Abstract
We reported that gene-selective formation of facultative heterochromatin silences transcription of acute inflammatory genes during endotoxin (LPS) tolerance, according to function. We discovered that reversal of the epigenetically silenced transcription restored mRNA levels but not protein synthesis. Here, we find that translation repression of tumor necrosis factor-α (TNFα) occurs independent of transcription silencing during LPS tolerance. The process required to disrupt protein synthesis followed Toll-like receptor 4 (TLR4)-dependent induction of microRNA (miR)-221, miR-579, and miR-125b, which coupled with RNA-binding proteins TTP, AUF1, and TIAR at the 3′-untranslated region to arrest protein synthesis. TTP and AUF1 proteins linked to miR-221, whereas TIAR coupled with miR-579 and miR-125b. Functional inhibition of miR-221 prevented TNFα mRNA degradation, and blocking miR-579 and miR-125b precluded translation arrest. The functional specificity of the TNFα 3′-untranslated region was demonstrated using luciferase reporter with mutations in the three putative miRNA binding sites. Post-transcriptional silencing was gene-specific, because it did not affect production of the IκBα anti-inflammatory protein. These results suggest that TLR4-dependent reprogramming of inflammatory genes is regulated at two separate and distinct levels. The first level of control is mediated by epigenetic modifications at the promoters that control transcription. The second and previously unrecognized level of control is mediated by TLR4-dependent differential expression of miRNAs that exert post-transcriptional controls. The concept of distinct regulation of transcription and translation was confirmed in murine sepsis. We conclude that transcription- and translation-repressive events combine to tightly regulate pro-inflammatory genes during LPS tolerance, a common feature of severe systemic inflammation.
Keywords: Chromatin, Cytokine, Epigenetics, Gene Silencing, Inflammation, Innate Immunity, Macrophage, MicroRNA, Translation Regulation
Introduction
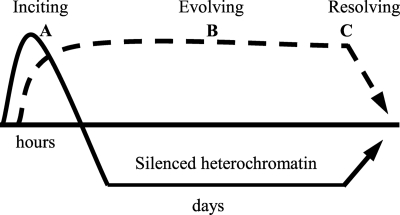
Inflammation development and progression are influenced by genetic, epigenetic, and environmental factors (1). Sensors such as Toll-like receptors and their cellular communication pathways couple inciters of inflammation to genes, after which epigenetic events reprogram gene expression patterns in a temporal sequence of defense and repair (2). In severe systemic inflammation (SSI)2 such as serious infection, a high magnitude response spreads systemically, which often results in death from cellular and organ failure. Such inflammation has distinct features and must be tightly controlled at transcriptional and post-transcriptional levels. Unlike in chronic inflammation, the inciting SSI phase is short-lived (lasts for hours) and progresses to an evolving phase typified by gene-selective reprogramming through histone and DNA methylation reactions that generate facultative heterochromatin (3–7). This gene-specific paradigm silences transcription of the acute pro-inflammatory genes responsible for the SSI autotoxicity, whereas sustaining transcription of distinctly functional genes like those encoding anti-inflammatory and antibiotic mediators (3–9) (see Fig. 1). Depending on the magnitude of the original inciting stimulus, the evolving phase of SSI can dissipate within days, or be sustained for weeks (2). Reversal of gene silencing correlates with resolution of SSI and survival. Our studies implicate nuclear factor κB (NF-κB) members p65 and RelB as essential regulators of the inciting, evolving, and restorative phases of SSI (3–5, 7, 8).
FIGURE 1.
Gene reprogramming during endotoxin tolerance and SSI. SSI is a germ line and epigenetically controlled gene-specific reprogramming process that evolves in stereotyped stages. This simplified scheme shows three functional and clinically significant gene expression patterns associated with SSI: A, inciting stage: immediate response pro-inflammatory gene set (e.g. TNFα, IL-1β, and others) that are activated by LPS and then become inactive. These mediators initiate SSI. B, evolving stage: feed-forward gene-specific epigenetic switches (e.g. RelB). This establishes transcription silencing. C, resolving stage: reversal of gene reprogramming and survival.
The NF-κB family is the prototypic master inflammation regulator (10). Toll-like receptors (TLRs) usually initiate SSI through NF-κB cytosolic activation with nuclear translocation and promoter binding of p65, which interacts with trans-acting factors. We discovered that temporally distinct de novo NF-κB RelB induction replaces promoter-bound NF-κB p65 after the inciting stage to generate gene set-specific transcription silencing. RelB interacts with and recruits the histone H3K9 methyltransferase, G9a, to dimethylate histone H3K9, leading to recruitment of the DNA methyltransferase Dnmt3a/b and subsequent methylation of promoter CpGs and chromatin condensation (4, 6, 7). Gene-selective facultative heterochromatin reverses to euchromatin during the SSI resolution/restorative phase. The evolving phase of SSI with gene-selective silencing is typified by the phenomenon of endotoxin tolerance observed in humans, animals, and cell models (2). We found that reversal of condensed heterochromatin to open euchromatin by inhibiting RelB or G9a restored transcription but not translation of pro-inflammatory TNFα and IL-1β (4–6), suggesting an additional silencing mechanism at the post-transcriptional level.
The inciting and evolving features of SSI conform to the network motif concept of type 1 incoherent feed-forward loops (11), in which a transcription activator X (e.g. NF-κB p65) controls a target gene Z (e.g. TNFα) and also sequentially activates a transcription repressor Y (e.g. RelB) of the target gene, thus producing physiologic adaptation, for example tolerance. Recent data support that such gene-encoded adaptive loops may involve microRNAs (miRNAs) (12). Feedback repression of inflammatory adhesion genes in endothelial cells by miRNAs recently provides an example (13). Thus transcription and post-transcription regulatory process may differentially influence the inflammatory phenotype.
miRNAs are a conserved and abundant class of endogenous non-coding RNAs of ∼22 nucleotides involved in post-transcriptional gene expression by controlling the stability and/or translation of target mRNAs (14, 15). miRNAs are processed from long primary transcripts that are transcribed from independent genes or from intronic sequences of protein-coding genes by RNA polymerase II (15). Primary transcripts are processed into ∼60- to 70-nucleotide hairpin precursors, which are exported to the cytoplasm and further processed into ∼22-nucleotide mature miRNAs by the RNase type III enzyme Dicer (15). Mature miRNAs are loaded onto a ribonucleoprotein complex, also known as miRNA-induced silencing complex (miRISC), where they act as guiding molecules to deliver the complex to target mRNA via binding to complementary sequences in the 3′UTR, resulting in mRNA degradation and/or translational repression (15). miRNAs usually base pair to target mRNA with imperfect complementarity, resulting in translational inhibition, whereas perfect base-pairing induces target mRNA degradation (15, 16). The most stringent requirement for miRNA function is a near perfect base-pairing within the miRNA nucleotides 2–7 (known as the seed region), which nucleates the interaction within the target mRNA 3′UTR (15). Recent studies in mammals implicate miRNAs in a number of biological processes, including regulation of normal immune function, cell differentiation, viral infection, inflammation, signal transduction, and cancer (14, 17, 18).
Here, we tested whether the SSI-inciting phase might induce an miRNA code that regulates translational events independent of the transcriptional silencing process of selective facultative heterochromatin formation. To do this, we used the THP-1 cell model of SSI as reflected in the generation of endotoxin (lipopolysaccharide (LPS)) tolerance by an inciting TLR4 LPS stimulus. We performed a computational miRNA target prediction algorithm and identified twenty miRNAs with sequences complementary to the TNFα 3′UTR, as a model of post-transcriptional repression of pro-inflammatory genes in LPS tolerance. Expression profiling distinguished three miRNAs (miR-221, miR-579, and miR-125b) whose levels were differentially altered after LPS stimulation in tolerant cells. Importantly, we show that these miRNAs assemble into the miRISC complex and target specific RNA-binding proteins (RBPs) to the TNFα 3′UTR. Functional inhibition revealed that miR-221 directs mRNA degradation, whereas miR-579 and miR-125b inhibit translation of TNFα mRNA. We also found that these miRNAs and RBPs may couple to disrupt TNFα protein synthesis in mice with sepsis. Our studies provide the first evidence linking differential changes in TLR-induced miRNA expression to LPS tolerance, which serves as a cell model of SSI gene reprogramming (2, 19), and suggest that both disruption of transcription and translation independently assure gene-specific silencing during the endotoxin tolerance/evolving phase of SSI.
