Abstract
Background
MicroRNAs (miRNAs) are involved in cancer pathogenesis, apoptosis and cell growth, thereby functioning as either tumor suppressors or oncogenes. However, expression alterations and roles of these miRNAs in pancreatic cancer are largely unknown. We hypothesize that pancreatic cancer may have a unique miRNA profile, which may play a critical role in pancreatic cancer development, progression, diagnosis and prognosis.
Methods
Differential expression of 95 miRNAs was analyzed by real time RT-PCR using the QuantiMir System. All 95 miRNAs chosen for the array are based on their potential functions related to cancer biology, cell development and apoptosis. The expression of miRNAs for pancreatic cancer tissue samples or cancer cell lines was normalized to U6 RNA and compared with those in the relatively normal pancreatic tissues or normal human pancreatic ductal epithelial (HPDE) cells. Human pancreatic tissue with chronic pancreatitis was also included for analysis.
Results
In the initial analysis, the expression of most 95 miRNAs was substantially changed in pancreatic cancer tissues (n=5) and cell lines (n=3) compared with relatively normal pancreatic tissues and HPDE cells. However, each pancreatic cancer tissue or cell type had a substantially different profiling pattern with other cases or cell types as well as chronic pancreatitis tissue, indicating the individual diversity of pancreatic cancer. Further analysis was performed on 10 pancreatic cancer cell lines and 17 pairs of pancreatic cancer/normal tissues. Eight miRNAs were significantly upregulated in most pancreatic cancer tissues and cell lines, including miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b and miR-95. The incidence of upregulation of these eight genes between normal controls and tumor cells or tissues was ranging from 70% to 100%. The magnitude of increase of these miRNAs in pancreatic cancer samples was ranging from 3 to 2018 fold of normal controls.
Conclusions
Pancreatic cancer tissues or cell lines have a unique miRNA profiling pattern at the individual basis as compared with relatively normal pancreatic tissues or cells as well as pancreatitis tissue. Upregulation of eight miRNAs occurs in the most of pancreatic cancer tissues and cell types. These miRNAs may share common pathways in pancreatic cancer pathogenesis. This study may provide useful information for further investigations of functional roles of miRNAs in pancreatic cancer development, progression, diagnosis and prognosis.
Keywords: microRNA, pancreatic cancer, real time PCR
Introduction
Non-coding RNAs are a class of RNAs that do not encode proteins, while possess regulatory functions in gene expression. Non-coding RNAs have drawn a great attention in recent years since.The discovery of small interfering RNAs (siRNAs) and microRNAs (miRNAs) has substantial impact on gene regulation. miRNAs are a novel class of short (typically 18–23 nucleotides) single stranded RNAs, which are identified as a new family of regulatory molecules involved in cancer development [1–4]. miRNAs cause posttranscriptional gene silencing by either inducing target mRNA degradation or by repressing the translation process upon binding to the 3’-untranslational region (UTR) of their target mRNAs [5]. Mature miRNAs are excised from stem-loop precursors, which are transcribed as part of longer primary transcripts. These primary miRNAs appear to be first processed by the RNase Drosha in the nucleus, after which the precursor miRNAs are exported to the cytoplasm where the RNase Dicer further processes them.
Regulation of miRNA expression has been demonstrated to play a key role in development, cell growth and differentiation processes in a variety of eukaryotic organisms [6,7]. Usually, miRNAs are dysregulated in cancers. Some miRNAs are temporally over-expressed in the early stage of cancer progression and they act like oncogenes by promoting proliferation and/or repressing apoptosis. Conversely, some miRNAs with tumor-repressor functions are downregulated in cancers. miRNA expression profiles may be unique in different tumors and from different origins. Both normal and malignant cancer tissues may have specific miRNA expression signatures and show differential expression across tumor types. Several studies have demonstrated altered miRNA expression profile in various hematological and solid tumor entities [1,2]. For example, a unique expression signature of only 13 miRNAs differentiated more aggressive form of chronic lymphocytic leukemia from the benign one and was found to be associated with the cancer progression [8]. Expression alterations of specific miRNAs appear to be correlated with clinically malignancy or metastatic phenotypes, and predict the clinical outcome even better than the mRNA expression data [9–11].
