreversibly ill during rotenone administration had an opposite metabolic transcriptional response in DA neurons and displayed a dramatic decrease in metabolic transcripts. Examination in a different species (mice) with a different complex I inhibitor (MPTP) revealed a qualitatively similar response in DA neurons, lending support to this finding as a general response to advanced complex I inhibition. Again, the response was a concerted one with an overall marked decrease in several metabolic transcripts involved in glycolysis (LDH, TPI, PDHC) and oxidative phosphorylation (ND1, COX1, ANT2) encoded by both nuclear (LDH, TPI, PDHC, ANT2) and mitochondrial (ND1, COX1) genes. A similar phenomenon has recently been described in SDNA neurons from postmortem PD cases, where there was a broad-based decrease in metabolism transcripts, indicating that both rotenone and MPTP intoxication in rodents accurately mimic the human disease in this respect (Simunovic et al., 2009).
The concerted decrease in metabolism transcripts could be a non-specific end-stage event prior to neuronal death or the result of a regulated cellular signaling cascade. Given that the majority of SNDA neurons do not typically degenerate in these model systems, we hypothesize that it may be the latter with potential intermediate signaling steps being production of mitochondrial free radicals, calcium dysregulation, or other events. For example, a decrease in PDHC mRNA after MPTP in mice has recently been hypothesized to be secondary to oxygen radical production (Lee et al., 2009). Dysfunctional dopamine neurotransmission may play an exacerbating role, as suggested by interesting proteomic data suggesting a similar downregulation of energy metabolism may occur in the striatum of PD mouse models (Chin et al., 2008).
Captured midbrain DA neurons in sick rotenone and MPTP-treated animals appeared healthy using the immunofluorescent LCM protocol; however the procedure emphasizes speed over morphology, so subtle changes could not be assessed. There was no consistent difference in metabolic transcripts between sick rotenone rats with or without a striatal lesion, indicating the decline in metabolism transcripts is likely not just secondary to injury or axotomy, but the sample sizes are small. Some of the sick rotenone-treated rats demonstrated agonal breathing at the end of the experiment raising concern that end-stage issues, such as respiratory status, may have contributed to the transcriptional response observed. However, MPTP-treated mice that were behaviorally normal at sacrifice exhibited a similar response.
It is unclear whether the biphasic response of energy metabolism transcripts during mitochondrial inhibition is part of a continuous response or the result of two independent mechanisms. One could hypothesize that rats that survive 4 weeks are ‘earlier’ in a process that will ultimately result in neurodegeneration or, alternatively, that those animals are a different ‘protected’ group as a result of an upregulation of energy metabolism. Our previous results investigating rotenone treatment of neuroblastoma cells in vitro suggest that upregulation of energy metabolism transcripts precedes cell damage, particularly to DNA, supporting the former hypothesis (Greene et al., 2008). Conversely, a recent paper suggests that a transcriptionally-mediated increase in mitochondrial density may protect against neurodegenerative insults (Wareski et al., 2009). There is no way to fully answer the question without longitudinal assessment of the animals, which is not technically feasible at this time.
Interestingly, the metabolic transcriptional response to complex I inhibition was more prominent (either up or down) in SNDA neurons than VTADA neurons. This confirms that SNDA neurons in vivo are intrinsically more vulnerable to several aspects of metabolic inhibition, not merely neuronal death. As we and others have previously reported and we have replicated in this study (Fig 4), SNDA neurons at baseline have a greater abundance of energy metabolism transcripts than VTADA neurons (Chung et al., 2005; Greene et al., 2005). We have interpreted this difference as a reflection of the greater metabolic demand under which SNDA neurons are operating. Our current results further support that conclusion. By decreasing metabolic capacity, complex I inhibition increases the ratio of metabolic demand to metabolic supply. In SNDA neurons that are more metabolically active to begin with (i.e., under greater demand), inhibiting mitochondrial output would produce a more profound relative metabolic deficit, resulting in a more dynamic transcriptional response. As we have previously maintained, knowledge of gene expression differences between neuron types under normal circumstances is critical for more complete interpretation of changes in response to stressors (Greene, 2006).
In conclusion, systemic complex I inhibition induces alterations in abundance of RNA transcripts encoding proteins related to energy metabolism selectively in SNDA neurons which may contribute significantly to the ultimate toxicity associated with mitochondrial inhibition. Understanding the mechanisms and consequences of transcriptional dysregulation in the face of mitochondrial inhibition may provide insight into the pathogenesis of PD and potential targets for neuroprotection.
Table 1.