EXPERIMENTAL PROCEDURES
Cell Culture and Tolerance
THP-1 cells, obtained from the American Type Culture Collection (Rockville, MD), were maintained in RPMI 1640 medium (Invitrogen) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, and 10% fetal bovine serum (HyClone Laboratories, Logan, UT) at 37 °C and 5% CO2. Log-phase cells were used in all experiments. Cells were made tolerant by overnight incubation with 1 μg/ml LPS (Sigma-Aldrich, 0111:B4, 1 μg/ml). The following morning, responsive (fresh cells; no preincubation with LPS) and tolerant cells were washed and resuspended in complete medium. All reagents were routinely tested for LPS contamination by Limulus amebocyte lysate assay (sensitivity of <1 pg/ml). Responsive and LPS-tolerant cells (1 × 106 cells/ml) were stimulated with LPS for various times. LPS used in these experiments was free of contaminating proteins that activate cells via a non-TLR4-dependent mechanism.
Construction of Plasmids
Renilla luciferase construct was obtained from Promega (Madison, WI). To create TNFα 3′UTR-luciferase reporter construct, the 3′UTR fragment (∼798 bp) of TNFα was cloned downstream of RPL10-driven firefly luciferase in pSGG vector (Switchgear Genomics, Menlo Park, CA). Mutated versions (mutTNFα 3′UTR-Luc) of this construct carrying 6-bp substitutions in the miR-221, miR-579, or miR-125b binding sites (either individually or combined) were obtained by site-directed mutagenesis (Fig. 10A). All mutations were verified by sequencing.
FIGURE 10.
Mutations in the miRNA binding sites in TNFα 3′UTR inhibit its repressive effects on a fused luciferase (Luc) reporter in LPS-tolerant cells. A, a luciferase reporter construct containing the 798-bp TNFα 3′UTR fragment. Mutations in the TNFα UTR sequences (corresponding to miRNA seed regions) were generated by site-directed mutagenesis and subcloned downstream of the luciferase gene in pSGG vector. Shown is a sequence alignment of the miRNAs and their target sites in the 3′UTR. Mutated sequences, with 6-nucleotide substitutions, disrupting base-pairing with the “seed region” of the miRNA are shown in lowercase letters. miRNA seed regions are underlined. The nucleotides in the 3′UTR corresponding to the miRNA seed regions are in boldface. B, luciferase reporter activity in tolerant cells. Cells were made tolerant and then transfected with the luc plasmids. After 24 h, cells were washed and left unstimulated or stimulated with 1 μg/ml LPS for 1 h. Doxycycline was added, and the incubation continued for an additional 1 h. Total RNA was isolated and Luc mRNA levels were determined by real-time PCR (top) using primers that amplify Luc sequences only. Cell lysate was prepared and firefly and Renilla luc activities were measured in triplicate. Firefly to Renilla Luc ratio (bottom) was calculated and further normalized to Luc mRNA (to obtain translation efficiency). RNA values were normalized to GAPDH RNA. Data are presented as % change relative to control Luc plasmid (without TNFα 3′UTR) transfection (set at 100%). Data represent the mean ± S.D. of at least three experiments. *, significant difference compared with wild-type TNFα 3′UTR-Luc. T, tolerant; wt, wild type; and mut, mutant.
Transfections
Log phase THP-1 cells were seeded at 0.5 × 106 cells/ml and made tolerant prior to transfection. Cells were transfected by electroporation using the Nucleofector system per the manufacturer's instructions (Lonza, Walkersville, MD). For knockdown studies, cells were transfected with pools of scrambled or target gene-specific siRNAs at 0.5 μm (final concentration). For miRNA studies, cells were transfected with 100 nm miRNA mimics or 2′-O-methyl antisense oligonucleotides (anti-miR221, anti-miR579, or anti-miR125b), either individually or in combination (Ambion, Austin, TX). For translation studies, miRNA inhibitor (anti-miR) was co-transfected with 100 ng of luciferase plasmids.
Luciferase Assays
Cells were harvested 24 h after transfection and lysed in 1× lysis buffer. Luciferase activities were measured using the Dual Luciferase Assay system per the manufacturer's instructions (Promega). The firefly to Renilla luciferase ratio was determined and further normalized for firefly mRNA.
Mice
C57BL/6 mice at 6–8 weeks were obtained from Harlan Sprague (Indianapolis, IN), housed under pathogen-free conditions at the Unit for Laboratory Animals at Wake Forest University School of Medicine, and treated in accordance with the guidelines set forth by the National Institutes of Health Committee on Care and Use of Laboratory Animals. Animal protocols were reviewed and approved by the Wake Forest University Animal Care Use Committee.
Cecal Ligation and Puncture
Cecal ligation and puncture surgery was performed as previously described (20), with some modifications. Briefly, mice were anesthetized by isoflurane inhalation (Halocarbon Laboratories, Riverridges, NJ). A 1-cm midline incision was made to the ventral surface of the abdomen, and the cecum was exposed. The cecum was partially ligated at its base with a 3-0 silk suture and punctured three times with a 20-gauge needle. The abdomen incision was closed using surgical staples. Mice were sacrificed after 24 h.
Isolation of Peritoneal Macrophages
Normal and septic mice were euthanized by cervical dislocation following anesthetization. Macrophages were collected from peritoneal lavage with Hanks' balanced salt solution. The lavage was then centrifuged at 2500 rpm for 5 min. Cells were washed two times with Hanks' balanced salt solution and then suspended in Dulbecco's modified Eagle's medium containing 10% FBS and antibiotics.
To obtain macrophage-enriched cell preparations, peritoneal cell suspensions were incubated in medium for 1 h to allow macrophages to adhere to plates. Non-adherent cells were removed by washing with Hanks' balanced salt solution. Cells were then cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum overnight at 37 °C and then stimulated with 100 ng/ml LPS.
Preparation of Cytoplasmic Extract
Because some RBPs have very rapid off-rates that lead to loss during extract preparation, we used in vivo formaldehyde cross-linking, in some experiments, to freeze RNA·RBP complexes (21). This cross-linking was reversed by incubating the immunoprecipitated complexes at 65 °C for 45 min before RNA was isolated and analyzed as described below. Immediately after harvest, cells were incubated with 0.2% formaldehyde in phosphate-buffered saline for 10 min at room temperature (21).
Cells were washed in phosphate-buffered saline and lysed according to published methods (22) with some modifications. Briefly, cells were incubated on ice for 10 min in lysis buffer (250 mm sucrose, 10 mm Tris-HCl (pH 7.5), 25 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, 0.1% Nonidet P-40, 30 units/ml RNase inhibitor plus 1× protease inhibitor mixture). Samples were centrifuged at 2,000 rpm for 5 min. The supernatants were cleared by centrifugation at 10,000 rpm for 5 min and saved as cytoplasmic extract. To prepare whole cell lysates, cells were lysed as described above except that the lysis buffer included 0.5% Nonidet P-40 and 0.5% deoxycholate.
Immunoprecipitation
Immunoprecipitation (IP) of Ago2 or RBP protein complexes was performed as described previously (23) with some modifications. Briefly, cell lysates or cytoplasmic extracts were pre-cleared by incubation with pre-blocked protein G-agarose beads for 1 h at 4 °C. Beads were pre-blocked by incubation for 1 h with 100 μg/ml bovine serum albumin. Pre-blocked beads were washed with buffer C (250 mm sucrose, 10 mm Tris-HCl (pH 7.5), 25 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, 30 units/ml RNase inhibitor, and 1× protease inhibitor mixture). Extract was centrifuged at 2000 rpm for 5 min, and supernatant (900 μl) was added to 100 μl of pre-blocked protein G-agarose beads that were coated with 10 μl of antibody against human Ago2 (clone =4G8, Wako, Richmond, VA), TTP, TIAR (Santa Cruz Biotechnology, Santa Cruz, CA), or AUF1 (Abcam, Cambridge, MA). After overnight incubation (with rotation) at 4 °C, the beads were centrifuged and washed three times with buffer C. Aliquots of bound protein complexes were taken for protein analysis by SDS-10% PAGE, and the remainder was subjected to RNA isolation (from the immunoprecipitated RBPs) using TRIzol reagent (Invitrogen) following the manufacturer's instructions. This RNA was used for mRNA and miRNA analysis.