Pancreatic cancer is the fourth leading cause of cancer death in the United States [12]. There was 37,170 new cases diagnosed, and approximately 33,370 deaths due to pancreatic adenocarcinoma in 2007 in the United States [13]. Although surgical resection provides a potential cure, about 70% patients still develop early recurrence within 6–12 months following surgery. Due to lack for reliable early detection markers, pancreatic tumors are usually in the advanced stage upon diagnosis. Moreover, pancreatic tumors have a predilection for early vascular dissemination and metastasis to distant organs. Clearly, the discovery of miRNA alterations in pancreatic cancer not only helps us to better understand the biology of this disease, but more importantly provides new prognostic and diagnostic strategies. Due to the high stability of miRNAs even in poorly preserved specimens, they are expected to be a valuable tool in clinical research and biomarkers discovery.
By Northern blotting analysis, several studies have shown that particular miRNAs were altered in pancreatic cancer tumor tissues [14–20]. However, these data are incomplete for many miRNAs or not consistent among studies due to limitations of methodologies and/or different conditions and sample sizes of cancer tissues and cell lines. In the current study, we used real time quantitative PCR, a more reliable detection method, to detect the expression levels of 95 cancer-related miRNAs in well controlled pancreatic cancer specimens and cell lines as well as pancreatitis tissues. This study may discover a unique miRNA profiling pattern for pancreatic cancer and identify important molecular targets for further functional investigations and for the developments of new diagnostic tools and therapeutic strategies.
Materials and methods
Cell cultures and tissue collections
Human pancreatic cancer cell lines, Panc-1, MIA PaCa-2, BxPC-3, Hs766T, ASPC-1, Capan-1, Capan-2, Panc3.27, HPAF-II, and PL45, were purchased from the American Type Culture Collection (ATCC, Rockville, MD). The human pancreatic ductal epithelium (HPDE) cells were provided as a generous gift from Dr. Ming-Sound Tsao [21,22]. All cells were cultured as previously described [23–26]. Human pancreatic adenocarcinoma specimens and their adjacent normal pancreatic tissues (17 pairs) and one pancreatic tissue sample with chronic pancreatitis were collected from patients who underwent surgery according to an approved human protocol at the Baylor College of Medicine (Houston, TX).
miRNA extraction and reverse-transcription
Total miRNAs of tissues and cultured cells were extracted and purified using mirVana miRNA Isolation kit (Applied Biosystems/Ambion, Austin, TX) following the manufacturer's instructions. Five µl of RNA was directly converted to cDNA with the QuantiMir™ RT System (SBI System Biosciences, Mountain View, CA).
Real time RT PCR
Differential expression of 95 miRNAs was analyzed by RT-PCR using the QuantiMir System (SBI System Biosciences). All 95 miRNAs chosen for the array are based on their potential roles in cancer, cell development and apoptosis. The array plate also included the U6 transcript as a normalization signal. The miRNA sequences and primer sequences used in RT-PCR were listed in Table 1. cDNAs from different cell lines and tissue samples were mixed with SYBR® Green Mastermix (Bio-Rad Laboratories, Hercules, CA) plus the universal reverse primer. Specific primers (1 µl) were added each well of the qPCR plate. Expression levels of each mature miRNA were evaluated using comparative threshold cycle (Ct) method as normalized to that of U6 (2−ΔCt). The fold change of each miRNA was calculated from the expression levels between tumor tissues/cells and normal tissues/cells.
Table 1.