Primer sequences for quantitative real-time RT-PCR
| Transcript | Primer | Rat (5’-3’ sequence) | Mouse (5’-3’ sequence) |
|---|---|---|---|
| HPRT | forward | CTCATGGACTGATTATGGACAGGAC | SAME |
| reverse | GCAGGTCAGCAAAGAACTTATAGCC | ||
| ND1 | forward | AAGCGGCTCCTTCTCCCTACAAAT | CCGGCCCATTCGCGTTATTCTTTA |
| reverse | GCTCGATTTGTTTCTGCGAGGGTT | AACGGAAGCGTGGATAAGATGCTC | |
| COX1 | forward | TCACTGCCAGTATTAGCAGCAGGT | n/a |
| reverse | TCTGGGTGGCCGAAGAATCAGAAT | ||
| LDHB | forward | CACGGGAGCTTATTTCTCCAGACT | TCGTGGTTTCCAACCCAGTGGATA |
| reverse | GATTGCATCCACTTCCAATCACGC | ATTGCATCCGCTTCCAATCACACG | |
| TPI | forward | GCTGCACAGAACTGCTACAAAGTG | SAME |
| reverse | GTCTTCTTTCAGAGTGTCCCAGCA | ||
| PDHC | forward | CATGGACACAGTATGAGCGATCCA | n/a |
| reverse | GAGCTGCTTCCTCGACTTCTTTCT | ||
| ANT2 | forward | ATGGGATTAACGGCCTGTACCAAG | SAME |
| reverse | TATTCTTGGGATCCGGGAGCATTC |
HPRT, hypoxanthine-guanine phosphoribosyltransferase; ND1, complex I subunit ND1; COX1, cytochrome oxidase subunit 1; LDHB, lactate dehydrogenase B; TPI, triose phosphate isomerase; PDHC, pyruvate dehydrogenase complex E1 subunit; ANT2, adenine nucleotide translocator 2
Acknowledgements
This work was supported by NINDS K08 NS048858 and a George C. Cotzias Memorial Fellowship from the American Parkinson Disease Association (JGG). We thank Georgia Taylor for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson G, et al. Loss of enteric dopaminergic neurons and associated changes in colon motility in an MPTP mouse model of Parkinson's disease. Exp Neurol. 2007;207:4–12. doi: 10.1016/j.expneurol.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Betarbet R, et al. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Bough KJ, et al. Mitochondrial biogenesis in the anticonvulsant mechanism of the ketogenic diet. Ann Neurol. 2006;60:223–235. doi: 10.1002/ana.20899. [DOI] [PubMed] [Google Scholar]
- Chan CS, et al. 'Rejuvenation' protects neurons in mouse models of Parkinson's disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- Chin MH, et al. Mitochondrial dysfunction, oxidative stress, and apoptosis revealed by proteomic and transcriptomic analyses of the striata in two mouse models of Parkinson's disease. J Proteome Res. 2008;7:666–677. doi: 10.1021/pr070546l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, et al. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14:1709–1725. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson T, et al. Animal models of PD: pieces of the same puzzle? Neuron. 2002;35:219–222. doi: 10.1016/s0896-6273(02)00780-8. [DOI] [PubMed] [Google Scholar]
- Dhar SS, et al. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke DC, et al. The medial and lateral substantia nigra in Parkinson's disease: mRNA profiles associated with higher brain tissue vulnerability. Neurogenetics. 2007;8:83–94. doi: 10.1007/s10048-006-0077-6. [DOI] [PubMed] [Google Scholar]
- Fearnley JM, Lees AJ. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- Fleming SM, et al. Behavioral and immunohistochemical effects of chronic intravenous and subcutaneous infusions of varying doses of rotenone. Exp Neurol. 2004;187:418–429. doi: 10.1016/j.expneurol.2004.01.023. [DOI] [PubMed] [Google Scholar]
- Gispert S, et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenamyre JT, et al. Quantitative autoradiography of dihydrorotenone binding to complex I of the electron transport chain. J Neurochem. 1992;59:746–749. doi: 10.1111/j.1471-4159.1992.tb09431.x. [DOI] [PubMed] [Google Scholar]
- Greene JG. Gene expression profiles of brain dopamine neurons and relevance to neuropsychiatric disease. J Physiol. 2006;575:411–416. doi: 10.1113/jphysiol.2006.112599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JG, et al. Gene expression profiling of rat midbrain dopamine neurons: implications for selective vulnerability in parkinsonism. Neurobiol Dis. 2005;18:19–31. doi: 10.1016/j.nbd.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Greene JG, et al. Sequential and concerted gene expression changes in a chronic in vitro model of parkinsonism. Neuroscience. 2008;152:198–207. doi: 10.1016/j.neuroscience.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JG, et al. Delayed gastric emptying and enteric nervous system dysfunction in the rotenone model of Parkinson's disease. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MA, et al. Expression profiling of substantia nigra in Parkinson disease, progressive supranuclear palsy, and frontotemporal dementia with parkinsonism. Arch Neurol. 2005;62:917–921. doi: 10.1001/archneur.62.6.917. [DOI] [PubMed] [Google Scholar]