In some experiments, re-immunoprecipitation of protein complexes was performed. First, immunoprecipitation was performed as described above. Beads were recovered and resuspended in 10 mm dithiothreitol for 25 min at 37 °C. The supernatant was recovered by centrifugation at 1000 rpm for 1 min, diluted to 900 μl in buffer C, and then re-immunoprecipitated by adding 100 μl of pre-blocked protein G-agarose beads coated with the second antibody. The rest of the procedures are as described above.
miRNA and mRNA Expression
To identify potential miRNA binding sites in target mRNA, we used computational target prediction algorithms utilized by the PicTar, miRanda, and TargetscanS 4.0 search engines (24, 25). miRNAs with sequences complementary to TNFα 3′UTR were then analyzed by expression profiling as shown below.
RNA was isolated from intact cells using an miRNA isolation and enrichment kit according to the manufacturer's instructions (Ambion). For immunoprecipitated RNA, immunoprecipitates were treated with DNase at 37 °C for 15 min and subsequently with proteinase K (35 μg/ml) for 15 min at 37 °C before RNA was purified by TRIzol reagent or RNeasy kit (Invitrogen).
miRNA expression in cell extracts or immunoprecipitated RNA was determined using an Ncode miRNA detection kit (Ambion) according to the manufacturer's instructions. This technique, unlike the conventional assay of random-primed reverse transcription (RT), consists of poly(A) tailing of 10 ng of miRNA-enriched RNA in a 25-μl reaction, followed by RT and a specific real-time PCR. The RT reaction consisted of 4 μl of polyadenylated miRNA, 3 μl of 25 μm universal RT primer, and 1 μl of annealing buffer. The reaction was incubated at 65 °C for 5 min followed by the addition of 10 μl of 2× first-strand synthesis reaction mix containing dNTP and 2 μl of SuperScript III RNase Out enzyme mix. The reaction (20 μl) was then incubated at 50 °C for 50 min, followed by 85 °C for 5 min to stop the reaction. The real-time PCR reaction consisted of 5 μl of 1:10 dilution of the RT product, 1 μl of 10 μm universal reverse primer, 1 μl of 10 μm miRNA-specific forward primer (an oligonucleotide identical to the entire mature miRNA sequence, in which U is replaced with T), and 25 μl of SYBR green qPCR Supermix-UDG (Ambion). PCR was run in triplicates at 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min using ABI Prism 7000 Sequence Detection System (Applied Biosystems). The relative expression of miRNA was calculated using the 2−ΔΔCt cycle threshold method after normalization to the endogenous U6 small RNA (as an internal control).
To analyze the expression of target gene mRNA, real-time RT-PCR was performed as described previously (5), using TaqMan gene-specific primer/probe sets (Applied Biosystems). The mRNA level of target genes was normalized to GAPDH RNA and calculated using the 2−ΔΔCt cycle threshold method. The 18 S RNA was also amplified as an internal control. For semiquantitative PCR, immunoprecipitated RNA was reverse transcribed and amplified by 25 cycles using gene-specific primers covering the 3′UTR sequence.
Immunoblotting
Proteins in fresh cytoplasmic extract or in immunoprecipitated complexes (IP) were detected by immunoblot analysis. Cytoplasmic extract or IP complexes was(were) mixed with 5× Laemmli gel loading buffer, resolved in gradient SDS-polyacrylamide gel, transferred to polyvinylidene difluoride membranes (Millipore), and incubated with primary antibodies diluted in 10% skim milk in Tris-buffered saline/Tween 20. After washing, membranes were incubated with secondary antibody conjugated to horseradish peroxidase. Proteins were visualized with the ECL detection system (Amersham Biosciences).
Data Analysis
All values are expressed as the mean ± S.D. of data obtained from at least three experiments. Comparison between groups was made with one-way analysis of variance, followed by unpaired Student's t test, with p ≤ 0.05 being statistically significant.
RESULTS
Transcription and Translation of Pro-inflammatory Genes Are Separately Regulated in LPS Tolerance
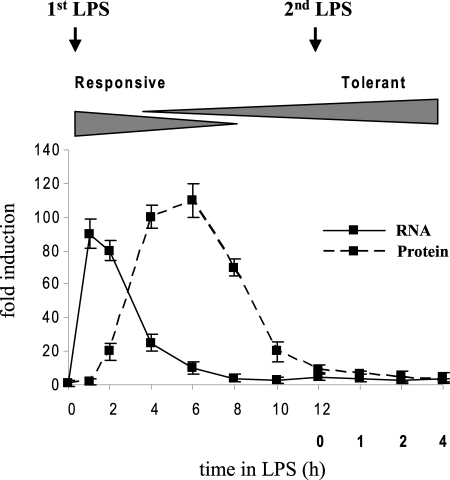
As shown in Fig. 2, TNFα mRNA and protein were markedly induced after THP-1 cell stimulation with LPS. Following this initial activation phase, which incites the “cytokine storm” observed at the onset of SSI (2), both mRNA and protein levels were rapidly decreased and reached background levels by 12 h. This coincides with the induction of LPS tolerance, as cells were unable to respond to a second dose of LPS.
FIGURE 2.
TNFα mRNA and protein are repressed in LPS-tolerant THP-1 cells. TNFα mRNA and protein analysis. Cells were made tolerant by pretreatment with 1 μg/ml LPS for 12 h. LPS-tolerant cells were washed and then re-stimulated with 1 μg/ml LPS for the indicated times. RNA levels were determined by real-time PCR. Sample data were normalized to GAPDH mRNA. Levels of total (intracellular and secreted) TNFα protein were measured by ELISA. The samples were assayed in triplicate. Data represent the mean ± S.D. from at least three independent experiments and are presented as -fold change relative to unstimulated cells (set at 1-fold).
The sustained repression of mRNAs of TNFα and IL-1β is reversible by depletion of RelB, which drives transcriptional epigenetic silencing (4–6) (Fig. 3). Cells were made tolerant and then transfected with RelB siRNA to inhibit RelB expression. Total RNA and proteins were isolated and analyzed at different time points. Although reversing transcription silencing by RelB knockdown restored TNFα mRNA to near normal levels seen in responsive (R) cells, we did not detect protein even after 4–6 h of re-stimulation. In addition, mRNA completely disappeared 1 h after blocking new RNA synthesis with doxycycline (Fig. 4). Actinomycin D at 5 μg/ml (our previous study (26)) exhibited similar results but reduced cell viability compared with doxycycline (not shown). In addition, previous studies reported the inhibitory effect of doxycycline on the expression of inflammatory mediators, including cytokines genes (27–30). These results suggested that mRNA was unstable in LPS-tolerant cells. Further analysis of mRNA decay showed that mRNA half-life was ∼20 min in LPS-tolerant cells compared with ∼50 min in responsive cells (Fig. 4).
FIGURE 3.
Knockdown of RelB protein in LPS-tolerant cells restores LPS-induced TNFα mRNA levels, but not TNFα protein. THP-1 cells were made tolerant by pretreatment with 1 μg/ml LPS for 12 h. Tolerant cells were washed and then transfected with pools of nonspecific (control) or RelB-specific siRNAs (to reverse transcription silencing). After 36 h, both responsive and tolerant cells were then washed and left unstimulated (0 h) or stimulated with 1 μg/ml LPS. A, total RNA was isolated after 1 h and analyzed by PCR (top). Sample data were normalized to GAPDH mRNA. B, total TNFα protein levels (bottom) were measured after 4 h by ELISA. Values from unstimulated cells (0 h) were subtracted, and the data represent presented relative to stimulated responsive cells (set at 100-fold). Data are the mean ± S.D. of at least three experiments. *, statistically significant (p ≤ 0.05). R, responsive; T, tolerant.
FIGURE 4.