Mature RNA sequences and real time PCR primers for 95 miRNAs.
| miRNA | MirBase # | miRNA Sequence(s) | RT-PCR Primer sequenc(s) |
|---|---|---|---|
| let-7-family | MIMAT0000062, MIMAT0000064, MIMAT0000065, MIMAT0000067 |
ugagguaguagguuguauaguu, ugagguaguagguuguaugguu, agagguaguagguugcauagu, ugagguaguagauuguauaguu |
tgaggtagtaggttgtatagtt, tgaggtagtaggttgtatggtt, agaggtagtaggttgcatagt, tgaggtagtagattgtatagtt |
| miR-7 | MIMAT0000252 | uggaagacuagugauuuuguug | tggaagactagtgattttgttg |
| miR-92 | MIMAT0000092 | uauugcacuugucccggccug | tattgcacttgtcccggcctg |
| miR-93 | MIMAT0000093 | aaagugcuguucgugcagguag | aaagtgctgttcgtgcaggtag |
| miR-9-1 | MIMAT0000441 | ucuuugguuaucuagcuguauga | tctttggttatctagctgtatga |
| miR-101-1 | MIMAT0000099 | uacaguacugugauaacugaag | tacagtactgtgataactgaag |
| miR-103 | MIMAT0000101 | agcagcauuguacagggcuauga | agcagcattgtacagggctatga |
| miR-106a | MIMAT0000103 | aaaagugcuuacagugcagguagc | aaaagtgcttacagtgcaggtagc |
| miR-106b | MIMAT0000680 | uaaagugcugacagugcagau | taaagtgctgacagtgcagat |
| miR-107 | MIMAT0000104 | agcagcauuguacagggcuauca | agcagcattgtacagggctatca |
| miR-10b | MIMAT0000254 | uacccuguagaaccgaauuugu | taccctgtagaaccgaatttgt |
| miR-1-1 | MIMAT0000416 | uggaauguaaagaaguaugua | tggaatgtaaagaagtatgta |
| miR-122a | MIMAT0000421 | uggagugugacaaugguguuugu | tggagtgtgacaatggtgtttgt |
| miR-125a | MIMAT0000443 | ucccugagacccuuuaaccugug | tccctgagaccctttaacctgtg |
| miR-125b | MIMAT0000423 | ucccugagacccuaacuuguga | tccctgagaccctaacttgtga |
| miR-126 | MIMAT0000444 | cauuauuacuuuugguacgcg | cattattacttttggtacgcg |
| miR-128b | MIMAT0000676 | ucacagugaaccggucucuuuc | tcacagtgaaccggtctctttc |
| miR-132 | MIMAT0000426 | uaacagucuacagccauggucg | taacagtctacagccatggtcg |
| miR-133a | MIMAT0000427 | uugguccccuucaaccagcugu | ttggtccccttcaaccagctgt |
| miR-134 | MIMAT0000447 | ugugacugguugaccagaggg | tgtgactggttgaccagaggg |
| miR-135b | MIMAT0000758 | uauggcuuuucauuccuaugug | tatggcttttcattcctatgtg |
| miR-136 | MIMAT0000448 | acuccauuuguuuugaugaugga | actccatttgttttgatgatgga |
| miR-137 | MIMAT0000429 | uauugcuuaagaauacgcguag | tattgcttaagaatacgcgtag |
| miR-140 | MIMAT0000431 | agugguuuuacccuaugguag | agtggttttaccctatggtag |
| miR-141 | MIMAT0000432 | uaacacugucugguaaagaugg | taacactgtctggtaaagatgg |
| miR-142-3p | MIMAT0000434 | uguaguguuuccuacuuuaugga | tgtagtgtttcctactttatgga |
| miR-143 | MIMAT0000435 | ugagaugaagcacuguagcuca | tgagatgaagcactgtagctca |
| miR-145 | MIMAT0000437 | guccaguuuucccaggaaucccuu | gtccagttttcccaggaatccctt |
| miR-146a | MIMAT0000449 | ugagaacugaauuccauggguu | tgagaactgaattccatgggtt |
| miR-149 | MIMAT0000450 | ucuggcuccgugucuucacucc | tctggctccgtgtcttcactcc |
| miR-150 | MIMAT0000451 | ucucccaacccuuguaccagug | tctcccaacccttgtaccagtg |
