Abstract

Slow μsec/msec dynamics involved in protein folding, binding, catalysis and allostery are currently detected using NMR dispersion experiments such as CPMG (Carr-Purcell-Meiboom-Gill) or spin-lock R1ρ. In these methods, protein dynamics are obtained by analyzing relaxation dispersion curves obtained from either changing the time-spacing between 180° pulses or by changing the effective spin-locking field strength. In this Communication, we introduce a new method to induce a dispersion of relaxation rates. Our approach relies on altering the shape of the adiabatic full passage pulse, and is conceptually different from existing approaches. By changing the nature of the adiabatic radiofrequency irradiation, we are able to obtain rotating frame R1 and R2 (R1ρ and R2ρ) dispersion curves that are sensitive to slow μsec/msec protein dynamics (demonstrated with ubiquitin). The strengths of this method are to (a) extend the dynamic range of the relaxation dispersion analysis, (b) avoid the need for multiple magnetic field strengths to extract dynamic parameters, (c) measure accurate relaxation rates that are independent of frequency offset, and (d) reduce the stress to NMR hardware (e.g., cryoprobes).
Protein dynamics is central to function. Specifically, conformational dynamics in the μsec-msec timescale is synchronous with phenomena such as allostery, folding, molecular recognition, catalysis and inhibition.1–11
NMR rotating frame (R1ρ) and Carr-Purcell-Meiboom-Gill (CPMG, R2) relaxation dispersion methods have been widely used to probe slow dynamics and discriminate ground and excited states in small and large proteins.12, 13 While these approaches have been critical in characterizing enthalpy and entropy in biomolecules, the methods are limited by (a) the small dynamic range of the relaxation dispersion phenomena, (b) difficulty in setting up the pulse sequences, (c) the low tolerance of cryogenic probes for continuous radiofrequency (RF) irradiation, and (d) the cost of performing relaxation measurements at multiple magnetic fields in order to extract reliable dynamic parameters. To expand the dynamic range of relaxation dispersions, sample conditions are often changed (pH, temperature, viscosity, etc.). While there have been some technical improvements to the CPMG method,14, 15 there have been no good remedies for the other drawbacks.
In this Communication, we present a conceptually different method for characterizing slow μsec/msec protein motions. Our approach consists of using adiabatic full passage (AFP) pulses16 to induce a dispersion of relaxation rates, which expands the dynamic range and substantially increases the ease for measuring relaxation dispersion experiments. Unlike the CPMG R2 or spin-lock R1ρ dispersion experiments (reviewed in Ref. 17), where relaxation dispersion is obtained by changing the duration between 180° pulses or the spinlocking field strength, we altered the shape of the adiabatic RF irradiation to obtain R1ρ and R2ρ dispersion curves. Contrast in relaxation rates can be generated by use of adiabatic RF pulses,16 which enables the coverage of large bandwidths with relatively low power. While adiabatic pulses have been incorporated in many biomolecular NMR experiments,18 the concept of using adiabatic RF pulses to induce relaxation dispersion has not been introduced.