TNFα mRNA is destabilized in LPS-tolerant cells. Figure shows the TNFα mRNA half-life. THP-1 cells were made tolerant by pretreatment with 1 μg/ml LPS for 12 h and then transfected with control or RelB-specific siRNA (to reverse transcription silencing). After 36 h, tolerant cells, together with responsive (fresh) cells, were washed and left unstimulated or stimulated with 1 μg/ml LPS (to induce TNFα expression). After 1 h (which corresponds to 0 min in doxycycline), 10 μg/ml doxycycline (in DMSO) was added for the indicated times (to stop RNA synthesis). Actinomycin D (5 μg/ml) exhibited similar results but reduced cell viability compared with doxycycline (not shown). RNA was isolated and analyzed by real-time PCR. Sample data were normalized to GAPDH mRNA, and values from unstimulated cells were subtracted. Data are presented as percent change relative to cells without doxycycline, i.e. 0 min (set at 100%). Data are the mean ± S.D. of three experiments. Cells without doxycycline received the same dose of DMSO. R, responsive; T, tolerant; and Doxy, doxycycline.
Thus, our experimental approach to investigating the role of miRNAs in the SSI phenotype is based on the observation that gene-specific repression in LPS-tolerant cells affects transcription and translation of selected inflammatory genes, that the two processes are regulated distinctly and separately, and that the post-transcriptional process does not require RelB induction. To study mRNA stability and translation, transcription and mRNA levels must be restored first. In this context, RelB was knocked down in tolerant cells in all subsequent experiments.
miRNAs Are Differentially Expressed in LPS Tolerance
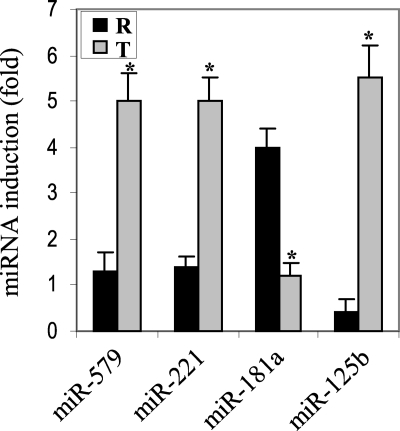
The TNFα 3′UTR region plays a role in its post-transcriptional regulation (31–33). miRNAs function by base-pairing with sequences in the 3′UTR of the targeted mRNA. To examine a potential role for miRNAs in translational repression of TNFα in LPS tolerance, we used a computational target prediction algorithm to screen for miRNAs with sequences complementary to the TNFα 3′UTR, which might then act to promote the translational repression. We identified 20 miRNAs with sequences complementary to the TNFα 3′UTR. Next, we performed miRNA expression profiling using miRNA quantitative RT-PCR. We observed increases or decreases in the levels of nine miRNAs. Interestingly, the levels of miR-150, miR-155, miR-146a, and miR-16 were increased, whereas miR-369–3p was decreased in both LPS-responsive and -tolerant cells after LPS stimulation (data not shown). Importantly, only four miRNAs (miR-181a, miR-221, miR-579, and miR-125b) showed a differential expression pattern when compared in both cell phenotypes (Fig. 5). Among the four miRNAs, miR-579 did not conform to a typical seed region match. miR-181a was significantly increased in responsive cells, whereas miR-221, miR-579, and miR-125b were significantly increased in tolerant cells. Further inspection of the data revealed that miR-221, miR-579, and miR-125b levels became elevated during the LPS-dependent evolving phase of tolerance; thereafter a second LPS stimulus markedly augmented their expression (data not shown). Because TNFα mRNA and protein were independently repressed in tolerant cells, we focused our analyses on the miRNAs that showed differential expression pattern in tolerant cells, because changes in their expression levels suggested them as candidates for post-transcriptional regulation.
FIGURE 5.
Differential changes in miRNA expression occurs in LPS tolerance. Profiling the expression of miRNAs that target TNFα 3′UTR. THP-1 cells were made tolerant as described before. Responsive and tolerant cells were left unstimulated or stimulated with 1 μg/ml LPS for 1 h. miRNA-enriched RNA was isolated using miRNA enrichment kit (Ambion). miRNAs were polyadenylated, reverse-transcribed, and amplified by real-time PCR, using a SYBR green Master Mix, a universal reverse primer (Ambion), and an miRNA-specific forward primer (the same miRNA sequence in which U is changed to T). Sample data were normalized to the U6 small nuclear RNA level (as an internal control) and are presented as -fold change relative to unstimulated cells. Data represent the mean ± S.D. of at least three experiments. *, statistically significant (p ≤ 0.05). R, responsive; T, tolerant.
miR-221 Accelerates TNFα mRNA Degradation, whereas miR-579 and miR-125b Block Its Translation
The rapid increase in miR-221, miR-579, and miR-125b 1 h after LPS stimulation in tolerant cells suggested that they may participate in disrupting protein synthesis. To examine the effects of the increase in miRNA expression in tolerant cells on TNFα mRNA stability, we used miRNA inhibitors (anti-miRNA; single-stranded and chemically modified oligonucleotides, known as antagomirs) (34) to inhibit their expression. Tolerant cells were transfected with RelB-specific siRNA (to reverse transcription silencing and enhance the level of TNFα mRNA that would be available for translation) together with anti-miRNAs (individually or in combinations). After 36 h, cells were washed and stimulated with LPS, without or with doxycycline to stop new transcription. Doxycycline was added after 1 h, at which time TNFα mRNA is significantly increased (Fig. 2). As shown in Fig. 6, TNFα mRNA was almost completely degraded in the presence of doxycycline. Anti-miR-221 transfection stabilized the mRNA but did not restore protein synthesis. Anti-miR-579 or anti-miR-125b had no effects on mRNA stability (not shown). In addition, miR-181a mimic (which increased miR-181a level in tolerant cells) was also co-transfected with any of the three miRNAs and showed no additional positive or negative effects on mRNA stability (data not shown). These results suggested that miR-221 targets TNFα mRNA for rapid decay in tolerant cells but does not regulate translation per se.
FIGURE 6.
Inhibition of miRNA-221 activity stabilizes TNFα mRNA in tolerant cells but does not recover protein levels. Cells were made tolerant as described in Fig. 3, washed, and then transfected with RelB siRNA (to reverse transcription silencing) together with control miRNA inhibitor or anti-miRNA-221 inhibitor (single-stranded chemically modified miRNA-221 oligonucleotide called antagomir) to inhibit miRNA-221 activity. After 36 h, cells were washed and left unstimulated or stimulated with 1 μg/ml LPS. After 1 h in LPS (which coincides with peak induction of mRNA; see Fig. 2), doxycycline or DMSO (−) was added (to stop new transcription), and the incubation was continued for 1 h. By this time, TNFα mRNA was barely detected in tolerant cells (data not shown). Total RNA (left) and protein (right) were extracted and analyzed by real-time PCR and ELISA, respectively. Sample data were normalized to GAPDH RNA, and values from unstimulated cells (0 h) were subtracted. Data are presented as percent change relative to cells without doxycycline (set at 100-fold). For protein expression, cells were harvested after 4 h and analyzed in triplicate by ELISA. Data represent the mean ± S.D. of at least three experiments.
Although inhibiting miR-221 expression in tolerant cells restored TNFα mRNA stability, protein synthesis remained blocked even after 4–6 h, suggesting that mRNA translation is independently disrupted in tolerant cells. To test this possibility, we transfected tolerant cells with anti-miR-221 together with anti-miR-579 and/or anti-miR-125b. Although transfection of anti-miR-221 with anti-miR-579 alone or anti-miR-125b alone had no additive effects on mRNA stability over that conferred by anti-miRNA transfection alone, each condition restored protein levels to ∼30–40% (data not shown). On the other hand, transfection of anti-miR-221 with anti-miR-579 plus anti-miR-125b restored TNFα protein levels to ∼80% of its levels detected in responsive cells (Fig. 7A). The levels of all miRNAs were markedly reduced after anti-miRNA transfections (Fig. 7B). Collectively, the data presented in Figs. 6 and 7 suggested that TNFα mRNA stability and translation are differentially regulated in tolerant cells and that miR-221 induces mRNA decay, whereas miR-579 and miR-125b arrest translation.
FIGURE 7.