| miR-151 | MIMAT0000757 | acuagacugaagcuccuugagg | actagactgaagctccttgagg |
| miR-153 | MIMAT0000439 | uugcauagucacaaaaguga | ttgcatagtcacaaaagtga |
| miR-154 | MIMAT0000452 | uagguuauccguguugccuucg | taggttatccgtgttgccttcg |
| miR-155 | MIMAT0000646 | uuaaugcuaaucgugauagggg | ttaatgctaatcgtgatagggg |
| miR-15a | MIMAT0000068 | uagcagcacauaaugguuugug | tagcagcacataatggtttgtg |
| miR-15b | MIMAT0000417 | uagcagcacaucaugguuuaca | tagcagcacatcatggtttaca |
| miR-16 | MIMAT0000069 | uagcagcacguaaauauuggcg | tagcagcacgtaaatattggcg |
| miR-17-3p | MIMAT0000071 | acugcagugaaggcacuugu | actgcagtgaaggcacttgt |
| miR-17-5p | MIMAT0000070 | caaagugcuuacagugcagguagu | caaagtgcttacagtgcaggtagt |
| miR-181a | MIMAT0000256 | aacauucaacgcugucggugagu | aacattcaacgctgtcggtgagt |
| miR-181b | MIMAT0000257 | aacauucauugcugucgguggg | aacattcattgctgtcggtggg |
| miR-181c | MIMAT0000258 | aacauucaaccugucggugagu | aacattcaacctgtcggtgagt |
| miR-181d | MIMAT0002821 | aacauucauuguugucgguggguu | aacattcattgttgtcggtgggtt |
| miR-183 | MIMAT0000261 | uauggcacugguagaauucacug | tatggcactggtagaattcactg |
| miR-185 | MIMAT0000455 | uggagagaaaggcaguuc | tggagagaaaggcagttc |
| miR-186 | MIMAT0000456 | caaagaauucuccuuuugggcuu | caaagaattctccttttgggctt |
| miR-188 | MIMAT0000457 | caucccuugcaugguggagggu | catcccttgcatggtggagggt |
| miR-18a | MIMAT0000072 | uaaggugcaucuagugcagaua | taaggtgcatctagtgcagata |
| miR-190 | MIMAT0000458 | ugauauguuugauauauuaggu | tgatatgtttgatatattaggt |
| miR-191 | MIMAT0000440 | caacggaaucccaaaagcagcu | caacggaatcccaaaagcagct |
| miR-192 | MIMAT0000222 | cugaccuaugaauugacagcc | ctgacctatgaattgacagcc |
| miR-194 | MIMAT0000460 | uguaacagcaacuccaugugga | tgtaacagcaactccatgtgga |
| miR-195 | MIMAT0000461 | uagcagcacagaaauauuggc | tagcagcacagaaatattggc |
| miR-196a | MIMAT0000226 | uagguaguuucauguuguugg | taggtagtttcatgttgttgg |
| miR-197 | MIMAT0000227 | uucaccaccuucuccacccagc | ttcaccaccttctccacccagc |
| miR-198 | MIMAT0000228 | gguccagaggggagauagg | ggtccagaggggagatagg |
|
miR- 199a+b |
MIMAT0000231, MIMAT0000263 |
cccaguguucagacuaccuguuc, cccaguguuuagacuaucuguuc |
cccagtgttcagactacctgttc, cccagtgtttagactatctgttc |
| miR-30b | MIMAT0000420 | uguaaacauccuacacucagcu | tgtaaacatcctacactcagct |
| miR-19a+b | MIMAT0000073, MIMAT0000074 |
ugugcaaaucuaugcaaaacuga, ugugcaaauccaugcaaaacuga |
tgtgcaaatctatgcaaaactga, tgtgcaaatccatgcaaaactga |
| miR-95 | MIMAT0000094 | uucaacggguauuuauugagca | ttcaacgggtatttattgagca |
| miR-20a | MIMAT0000075 | uaaagugcuuauagugcagguag | taaagtgcttatagtgcaggtag |
| miR-200a | MIMAT0000682 | uaacacugucugguaacgaugu | taacactgtctggtaacgatgt |
| miR-200b | MIMAT0000318 | uaauacugccugguaaugaugac | taatactgcctggtaatgatgac |
| miR-200c | MIMAT0000617 | uaauacugccggguaaugaugg | taatactgccgggtaatgatgg |
| miR-202 | MIMAT0002811 | agagguauagggcaugggaaaa | agaggtatagggcatgggaaaa |