We tested this approach with ubiquitin, a globular protein whose dynamics has been recognized to be crucial for proteinprotein recognition.19 Adiabatic R1ρ and R2ρ relaxation rates 20–22 were measured at 14.1 T (600 MHz 1H frequency) on a sample of [U-15N] labeled ubiquitin at 5 °C and 25 °C. A standard 2D HSQC-type pulse sequence (Figure 1)23 was modified by inserting 15N pulse trains of 4, 8, 12, 16, 20, 24 and 28 AFP pulses. The relaxation experiments were conducted such that the magnetization rotated in a perpendicular (R2ρ) direction around the effective field or was aligned with it (R1ρ). Adiabatic hyperbolic secant (HS) pulses were designed with different “stretching” factors (HSn: n = 1, 2, 4, 6 and 8).24 The applied RF amplitude (ω1(t)) and frequency sweep (ωRF(t) - ωc) of the HSn AFP pulses are:
| (1) |
| (2) |
where ω1max is the maximum amplitude of the pulse, β is a truncation factor (sech(β)=0.01), Tp is the length of the AFP pulse, ωc is the carrier frequency, BW is the bandwidth of the pulse, and t and t′ represent time. Note that ω1, ω1max, ωRF, ωc and BW are expressed in units of rad/s. Each AFP pulse was 4-msec and implemented in groups of four (phase-cycled using MLEV-425), leading to total relaxation delays of 16, 32, 48, 64, 80, 96 and 112 msec (no 112 msec for R2ρ). The maximum amplitude of the AFP pulse was ω1max/(2π) = 3.5 kHz, and the bandwidth, BW/(2π), was 10 kHz. This choice of parameters guaranteed full adiabatic inversion of all residues within a range of ~3 kHz for every HSn pulse. Relaxation rates were obtained in a residue dependent manner by fitting a mono-exponential decay function to the signal intensities versus AFP-train duration. No proton decoupling scheme was used, since net evolution due to heteronuclear scalar coupling was refocused by the 15N AFP pulses in the R1ρ and R2ρ experiments.26
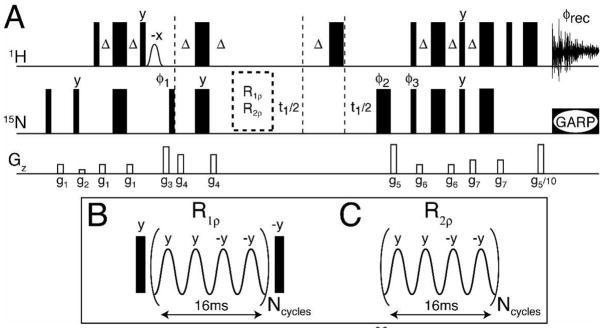
Figure 1.
HSQC-type pulse sequence23 (A) used to measure R1ρ (B) and R2ρ (C). In the R1ρ experiment, 15N magnetization is positioned longitudinally, and then subjected to a train of adiabatic pulses. For the R2ρ experiment, 15N magnetization is positioned in the transverse plane, and then subjected to a train of adiabatic pulses. Note that Ncycles needs to be an integer value, and differs from n, the stretching factor (see Eqs. 1–2). Phases are φ1 = x, −x, x, −x, φ2 = x, x, y, y, φ3 = x, φrec = x, −x, x, −x. The phases of ϕ3 and ϕrec are both inverted (shifted by 180°) in alternate t1 increments. Gradient magnitudes for G1–G7 are 1.8, 1.3, 26.6, 14.2, 42.6, 3.6 and 5.3 G/cm with lengths of 0.5, 0.5, 1, 1, 2, 0.5 and 0.5 msec, respectively. To achieve phase-sensitive t1 detection, the phases of φ1 and the magnitude of the second G5 gradient are inverted (−42.6 G/cm).
Several relaxation studies have been conducted on ubiquitin, which has a compact fold with nearly all of the residues displaying similar relaxation rates. The relaxation rates measured with our methods (Figure 2) are in agreement with previous data,27 showing essentially constant R1ρ and R2ρ rates under each of the HSn pulses. Note that adiabatic pulses cover the entire bandwidth for the 15N chemical shifts. This avoids the cumbersome back-calculation of the “nominal” effective frequency and tilt angle for each residue of interest, a procedure that requires extremely accurate power calibration when using the spin-lock R1ρ dispersion approach.
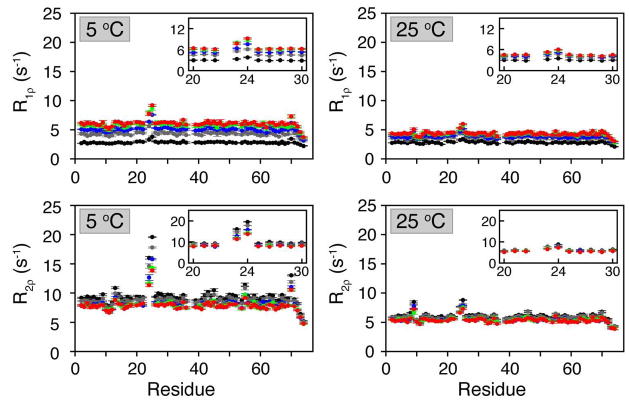
Figure 2.