Concurrent inhibition of miR-221, miR-125b, and miR-579 activities reverses TNFα translational repression and recovers TNFα mRNA and protein in LPS-tolerant cells. Tolerant THP-1 cells were transfected and treated exactly as described in Fig. 6, except that cells were transfected with a mixture of anti-miR-221, anti-miR-125b, and anti-miR-579 (miRNA inhibitors). A, data from responsive LPS-stimulated cells are included as a reference, to show the relative extent of TNFα mRNA and protein recovery. Responsive cells also received control siRNA. For mRNA, RNA was isolated after 1 h and analyzed by real-time PCR (left). Sample data were normalized to GAPDH and are presented as -fold change relative to responsive cells (set at 100-fold). For TNFα protein, cells were harvested after 4 h, and the absolute values from ELISA are shown (right). Data are the mean ± S.D. of at least three experiments. R, responsive; T, tolerant. In addition, real-time PCR analysis showed a significant decrease in the levels of miRNA-221, miRNA-125b, and miRNA-579 after inhibiting their expression by the miRNA inhibitors (B).
miRNAs Bind to and Recruit Specific RNA-binding Proteins to TNFα 3′UTR to Mediate Translational Repression
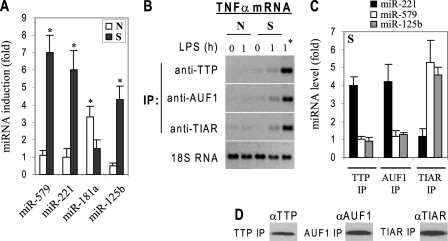
RBPs control stability and translation by binding to sequences within the 3′UTRs of target mRNA (35, 36). The TNFα 3′UTR region plays a major role its in gene expression and in the inflammation code and is a target for binding by RBPs, including mRNA destabilizers TTP and AUF1 and translational inhibitor TIAR (31, 36–38). To test the possibility that miRNAs might recruit these proteins to the TNFα 3′UTR, we performed RNA immunoprecipitation using cytoplasmic extracts and specific antibodies to TTP, AUF1, and TIAR. As shown in Fig. 8A, TNFα mRNA was enriched in TTP, AUF1, and TIAR immunoprecipitates from tolerant but not responsive cells. The protein binding to the mRNA was markedly increased after LPS stimulation of the tolerant state, suggesting the involvement of TLR4 signal in translation repression. The lack of this protein binding in responsive cells was not due to a decrease in their expression, because Western blot analysis showed no significant changes in their expression levels compared with tolerant cells (data not shown). These results demonstrate that inhibitory RBPs are specifically and actively recruited to TNFα 3′UTR, likely through miRNAs, to mediate its translational repression in LPS tolerance.
FIGURE 8.
TTP, AUF1, and TIAR proteins bind to the TNFα 3′UTR sequences and contribute to its translational repression in LPS-tolerant cells. THP-1 cells were made tolerant and then transfected with RelB-specific siRNA (to reverse transcription silencing). After 36 h, tolerant cells (together with responsive (fresh) cells) were left unstimulated or stimulated with 1 μg/ml LPS for 1 h (by which time TNFα mRNA expression is peaked in responsive cells; see Fig. 2). Cytoplasmic extract was prepared and immunoprecipitated with TTP-, AUF1-, or TIAR-specific antibody or IgG control. A, RNA was extracted from the immunoprecipitates (IP) and analyzed by PCR using specific primers to amplify the TNFα 3′UTR region. This method measures the extent of TNFα mRNA enriched in the IP. Total RNA was also isolated and analyzed for the 18 S RNA expression level (as an internal control). These results are representative of three independent experiments. The IgG-immunoprecipitated control RNA was also assayed for the presence of the TNFα mRNA and exhibited a background binding (not shown). B, miRNA immunoprecipitation. miR-221 co-immunoprecipitates with TTP and AUF1, whereas miR-579 and miR-125b co-immunoprecipitate with TIAR in tolerant cells after RelB KD. miRNA levels in the immunoprecipitated RNA from tolerant cells was amplified by real-time PCR. Data are the mean ± S.D. of at least three independent experiments. Sample data were normalized to endogenous U6 small nuclear RNA and are presented as -fold change relative to unstimulated tolerant cells (set at 1-fold). C, aliquots of TTP, AUF1, TIAR, and IgG control IPs were immunoblotted with the respective antibody to confirm specificity. R, responsive; T, tolerant.
miRNAs recruit RBPs to target mRNAs (15). To determine whether miRNAs directly associate with RBPs at the TNFα 3′UTR, we assessed miR-221, miR-579, and miR-125b levels in TTP, AUF1, and TIAR immunoprecipitates from tolerant cells. RNA extracted from the immunoprecipitates was analyzed by quantitative RT-PCR using miRNA-specific primers. miR-221 was detected in both TTP and AUF1 immunoprecipitates, whereas miR-579 and miR-125b were detected in TIAR immunoprecipitate only (Fig. 8B). In addition, Western blot analysis confirmed the specific immunoprecipitation of TTP, AUF1, and TIAR proteins complexes (Fig. 8C). Next, we determined the functional relevance of RBP recruitment to TNFα 3′UTR using siRNA. Knockdown of TTP or AUF1 partially increased mRNA stability, whereas combined knockdown completely restored mRNA stability but not translation (data not shown). In addition, TIAR knockdown did not affect mRNA stability. Importantly, concurrent knockdown of the three proteins completely restored mRNA stability and translation (Fig. 9). Together, these results demonstrate that miRNAs specifically bind to and recruit RBPs to the TNFα 3′UTR and suggest that they couple in a combinatorial fashion to induce translational repression of TNFα in LPS-tolerant cells.
FIGURE 9.

Combined knockdown of TTP, AUF1, and TIAR restores both TNFα mRNA and protein in LPS-tolerant cells. Cells were made tolerant by pretreatment with 1 μg/ml LPS for 12–16 h and then transfected with RelB siRNA (to reverse transcription silencing) together with control siRNA or a mixture of TTP, AUF1, and TIAR siRNAs. After 36 h, tolerant cells (along with responsive (fresh) cells) were washed and left unstimulated or stimulated with 1 μg/ml LPS. After 1 h, 1 μg/ml doxycycline was added, and the incubation continued for an additional 1 h (at which time TNFα mRNA was almost completely degraded in tolerant cells; see Fig. 2). TNFα mRNA and protein were analyzed by real-time PCR and ELISA, respectively. Sample data were normalized to GAPDH RNA. Values from unstimulated cells were then subtracted. Data are the mean ± S.D. of three independent experiments. RNA data are presented relative to stimulated responsive cells (set at 100-fold and are shown here as a reference for the extent of RNA and protein recovery in T cells). For TNFα protein, cells were harvested after 4 h. Western blotting showed a marked decrease in RelB, TTP, AUF1, and TIAR protein expression after siRNA transfection (not presented). R, responsive; T, tolerant.
The miRNA Binding Sites in the TNFα 3′UTR Mediate Decay and Arrest Translation in a Linked Luciferase Reporter mRNA Introduced into Tolerant Cells
To verify the translational repressive effects of miRNAs on TNFα mRNA in tolerant cells, we transfected mutants of TNFα 3′UTR linked to a luciferase reporter mRNA and then measured their effects on luciferase mRNA stability and translation. The luciferase gene (Luc2P, Promega) is constitutively active under the control of RPL10 promoter in pSGG vector, meaning it is highly expressed when not attached to any heterologous 3′UTR. As shown in Fig. 10B, transfection of the luciferase plasmid alone expressed both mRNA and protein, whereas a plasmid containing the luciferase gene fused to the wild-type TNFα 3′UTR reduced both luciferase mRNA and protein to a background levels in tolerant cells, demonstrating that the TNFα 3′UTR exert inhibitory effects on the reporter gene translation. Transfection of luciferase plasmid bearing mutation in the miR-221 binding site within the linked TNFα 3′UTR restored luciferase mRNA to levels close to those observed with luciferase plasmid without the TNFα 3′UTR (Luc). However, this mutation did not restore translation, indicating that miR-221 affects mRNA stability in tolerant cells. On the other hand, mutation in the miR-579 or miR-125b binding site alone did not restore mRNA stability or translation. In addition, combined double mutations in miR-221 plus miR-579 or miR-125 completely restored mRNA stability but partially increased translation efficiency. In contrast, a triple mutation in the three miRNA binding sites restored both mRNA stability and translation. Together, these results support that the miR-221 binding site in the TNFα 3′UTR mediates mRNA decay, whereas miR-579 and miR-125b binding sites mediate translation arrest.