| miR-203 | MIMAT0000264 | gugaaauguuuaggaccacuag | gtgaaatgtttaggaccactag |
| miR-204 | MIMAT0000265 | uucccuuugucauccuaugccu | ttccctttgtcatcctatgcct |
| miR-205 | MIMAT0000266 | uccuucauuccaccggagucug | tccttcattccaccggagtctg |
| miR-206 | MIMAT0000462 | uggaauguaaggaagugugugg | tggaatgtaaggaagtgtgtgg |
| miR-21 | MIMAT0000076 | uagcuuaucagacugauguuga | tagcttatcagactgatgttga |
| miR-210 | MIMAT0000267 | cugugcgugugacagcggcuga | ctgtgcgtgtgacagcggctga |
| miR-214 | MIMAT0000271 | acagcaggcacagacaggcag | acagcaggcacagacaggcag |
| miR-215 | MIMAT0000272 | augaccuaugaauugacagac | atgacctatgaattgacagac |
| miR-372 | MIMAT0000724 | aaagugcugcgacauuugagcgu | aaagtgctgcgacatttgagcgt |
| miR-373 | MIMAT0000726 | gaagugcuucgauuuuggggugu | gaagtgcttcgattttggggtgt |
| miR-218 | MIMAT0000275 | uugugcuugaucuaaccaugu | ttgtgcttgatctaaccatgt |
| miR-219 | MIMAT0000276 | ugauuguccaaacgcaauucu | tgattgtccaaacgcaattct |
| miR-22 | MIMAT0000077 | aagcugccaguugaagaacugu | aagctgccagttgaagaactgt |
| miR-488 | MIMAT0002804 | cccagauaauggcacucucaa | cccagataatggcactctcaa |
| miR-221 | MIMAT0000278 | agcuacauugucugcuggguuuc | agctacattgtctgctgggtttc |
| miR-222 | MIMAT0000279 | agcuacaucuggcuacugggucuc | agctacatctggctactgggtctc |
| miR-223 | MIMAT0000280 | ugucaguuugucaaauacccc | tgtcagtttgtcaaatacccc |
| miR-224 | MIMAT0000281 | caagucacuagugguuccguuua | caagtcactagtggttccgttta |
| miR-23a | MIMAT0000078 | aucacauugccagggauuucc | atcacattgccagggatttcc |
| miR-24 | MIMAT0000080 | uggcucaguucagcaggaacag | tggctcagttcagcaggaacag |
| miR-25 | MIMAT0000081 | cauugcacuugucucggucuga | cattgcacttgtctcggtctga |
| miR-26a | MIMAT0000082 | uucaaguaauccaggauaggc | ttcaagtaatccaggataggc |
| miR-26b | MIMAT0000083 | uucaaguaauucaggauagguu | ttcaagtaattcaggataggtt |
| miR-27a+b | MIMAT0000084, MIMAT0000419 |
uucacaguggcuaaguuccgc, uucacaguggcuaaguucugc |
ttcacagtggctaagttccgc, ttcacagtggctaagttctgc |
| miR-30c | MIMAT0000244 | uguaaacauccuacacucucagc | tgtaaacatcctacactctcagc |
|
miR- 29a+b+c |
MIMAT0000086, MIMAT0000100, MIMAT0000681 |
uagcaccaucugaaaucgguu, uagcaccauuugaaaucaguguu, uagcaccauuugaaaucggu |
tagcaccatctgaaatcggtt, tagcaccatttgaaatcagtgtt, tagcaccatttgaaatcggt |
| miR-30a-3p | MIMAT0000088 | cuuucagucggauguuugcagc | ctttcagtcggatgtttgcagc |
| miR-30a-5p | MIMAT0000087 | uguaaacauccucgacuggaag | tgtaaacatcctcgactggaag |
| miR-296 | MIMAT0000690 | agggcccccccucaauccugu | agggccccccctcaatcctgt |
| U6 snRNA | NCBI: X07425.1 | caccacguuuauacgccggug | caccacgtttatacgccggtg |
Statistical analysis
The expressions of 8 miRNAs in cancer tissues or cells and normal tissues or cells were compared with paired Student’s t-test. Data are presented as means ± standard deviation (SD). A p value less than 0.05 was considered statistically significant.