R1ρ and R2ρ relaxation rates at 5 °C and 25 °C. The trends in the relaxation rates for R1ρ are HS1 (black) < HS2 (grey) < HS4 (blue) < HS6 (green) < HS8 (red) and for R2ρ are HS1 > HS2 > HS4 > HS6 > HS8, are consistent with anticipated results. 22 Residues that are known to undergo chemical exchange at 5 °C (Glu24, Asn25, Thr55 and Val70) displayed a pronounced dispersion between the HSn pulse types, which is not observed at 25 °C (i.e., exchange becomes too fast).
The utilization of different modulation functions induced a dispersion of relaxation rates for all residues, with the smallest adiabatic R1ρ given by HS1 pulses, followed by HS2, HS4, HS6, and HS8. The opposite trend was seen for R2ρ. Notably, residues undergoing conformational exchange (e.g., Asn25) exhibited faster adiabatic R1ρ and R2ρ and larger dispersions (i.e., the difference in relaxation rates between HS1 and HS8) as compared to residues with no conformational exchange (e.g., Lys48). Residues not undergoing exchange displayed larger R1ρ dispersion as compared to R2ρ, while the opposite trend is detected for residues with a contribution from chemical exchange. The dispersion rates become considerably smaller upon increasing the temperature (Figure 2), implying that adiabatic R1ρ and R2ρ are mainly sensitive to slow exchange dynamics. Notably, no dispersion of relaxation rates (above 1 Hz) was observed for any residue at 5 °C when measured using the classical CPMG relaxation dispersion method (Figure 3). A small relaxation dispersion was observed in Asn25 using the off-resonance R1ρ experiment previously used to measure μsec-msec motions in ubiquitin (see Figure 3A).27
Figure 3.
Relaxation dispersion results for Asn25 (black circles) and Lys48 (grey circles) of ubiquitin collected at 5 °C. (A) R1ρ and (B) CPMG relaxation dispersion experiments. (C) R1ρ and (D) R2ρ relaxation rates plotted as a function of HSn adiabatic pulse. Chemical exchange contributions to R1ρ (E) and R2ρ (F) for Asn25 (open circles) obtained by subtracting the rates of Lys48 from those of Asn25. Best fits of the data to Eqs 3 and 4 give Δω2pApB = 29 × 104 [rad/s]2 and 1/τex ~ 2.5 × 104 s−1.
Although the complete description of the relaxation during adiabatic pulses would require the evaluation of chemical shift anisotropy, cross-correlated relaxation and interference effects, we assumed that relaxation phenomena not related to chemical exchange are approximately similar across the protein sequence (excluding terminal residues). Under these approximations, the difference of rates between the two kinds of residues represents purely the exchange contribution [e.g., for each HSn pulse: R1,2, ρ,ex=R1,2ρ(Asn25)-R1,2ρ(Lys48)]. We analyzed the exchange-induced relaxations during the AFP pulses using the classical formalism developed for the fast exchange regime (1/τex > Δω).20, 28, 29 According to this treatment, the R1ρ and R2ρ result from an average of instantaneous time-dependent contributions, arising from the inherent time evolution of the effective frequency (ωeff) and the tilt angle, α, during the pulse:
| (3) |
| (4) |
pA and pB are the populations of the two exchanging sites, Δω is the chemical shift difference, and τex is the exchange correlation time. For Asn25 of ubiquitin, the exchange parameters found using Eqs 3 and 4 gave Δω2pApB = 29 × 104 [rad/s]2 and 1/τex ~ 2.5 × 104 s−1, which are in quantitative agreement with the corresponding values reported previously.27
In conclusion, adiabatic rotating frame relaxation measurements, R1ρ and R2ρ, provide a solid approach for characterizing μsec-msec protein dynamics. These pulse sequences are straightforward to set-up and implement on any modern spectrometer that can perform phase and amplitude RF modulation. Unlike currently available methods, this approach gives frequency offset independent relaxation rates without the need to apply residue-specific corrections. More importantly, the use of AFP extends the dynamic range of the relaxation dispersion analysis, avoiding troublesome variations of sample conditions and/or use of multiple magnetic field strengths to extract dynamic parameters. The relatively low power utilized for these pulses avoids excess stress to the hardware (probes), and at the same time substantially reduces sample heating. This approach can be applied to a wide range of relaxation measurements from small to large soluble proteins as well as membrane proteins reconstituted in detergent micelles,30 taking advantages of TROSY schemes.31 The implementation to other nuclei (1H, 13C, 2H) will be highly beneficial in the characterization of both backbone and side chain dynamics in macromolecular complexes.