Epigenetic-based Transcription Silencing Occurs during Animal SSI and Is Dissociated from Post-transcriptional Events
The stereotypical features of SSI, as demonstrated by the decreased production of pro-inflammatory mediators, temporally follow within several hours of the initial inciting phase in animal and human (2, 39). We used ex vivo preparation of peritoneal macrophages from mice with SSI to examine whether an epigenetic gene reprogramming occurs during animal SSI. We used the cecal ligation and puncture model, which has been described previously (20). Peritoneal macrophages were recovered from normal (sham) and septic mice and then stimulated with LPS for 1–6 h. Because macrophages are sensitive to LPS, we used 0.1 μg/ml doses. TNFα mRNA and protein were significantly induced after LPS stimulation of normal macrophages (Fig. 11). In the meantime, macrophages from septic mice exhibited background expression of mRNA and protein, even with longer stimulation of 2–8 h (data not shown), suggesting that transcription is silenced in mice with sepsis.
FIGURE 11.
TNFα expression is silenced in macrophage of septic mice, and treatment with the G9a methyltransferase inhibitor BIX-01294 restores mRNA but not protein levels. Sepsis was induced in C57BL/6 mice by cecal ligation and puncture. Mice (n = 5 mice per group) were sacrificed after 24 h. Peritonium was flushed with Hanks' balanced salt solution to collect macrophages. Macrophages were then washed and cultured in Dulbecco's modified Eagle's medium at 2 × 105 cells/ml. Cells were incubated with 5 μm BIX-01294 or DMSO. After 6 h, 0.1 μg/ml LPS was added to the cultures, and the incubation was continued for the indicated times. RNA (top) and protein (bottom) were extracted and assayed by real-time PCR and ELISA, respectively. Sample data were normalized to GAPDH mRNA. Data are the mean ± S.D. of three experiments. Data are presented as -fold change relative to the values from normal unstimulated macrophages (set as 1-fold). N, normal; S, septic.
We have previously shown that G9a promotes chromatin condensation and transcription silencing of TNFα and IL-1β by a complex of histone and DNA modifiers (4, 6). G9a knockdown (similar to RelB knockdown) restored TNFα mRNA but not protein levels in tolerant THP-1 cell model of SSI (5, 6). To test the possibility that G9a may induce epigenetic silencing in septic mice, macrophages were incubated with the G9a inhibitor BIX-01294, which has been shown to inhibit H3K9 methylation and chromatin condensation (40) (and our previous studies (6)). Pretreatment of septic macrophages with BIX for 6 h before LPS stimulation resulted in a marked recovery of TNFα mRNA but not protein (Fig. 11). Longer stimulation up to 8 h did not exhibit noticeable changes in protein levels (data not shown). These results suggested that the epigenetic reprogramming of pro-inflammatory genes also occurs during animal SSI and further indicate that transcription and translation events are separately regulated.
Negative Regulatory miRNA Code Also Exists in Septic Macrophages and Couples with RBPs at the TNFα mRNA
Our results showed that miR-221, miR-579, and miR-125b expression was significantly increased in tolerant THP-1 cells. Next, we determined the expression of these miRNAs in normal and septic macrophages by RT-PCR. We detected a significant increase in miR-221, miR-579, and miR-125b levels after LPS stimulation in septic macrophages (Fig. 12A).
FIGURE 12.
miR-221, miR-579, and miR-125b are significantly induced in septic macrophages and couple with TNFα mRNA after reversal of transcription silencing by BIX. A, normal and septic macrophages were isolated and left unstimulated or stimulated with 0.1 μg/ml LPS for 1 h. miRNA-enriched RNA was isolated and analyzed by RT-PCR. Sample data were normalized to endogenous U6 small nuclear RNA (as an internal control). Data represent the mean ± S.D. of three experiments and are presented as -fold change relative to unstimulated normal cells (set at 1-fold).*, significant difference (p ≤ 0.05). B, analysis of RBPs binding to the TNFα 3′UTR. Septic macrophages were treated with 5 μm BIX-01294 or DMSO for 6 h. Septic and normal macrophages were then left unstimulated or stimulated with 0.1 μg/ml LPS for 1 h. Cytoplasmic extract was prepared and immunoprecipitated (IP) with the indicated antibodies or IgG control. TNFα mRNA enriched in the IP was determined by PCR. The IgG-immunoprecipitated control RNA exhibited a background binding (not shown). The results are representative of three experiments. 1*, septic macrophages treated with BIX prior to stimulation. C, miRNA levels in TTP, AUF1, and TIAR immunoprecipitates was determined by RT-PCR. Because TNFα mRNA was markedly enriched in the IP of septic cells that were treated with BIX prior to stimulation, we used these conditions only to measure the levels of miRNAs associated with RBPs. Cytoplasmic extract was immunoprecipitated with antibody against TTP, AUF1, or TIAR. RNA was extracted from the IP and analyzed by RT-PCR. Sample data were normalized to endogenous U6 RNA. Data represent the mean ± S.D. of three experiments and are presented as -fold change relative to unstimulated septic macrophages (set at 1-fold). D, aliquots of TTP, AUF1, TIAR, and IgG control IPs were immunoblotted with the respective antibody to confirm specificity. *, statistically significant (p ≤ 0.05). N, normal; S, septic.
miRNAs function in the context of miRISC complex with its associated RBPs. To examine whether these miRNAs contribute to the post-transcriptional repression observed in septic macrophages, we first determined the binding pattern of the RBPs (identified in THP-1 cells) at the TNFα 3′UTR. Cytoplasmic extract was isolated and immunoprecipitated with antibodies against TTP, AUF1, and TIAR proteins. RNA was extracted from the immunoprecipitates and analyzed for the presence of TNFα mRNA. RNA from normal mice showed a background binding, whereas septic macrophages showed a marked increase in binding to TTP, AUF1, and TIAR after LPS stimulation (Fig. 12B).
Next, we measured miRNA levels in TTP, AUF1, and TIAR immunoprecipitates. We detected significantly higher levels of miR-221 associated with TTP and AUF1 proteins in septic compared with normal macrophages (Fig. 12C). In the meantime, miR-579 and miR-125b were co-immunoprecipitated in significant amounts with TIAR protein. Western blot analysis confirmed the specific immunoprecipitation of TTP, AUF1, and TIAR protein complexes (Fig. 12D). Together, the results presented in Fig. 12 show a differentially induced negative miRNA and RBP profile in septic mice and suggest that they may couple to disrupt TNFα protein synthesis.
DISCUSSION
Innate immunity employs a multilayered regulatory system to control inflammation through inciting, evolving, and resolving phases (2). The distinct and consistent pro- and anti-inflammatory physiologic phases of SSI suggest that gene reprogramming and not TLR specificity ultimately controls host response to infection (9, 39, 41). Our and other reports on epigenetically controlled chromatin structural modifications during SSI in humans and animals support this concept (4–6, 9, 42). The formation of facultative heterochromatin at promoters of acute pro-inflammatory genes and not anti-inflammatory genes reversibly silences a set of inflammation inducing genes to switch phenotype during the evolving phase of SSI (2). The reversible nature of facultative heterochromatin parallels a return to homeostasis when SSI resolves. Here, we identify an additional, indispensable, and distinct negative regulatory mechanism by which differentially expressed miRNAs disrupt protein synthesis of acute pro-inflammatory TNFα during the evolving anti-inflammatory stage of SSI that is typified by LPS tolerance.
We identified miRNAs that base pair with sequences in the TNFα 3′UTR. Expression profiling revealed that miR-221, miR-579, and miR-125b were selectively induced in LPS-tolerant cells and assembled with RBPs that target TNFα mRNA for translational repression. We also showed that these miRNAs induced translational repression through a combinatorial effect. miR-221 promoted degradation of endogenous as well as a luciferase reporter mRNA fused to TNFα 3′UTR, whereas miR-579 and miR-125b promoted translation arrest. The selective LPS-mediated induction of miRNAs in LPS-tolerant cells strongly suggests that differential induction of target-specific miRNAs rather than a global change in TLR-mediated pathway is a primary mechanism for translational repression in SSI.
mRNA instability and translation are mainly regulated by AU-rich elements (AREs) located in the 3′UTR of a variety of short-lived mRNAs, including cytokines (15, 43). Indeed, a recent report indicates that the number of AU sequences correlates with differential expression patterns in the inflammation gene code (1). The repressive function of AREs is regulated by protein factors that bind to the 3′UTR. The TNFα 3′UTR contains a ∼70-bp ARE sequence composed of several repeats of AUUUA pentamer (32). The 3′UTR of TNFα influences its synthesis in vivo. Mice expressing TNFα gene lacking the 3′UTR overexpress TNFα protein (44), whereas mice lacking RBPs that target the 3′UTR (e.g. TTP and AUF1) overproduce TNFα protein (36, 45). The three miRNA binding sites identified in this study are located within overlapping distances in the TNFα ARE element (see Fig. 13). We found that miR-221 increased mRNA degradation and was co-immunoprecipitated with TTP and AUF1 proteins that are implicated in mRNA instability. In addition, miR-579 and miR-125b induced translation arrest and were co-immunoprecipitated with the translation silencer TIAR. Thus, our results indicate that LPS tolerance is not only associated with differential alteration in miRNAs profile, but also with distinct post-transcriptional mechanisms of mRNA stability and translation. Also, functional analysis showed that TNFα 3′UTR conferred instability and translational arrest on a fused luciferase reporter mRNA in LPS-tolerant cells. Combined mutations in the 3′UTR that disrupted binding by miR-221, miR-579, and miR-125b restored mRNA stability and translation, demonstrating a critical role for the TNFα 3′UTR in its translational repression by miRNAs.