Results
The expression of 95 miRNAs in chronic pancreatitis, pancreatic cancer cell lines and surgical specimens
Initially, the expression of 95 miRNAs in 1 pancreatitis tissue, 5 pancreatic cancer tissues and their adjacent benign tissues, 3 human pancreatic cancer cell lines (MIA PaCa-2, Panc-1 and BxPC-3) and HPDE cells was determined by real-time PCR. After normalization to the control U6 expression, the differential expression of miRNAs of pancreatitis tissue compared with normal pancreatic tissues, pancreatic cancer tissues compared with normal pancreatic tissues, and pancreatic cancer cell lines compared with HPDE cells was determined and shown in Fig. 1 and Table 2. Substantial differences of the expression profile of 95 mRNAs were observed between cancer and normal tissues or between cancer cell lines and normal HPED cells at the individual basis, indicating potential roles of miRNAs in the cancer formation. These differences indicate the individual characteristics and variability of each case compared other cases. The relative expression values for these mature miRNAs spanned 6-logs (from 0.01 to 10000). A number of miRNAs were increased in the most of pancreatic cancer tissues and cell types, but not in normal tissues and cells as well as the pancreatitis sample.
Fig. 1.
The expression pattern of 95 miRNAs in chronic pancreatitis, pancreatic cancer cell lines and surgical specimens. MiRNAs of tissues and cultured cells were extracted and purified using mirVana miRNA Isolation kit and converted to cDNAs with the QuantiMir™ RT System. Differential expression was analyzed by RT-PCR using QuantiMir 95 microRNAs array System. U6 primer was also included in the array as a normalization control. After normalizing to the control U6 in all samples, the fold change in 95 miRNAs was calculated by comparing the pancreatic cancer tissue or cell lines with normal pancreatic tissues or HPDE cells. A. Chronic pancreatitis versus normal pancreatic tissue (n=1). B. Pancreatic cancer cell lines versus HPDE cells (n=3). C. Surgical specimens of pancreatic cancer tissues versus their adjacent normal pancreatic tissues (n=5).
Table 2.