Supplementary Material
Acknowledgments
NIH Grants GM64742, HL80081, GM072701 (to G. Veglia); NIH P41 RR008079 (to CMRR); R01NS061866 and R21NS059813 (to S. Michaeli).
Footnotes
Supporting Information Available: The pulse shape profiles (frequency and amplitude modulation) as a function of the AFP pulse duration are given for the five HSn pulses that were employed in this study. The adiabatic pulses and pulse sequence code (for Varian spectrometers) are available for download at our website: www.chem.umn.edu/groups/veglia.
References
- 1.Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 2.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 3.Korzhnev DM, Kay LE. Acc Chem Res. 2008;41:442–451. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 4.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beach H, Cole R, Gill ML, Loria JP. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- 7.Das R, Chowdhury S, Mazhab-Jafari MT, Sildas S, Selvaratnam R, Melacini G. J Biol Chem. 2009;284:23682–23696. doi: 10.1074/jbc.M109.011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishima R, Louis JM, Torchia DA. J Mol Biol. 2001;305:515–521. doi: 10.1006/jmbi.2000.4321. [DOI] [PubMed] [Google Scholar]
- 9.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson BD, Wilson KA, Etzkorn FA, Peng JW. J Am Chem Soc. 2010;132:5607–5609. doi: 10.1021/ja9096779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kern D, Zuiderweg ER. Curr Opin Struct Biol. 2003;13:748–57. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Igumenova TI, Lee AL, Wand AJ. Biochemistry. 2005;44:12627–12639. doi: 10.1021/bi050832f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Nat Struct Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 13.Palmer AG, 3rd, Kroenke CD, Loria JP. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 14.Long D, Liu M, Yang D. J Am Chem Soc. 2008;130:2432–2433. doi: 10.1021/ja710477h. [DOI] [PubMed] [Google Scholar]
- 15.Yip GN, Zuiderweg ER. J Magn Reson. 2004;171:25–36. doi: 10.1016/j.jmr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Garwood M, DelaBarre L. J Magn Reson. 2001;153:155–177. doi: 10.1006/jmre.2001.2340. [DOI] [PubMed] [Google Scholar]
- 17.Loria JP, Berlow RB, Watt ED. Acc Chem Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 18.Zweckstetter M, Holak TA. J Magn Reson. 1998;133:134–147. doi: 10.1006/jmre.1998.1437. [DOI] [PubMed] [Google Scholar]
- 19.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 20.Michaeli S, Sorce DJ, Idiyatullin D, Ugurbil K, Garwood M. J Magn Reson. 2004;169:293–299. doi: 10.1016/j.jmr.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Michaeli S, Sorce DJ, Springer CS, Jr, Ugurbil K, Garwood M. J Magn Reson. 2006;181:135–147. doi: 10.1016/j.jmr.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Mangia S, Liimatainen T, Garwood M, Michaeli S. Magn Reson Imaging. 2009;27:1074–1087. doi: 10.1016/j.mri.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 24.Tannus A, Garwood M. NMR Biomed. 1997;10:423–434. doi: 10.1002/(sici)1099-1492(199712)10:8<423::aid-nbm488>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 25.Levitt M, Freeman R, Frenkel T. J Magn Reson. 1982;47:328–30. [Google Scholar]
- 26.Bendall MR. J Magn Reson A. 1995;116:46–58. [Google Scholar]
- 27.Massi F, Grey MJ, Palmer AG., 3rd Protein Sci. 2005;14:735–742. doi: 10.1110/ps.041139505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abergel D, Palmer AG. Conc Magn Reson. 2003;19A:134–48. [Google Scholar]
- 29.Sorce DJ, Michaeli S, Garwood M. J Magn Reson. 2006;179:136–139. doi: 10.1016/j.jmr.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Traaseth NJ, Veglia G. Biochim Biophys Acta. 2010;1798:77–81. doi: 10.1016/j.bbamem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pervushin K, Riek R, Wider G, Wuthrich K. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.