FIGURE 13.
A model of TNFα post-transcriptional repression during SSI. LPS stimulation results in increased expression of miR-221, miR-579, and miR-125b in THP-1-tolerant cells. These miRNAs are then loaded into a ribonucleoprotein complex (miRISC) where they help recruit the complex to the TNFα 3′UTR through binding to complementary sequences (mostly AREs) within this region. RNA-binding proteins TTP, AUF1, and TIAR, which are included in the RISC complex, will bind to the ARE (likely through one or more of these miRNAs) and, jointly with the rest of RISC components, will induce TNFα repression by three possible mechanisms: either mRNA deadenylation (1) and/or decapping (2) resulting in mRNA degradation OR translational repression by inhibiting translation initiation or elongation (3). Note that the constitutive decay element (CDE) has been implicated in mediating mRNA instability, but the one or more proteins involved have not been identified. Our results suggested that this CDE element is targeted by AUF1.
miRNAs function by recruiting RBPs to target mRNA for translational repression (15, 46). We detected TTP, AUF1, and TIAR proteins bound at the TNFα 3′UTR in tolerant, but not LPS-responsive cells. The binding of these proteins was dependent on miRNAs induction in tolerant cells, because they did not bind in responsive cells despite their normal protein levels (dad not shown). Previous reports indicated that TTP and AUF1 enhance TNFα and IL-1β mRNA degradation in activated macrophages and other cell types (36, 47). TIAR, and the related protein TIA-1, down-regulates TNFα translation (37, 48). Our co-immunoprecipitation experiments demonstrated that miR-221 associated with TTP and AUF1 proteins, whereas miR-579 and miR-125b associated with TIAR. These results, together with our finding that functional inhibition of miR-221 stabilized mRNA while miR-579 and miR-125b inhibition enhanced translation of TNFα, suggest that these miRNAs directly interact with and recruit specific RBPs to TNFα 3′UTR, thereby mediating translational repression. In addition, siRNA-mediated knockdown of TTP and AUF1 in tolerant cells stabilized TNFα mRNA, whereas TIAR knockdown increased mRNA translation to almost the same extent seen upon functional inhibition of miRNA expression, indicating that these miRNAs promoted translational repression of TNFα in tolerant cells by recruiting inhibitory RBPs. A recent study reported that a constitutive decay element in the TNFα 3′UTR, downstream of the ARE (see Fig. 13) targeted TNFα mRNA for rapid decay (38). Interestingly, miR-221 binding site is located within this element. Because miR-221 co-immunoprecipitated with AUF1, it is possible that miR221 recruits AUF1 to this decay element. Thus, our results suggest that differentially induced miRNAs couple with specific RBPs to mediate TNFα translational repression in LPS-tolerant phenotype.
Some RBPs may protect certain mRNAs from miRNA-mediated repression (31, 49) by facilitating mRNA targeting to polysomes for translation rather than to p-bodies (the sites of mRNA decay and translation arrest) for translational repression (15). For example, HuR protein stabilizes TNFα mRNA in macrophages under normal physiologic conditions (31), and Dnd1, another RBP, may increase mRNA stability and translation of some tumor suppressor mRNAs by preventing negative regulatory miRNA binding (49, 50). In this study, we observed that HuR and Dnd1 bound to TNFα 3′UTR in responsive but not LPS-tolerant cells (data not shown). This is consistent with these regulators stabilizing TNFα mRNA and translation during the inciting phase of SSI (i.e. in responsive cells). In contrast, during the evolving phase of endotoxin tolerance and gene silencing, HuR and Dnd1 did not bind to TNFα mRNA despite their normal expression in the cytoplasm. Thus, the specific repressive effect conferred by miRNAs on 3′UTR may displace HuR or Dnd1 and override their positive effects.
Recent studies reported that miR-146a and miR-155 are inflammation modifiers and are induced by LPS in normal monocytes/macrophages (51, 52). miR-146a down-regulates TLR signaling proteins TRAF6 and IRAK1 (52) and thus may reduce TNFα expression and indirectly prevent the generation of endotoxin tolerance (51). The prevention context of gene silencing should be distinguished from the reversal concept that would be needed to intervene in SSI. miR-155 can attenuate TLR4 signal by targeting IKKϵ kinase (53) but can also activate LPS signaling pathways by enhancing TNFα expression (53, 54). Although miR-146a and miR-155 can attenuate TLR signaling, they do not appear to directly affect pro-inflammatory gene translational repression in LPS-tolerant cells. Our data indicate that miR-146a and miR-155 expression increases in both responsive and LPS-tolerant cells. Our study does not exclude the possibility that these miRNAs may directly or indirectly influence tolerance.
Although there are no previous reports to our knowledge implicating miR-221 or miR-579 in regulating TNFα expression, a recent study reported that miR-125b targets TNFα mRNA transcripts for degradation in murine macrophages, which may limit TNFα production under basal normal conditions (53). After LPS stimulation, however, the miR-125b level decreases to allow TNFα production in the inciting phase. Our finding of increased miR-125b expression in LPS-tolerant THP-1 cells, which follows the earlier decrease, is consistent with this report in that a reduction of miR-125b during the LPS-inciting phase would support TNFα translation, whereas a subsequent increase in the evolving tolerant phase would feed forward to repress TNFα translation. A recent report further supports that feed-forward repressor loops can limit innate immune responses (13). TNFα-Induced miRNAs miR-31 and miR-17–3p reduced expression of E-selectin and intercellular adhesion molecule 1 in human endothelial cells to limit endothelial and neutrophil adhesion events after the inciting phase.
We showed that epigenetic gene reprogramming also occurred in mice with SSI and that this process is reversible by returning silent heterochromatin to euchromatin through G9a inhibition (Fig. 11). The finding, that septic macrophages with reversed epigenetic silencing expressed TNFα mRNA but not protein, suggested a post-transcriptional repression mechanism likely promoted by the induction of negative regulatory miRNAs, similar to that observed in THP-1 cell model of SSI. Recent studies reported selective epigenetic-based gene repression mechanisms using an animal model of inflammation (9, 20). LPS-tolerant macrophages derived from bone marrow exhibited a differential gene-silencing profile after a second LPS stimulation ex vivo. Such silencing was associated with a loss of H3K4 trimethylation, which usually marks transcriptionally active genes (9, 55). A recent study (20), using the cecal ligation and puncture model of sepsis, demonstrated that IL-12 gene transcription is associated with a shift from H3K27 trimethylation to monomethylation, which associates with gene silencing (56). Thus, epigenetic silencing seems as a primary process of transcriptional repression in animal systemic and local inflammation. Our studies add on to these findings and, further, reveal an additional miRNA-dependent post-transcriptional silencing mechanism during SSI.