The expression of 95 miRNAs in chronic pancreatitis, pancreatic cancer cell lines* and surgical specimens**
| miRNA | P2 | MIA | Panc-1 | BxPC-3 | T2 | T7 | T22 | T33 | T35 |
|---|---|---|---|---|---|---|---|---|---|
| let-7-family | 0.476319 | 0.3737123 | 1.494849 | 1.328686 | 0.186856 | 0.707107 | 9.3178687 | 146.0178 | 106.89125 |
| miR-7 | 0.065154 | 0.6328783 | 0.432269 | 5.098243 | 0.151774 | 1.156688 | 39.396621 | 0.210224 | 1.1095695 |
| miR-92 | 0.721965 | 0.4537596 | 1.292353 | 2.789487 | 0.339151 | 2.158456 | 0.5212329 | 4.287094 | 6.2333166 |
| miR-93 | 1.827663 | 1.3755418 | 3.24901 | 4.40762 | 4.69134 | 2.948538 | 6.6345564 | 58.89201 | 10.556063 |
| miR-9-1 | 1.569168 | 0.0245183 | 1.717131 | 0.008201 | 0.358489 | 0.82932 | 0.1780063 | 6.233317 | 51.268472 |
| miR-101-1 | 3.226567 | 0.9930925 | 0.823591 | 0.835088 | 1.265757 | 1.337928 | 0.4863275 | 1.148698 | 1.4948492 |
| miR-103 | 3.5801 | 1.4742692 | 3.89062 | 6.276673 | 4.316913 | 1.853176 | 2.9690471 | 11.87619 | 3.810552 |
| miR-106a | 2.514027 | 0.7120251 | 1.569168 | 2.828427 | 2.297397 | 3.317278 | 21.406841 | 27.47409 | 12.996038 |
| miR-106b | 2.42839 | 1.2483305 | 3.031433 | 3.506423 | 3.630077 | 1.337928 | 0.8010699 | 2.86791 | 0.463294 |
| miR-107 | 3.810552 | 1.6245048 | 4.823231 | 7.674113 | 4.027822 | 1.79005 | 2.5315132 | 14.82541 | 5.464161 |
| miR-10b | 3.363586 | 6.7739625 | 153.2773 | 10.33882 | 1.777685 | 5.063026 | 3.732132 | 95.00951 | 10.126053 |
| miR-1-1 | 17.87659 | ∞ | 11268.44 | 2.657372 | 10.26741 | 165.4212 | 0.016176 | 286.0255 | 28724.616 |
| miR-122a | 0.19751 | 0.2793218 | 1.347234 | 1.494849 | 0.047696 | 1.918528 | ∞ | 3.680751 | 3.4822023 |
| miR-125a | 0.784584 | 0.8408964 | 4.756828 | 4.594793 | 1.265757 | 0.632878 | 2.4794154 | 70.0348 | 36.758347 |
| miR-125b | 1.905276 | 0.1088188 | 4.198867 | 2.770219 | 2.80889 | 0.907519 | 33.128478 | 11.00433 | 12.295001 |
| miR-126 | 2.514027 | 3.4822023 | 6.868523 | 3.458149 | 0.126745 | 0.946058 | 100.42676 | 4.112455 | 5.5021673 |
| miR-128b | 0.432269 | 2.3949574 | 8.339726 | 2.989698 | 0.876606 | 2.751084 | 0.283221 | 2.17347 | 4.5630549 |
| miR-132 | 2.639016 | 0.3977682 | 1.729074 | 5.61778 | 0.61132 | 0.707107 | 0.3391511 | 3.732132 | 1.9724654 |
| miR-133a | 1.385109 | 0.5864175 | 3.758091 | 1.840375 | 1.827663 | 2.07053 | 9.9176616 | 0.959264 | 0.4506252 |
| miR-134 | 0.316439 | 0.5358867 | 7.110741 | 2.462289 | 0.211686 | 0.578344 | 2.1584565 | 2.80889 | 0.5471469 |
| miR-135b | 1.375542 | 1.2570134 | 0.384219 | 1.265757 | 9.781122 | 7.012846 | 0.3321715 | 11.23556 | 37.271475 |
| miR-136 | 0.80107 | 1.1647336 | 23.58831 | 3.052518 | 0.225313 | 0.41466 | 0.0133224 | 0.406126 | ∞ |
| miR-137 | 0.042689 | 23.917588 | 0.503478 | 0.795536 | 0.570382 | 1.658639 | 0.0025772 | 1.140764 | 261.3791 |