- Hirsch E, et al. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson's disease. Nature. 1988;334:345–348. doi: 10.1038/334345a0. [DOI] [PubMed] [Google Scholar]
- Lee DW, et al. Inhibition of prolyl hydroxylase protects against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced neurotoxicity: model for the potential involvement of the hypoxia-inducible factor pathway in Parkinson disease. J Biol Chem. 2009;284:29065–29076. doi: 10.1074/jbc.M109.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss B, et al. K-ATP channels promote the differential degeneration of dopaminergic midbrain neurons. Nat Neurosci. 2005;8:1742–1751. doi: 10.1038/nn1570. [DOI] [PubMed] [Google Scholar]
- Loeb V, et al. The transgenic over expression of {alpha}-Synuclein and not its related pathology, associates with complex I inhibition. J Biol Chem. 2010 doi: 10.1074/jbc.M109.061051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurers BH, et al. Low dose rotenone treatment causes selective transcriptional activation of cell death related pathways in dopaminergic neurons in vivo. Neurobiol Dis. 2009;33:182–192. doi: 10.1016/j.nbd.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, et al. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas WJ, et al. Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenyl-pyridine, a metabolite of the neurotoxin, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci. 1985;36:2503–2508. doi: 10.1016/0024-3205(85)90146-8. [DOI] [PubMed] [Google Scholar]
- Okamura H, et al. Lateromedial gradient of the susceptibility of midbrain dopaminergic neurons to neonatal 6-hydroxydopamine toxicity. Exp Neurol. 1995;136:136–142. doi: 10.1006/exnr.1995.1090. [DOI] [PubMed] [Google Scholar]
- Papapetropoulos S, et al. Optimizing human post-mortem brain tissue gene expression profiling in Parkinson's disease and other neurodegenerative disorders: from target "fishing" to translational breakthroughs. J Neurosci Res. 2007;85:3013–3024. doi: 10.1002/jnr.21355. [DOI] [PubMed] [Google Scholar]
- Parker WD, Jr, et al. Abnormalities of the electron transport chain in idiopathic Parkinson's disease. Ann Neurol. 1989;26:719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- Rinne JO. Nigral degeneration in Parkinson's disease. Mov Disord. 1993;8 Suppl 1:S31–S35. doi: 10.1002/mds.870080507. [DOI] [PubMed] [Google Scholar]
- Schapira AH, et al. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- Schapira AH, et al. Anatomic and disease specificity of NADH CoQ1 reductase (complex I) deficiency in Parkinson's disease. J Neurochem. 1990;55:2142–2145. doi: 10.1111/j.1471-4159.1990.tb05809.x. [DOI] [PubMed] [Google Scholar]
- Schneider JS, et al. Selective loss of subpopulations of ventral mesencephalic dopaminergic neurons in the monkey following exposure to MPTP. Brain Res. 1987;411:144–150. doi: 10.1016/0006-8993(87)90691-3. [DOI] [PubMed] [Google Scholar]
- Sherer TB, et al. Subcutaneous rotenone exposure causes highly selective dopaminergic degeneration and alpha-synuclein aggregation. Exp Neurol. 2003;179:9–16. doi: 10.1006/exnr.2002.8072. [DOI] [PubMed] [Google Scholar]
- Simunovic F, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson's disease pathology. Brain. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teismann P, et al. Cyclooxygenase-2 is instrumental in Parkinson's disease neurodegeneration. Proc Natl Acad Sci U S A. 2003;100:5473–5478. doi: 10.1073/pnas.0837397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varastet M, et al. Chronic MPTP treatment reproduces in baboons the differential vulnerability of mesencephalic dopaminergic neurons observed in Parkinson's disease. Neuroscience. 1994;63:47–56. doi: 10.1016/0306-4522(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Wareski P, et al. PGC-1{alpha} and PGC-1{beta} regulate mitochondrial density in neurons. J Biol Chem. 2009;284:21379–21385. doi: 10.1074/jbc.M109.018911. [DOI] [PMC free article] [PubMed] [Google Scholar]