Based on our results, we propose a model as depicted in Fig. 13, where differentially induced miRNAs bind specifically to TNFα 3′UTR. These miRNAs recruit the RNA-binding proteins TTP, AUF1, and TIAR, which in turn could interact with Ago2 protein, resulting in the assembly of miRISC repressor complexes. Such complexes are then targeted to p-bodies, where mRNA degradation by deadenylation or decapping, and/or translational arrest due to inhibition of translation initiation, occur. This silencing process is gene-specific, allowing for sustained expression of other inflammation modifiers such as IκBα. We found that RBPs and miRNAs were associated with Ago2 at the TNFα 3′UTR in LPS-tolerant but not -responsive cells, and knockdown of Ago2 resulted in dissociation of the complex and relieved translation repression (data not shown). Thus, we conclude that epigenetic modifications generating gene-specific formation of transcription silencing facultative heterochromatin combine with the post-transcriptional repressor events shown herein to direct the physiology of evolving SSI through tightly regulated feed-forward loops. The reversible nature of facultative heterochromatin and the post-transcriptional silencing described in this report and the recent success in using LAN-based antagomirs to silence miRNA expression in mice and primates (57), if applied to SSI, might provide a novel approach for therapeutic interventions.
Acknowledgments
We thank A. Church, J. Hu, and S. Cousart for technical assistance, T. Liu for Western blotting, and B. Yoza for critical discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI065791 and R01AI079144.
- SSI
- severe systemic inflammation
- TLR
- Toll-like receptor
- IL-1
- interleukin-1
- LPS
- lipopolysaccharide
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ELISA
- enzyme-linked immunosorbent assay
- ARE
- AU-rich element
- miRNA
- microRNA (also miR)
- miRISC
- miRNA-induced silencing complex
- UTR
- untranslated region
- TNFα
- tumor necrosis factor α
- siRNA
- small interfering RNA
- IP
- immunoprecipitation
- RBP
- RNA-binding protein.
REFERENCES
- 1.Hao S., Baltimore D. (2009) Nat. Immunol. 10, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCall C. E., Yoza B. K. (2007) Am. J. Respir. Crit Care Med. 175, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan C., Li L., McCall C. E., Yoza B. K. (2005) J. Immunol. 175, 461–468 [DOI] [PubMed] [Google Scholar]
- 4.Chen X., El Gazzar M., Yoza B. K., McCall C. E. (2009) J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Gazzar M., Yoza B. K., Hu J. Y., Cousart S. L., McCall C. E. (2007) J. Biol. Chem. 282, 26857–26864 [DOI] [PubMed] [Google Scholar]
- 6.El Gazzar M., Yoza B. K., Chen X., Hu J., Hawkins G. A., McCall C. E. (2008) J. Biol. Chem. 283, 32198–32208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Gazzar M., Liu T., Yoza B. K., McCall C. E. (2010) J. Biol. Chem. 285, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Yoza B. K., El Gazzar M., Hu J. Y., Cousart S. L., McCall C. E. (2009) Clin. Vaccine Immunol. 16, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster S. L., Hargreaves D. C., Medzhitov R. (2007) Nature 447, 972–978 [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S., Karin M. (2002) Cell 109, (suppl.) S81–S96 [DOI] [PubMed] [Google Scholar]
- 11.Mangan S., Alon U. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005) Nature 435, 839–843 [DOI] [PubMed] [Google Scholar]
- 13.Suárez Y., Wang C., Manes T. D., Pober J. S. (2010) J. Immunol. 184, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 15.Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 16.Bushati N., Cohen S. M. (2007) Annu. Rev. Cell Dev. Biol. 23, 175–205 [DOI] [PubMed] [Google Scholar]
- 17.Sonkoly E., Ståhle M., Pivarcsi A. (2008) Semin. Cancer Biol. 18, 131–140 [DOI] [PubMed] [Google Scholar]
- 18.Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J., Ruan Q., Johnson D. P., Chen Y., O'Neill L. A. (2010) Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 19.LaRue K. E., McCall C. E. (1994) J. Exp. Med. 180, 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen H., Dou Y., Hogaboam C. M., Kunkel S. L. (2008) Blood 111, 1797–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M. A. (2002) Methods 26, 182–190 [DOI] [PubMed] [Google Scholar]
- 22.Landthaler M., Gaidatzis D., Rothballer A., Chen P. Y., Soll S. J., Dinic L., Ojo T., Hafner M., Zavolan M., Tuschl T. (2008) RNA 14, 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. (2005) Science 309, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 24.Asirvatham A. J., Gregorie C. J., Hu Z., Magner W. J., Tomasi T. B. (2008) Mol. Immunol. 45, 1995–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethupathy P., Megraw M., Hatzigeorgiou A. G. (2006) Nat. Methods 3, 881–886 [DOI] [PubMed] [Google Scholar]
- 26.Yoza B. K., Wells J. D., McCall C. E. (1998) Clin. Diagn. Lab. Immunol. 5, 281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung D. R., Lee Y. S., Lee S. S. (2008) J. Infect. 56, 44–50 [DOI] [PubMed] [Google Scholar]
- 28.Amin A. R., Attur M. G., Thakker G. D., Patel P. D., Vyas P. R., Patel R. N., Patel I. R., Abramson S. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14014–14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoyt J. C., Ballering J., Numanami H., Hayden J. M., Robbins R. A. (2006) J. Immunol. 176, 567–572 [DOI] [PubMed] [Google Scholar]
- 30.Lindeman J. H., Abdul-Hussien H., van Bockel J. H., Wolterbeek R., Kleemann R. (2009) Circulation 119, 2209–2216 [DOI] [PubMed] [Google Scholar]
- 31.Dean J. L., Wait R., Mahtani K. R., Sully G., Clark A. R., Saklatvala J. (2001) Mol. Cell. Biol. 21, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J., Beutler B. (1990) Eur. Cytokine Netw. 1, 71–75 [PubMed] [Google Scholar]
- 33.Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G. (1999) Immunity 10, 387–398 [DOI] [PubMed] [Google Scholar]
- 34.Mattes J., Yang M., Foster P. S. (2007) Am. J. Respir. Cell Mol. Biol. 36, 8–12 [DOI] [PubMed] [Google Scholar]
- 35.Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. (2001) Cell 107, 451–464 [DOI] [PubMed] [Google Scholar]
- 36.Lu J. Y., Sadri N., Schneider R. J. (2006) Genes Dev. 20, 3174–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. (1999) J. Biol. Chem. 274, 2322–2326 [DOI] [PubMed] [Google Scholar]
- 38.Stoecklin G., Lu M., Rattenbacher B., Moroni C. (2003) Mol. Cell. Biol. 23, 3506–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavaillon J. M., Adib-Conquy M. (2006) Crit. Care 10, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubicek S., O'Sullivan R. J., August E. M., Hickey E. R., Zhang Q., Teodoro M. L., Rea S., Mechtler K., Kowalski J. A., Homon C. A., Kelly T. A., Jenuwein T. (2007) Mol. Cell 25, 473–481 [DOI] [PubMed] [Google Scholar]
- 41.Foster S. L., Medzhitov R. (2009) Clin. Immunol. 130, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medzhitov R., Horng T. (2009) Nat. Rev. Immunol. 9, 692–703 [DOI] [PubMed] [Google Scholar]
- 43.Chen C. Y., Shyu A. B. (1995) Trends Biochem. Sci. 20, 465–470 [DOI] [PubMed] [Google Scholar]
- 44.Keffer J., Probert L., Cazlaris H., Georgopoulos S., Kaslaris E., Kioussis D., Kollias G. (1991) EMBO J. 10, 4025–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carballo E., Lai W. S., Blackshear P. J. (1998) Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 46.Peters L., Meister G. (2007) Mol. Cell 26, 611–623 [DOI] [PubMed] [Google Scholar]
- 47.Hau H. H., Walsh R. J., Ogilvie R. L., Williams D. A., Reilly C. S., Bohjanen P. R. (2007) J. Cell. Biochem. 100, 1477–1492 [DOI] [PubMed] [Google Scholar]
- 48.Piecyk M., Wax S., Beck A. R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Anderson P. (2000) EMBO J. 19, 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ketting R. F. (2007) Cell 131, 1226–1227 [DOI] [PubMed] [Google Scholar]
- 50.Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Ørom U. A., Lund A. H., Perrakis A., Raz E., Agami R. (2007) Cell 131, 1273–1286 [DOI] [PubMed] [Google Scholar]
- 51.Nahid M. A., Pauley K. M., Satoh M., Chan E. K. (2009) J. Biol. Chem. 284, 34590–34599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 54.O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 56.Smale S. T. (2003) Nat. Immunol. 4, 607–615 [DOI] [PubMed] [Google Scholar]
- 57.Stenvang J., Lindow M., Kauppinen S. (2008) Biochem. Soc. Trans. 36, 1197–1200 [DOI] [PubMed] [Google Scholar]