| miR-140 | 2.234574 | 2.0562277 | 3.160165 | 0.628507 | 5.540438 | 0.993092 | 0.0066612 | 14.92853 | 4.9588308 |
| miR-141 | 0.246558 | 0.0113592 | 0.028756 | 1.375542 | 0.493116 | 0.309927 | 0.6285067 | 0.323088 | 0.4413515 |
| miR-142-3p | 45.25483 | 0.2812646 | 0.406126 | 1.205808 | 19.83532 | 2.657372 | 0.0418102 | 10.12605 | 19.835323 |
| miR-143 | 3.917681 | 0.5212329 | 1.214195 | 1.526259 | 13.73705 | 8.168097 | 3.4105396 | 23.75238 | 22.943284 |
| miR-145 | 10.48315 | 0.289172 | 1.319508 | 1.231144 | 10.12605 | 4.316913 | 2.6390158 | 67.64915 | 38.054628 |
| miR-146a | 16.56424 | 0.1486509 | 2.013911 | 1.958841 | 3.271608 | 1.214195 | 11.551434 | 24.93327 | 31.77896 |
| miR-149 | 1.356604 | 2.250117 | 6.773962 | 5.028053 | 0.376312 | 1.057018 | 2.0139111 | 5.169411 | 0.1582196 |
| miR-150 | 15.03236 | 0.1396609 | 0.63728 | 6.408559 | 0.784584 | 1.717131 | 0.4444213 | 17.14838 | 5.5789747 |
| miR-151 | 0.806642 | 0.9726549 | 4.469149 | 3.97237 | 0.441351 | 0.920188 | 0.0674518 | 3.07375 | ∞ |
| miR-153 | 0.473029 | 25.457167 | 18.12614 | 0.790041 | 0.144586 | 1.580083 | 0.0083732 | 0.687771 | ∞ |
| miR-154 | 0.673617 | 1.0352649 | 5.278032 | 2.969047 | 0.395021 | 0.554785 | 0.7219646 | 4.112455 | 0.4383029 |
| miR-155 | 15.03236 | 0.0060872 | 0.395021 | 0.835088 | 2.158456 | 2.732081 | 5.6177795 | 52.34573 | 48.840295 |
| miR-15a | 1.375542 | 2.6026837 | 4.789915 | 5.426417 | 1.802501 | 0.870551 | 18.507011 | 5.979397 | 7.674113 |
| miR-15b | 0.90125 | 1.5583292 | 5.388934 | 4 | 0.395021 | 1.547565 | 8.6338259 | 29.04061 | 28.442966 |
| miR-16 | 0.403321 | 3.6807506 | 6.020987 | 4.563055 | 0.411796 | 0.721965 | 27.09585 | 7.568461 | 4.4382779 |
| miR-17-3p | 1.729074 | 0.4537596 | 1.729074 | 2.828427 | 0.566442 | 1.729074 | 1.0352649 | 1.866066 | 1.1095695 |
| miR-17-5p | 1.301342 | 0.8066418 | 1.591073 | 2.751084 | 2.281527 | 3.482202 | 7.061624 | 20.11221 | 13.269113 |
| miR-181a | 0.463294 | 0.2660925 | 0.532185 | 0.876606 | 0.535887 | 0.320856 | 1.3286858 | 2.549121 | 7.1107414 |
| miR-181b | 1.071773 | 0.2016604 | 1.717131 | 3.271608 | 0.888843 | 0.632878 | 18.635737 | 10.62949 | 12.728584 |
| miR-181c | 0.450625 | 0.2284579 | 0.607097 | 4.346939 | 0.316439 | 3.317278 | 2.5847057 | 6.868523 | 6.4531341 |
| miR-181d | 1.257013 | 0.25 | 2.12874 | 3.5801 | 1.802501 | 0.632878 | 2.8088898 | 21.70567 | 11.235559 |
| miR-183 | 0.05366 | 1.5052467 | 6.821079 | 8.168097 | 0.075363 | 0.913831 | 0.1111053 | 6.19026 | 6.1475007 |
| miR-185 | 2.34567 | 0.7169776 | 2.828427 | 2.969047 | 4.626753 | 2.313376 | 0.3634931 | 13.73705 | 2.6390158 |
| miR-186 | 0.97942 | 2.907945 | 7.727491 | 9.38268 | 2.361985 | 1.109569 | ∞ | 3.09513 | 1.4640857 |
| miR-188 | 0.539614 | 0.7474246 | 2.445281 | 3.97237 | 0.10083 | 0.779165 | 14.928528 | 0.246558 | 0.4600938 |
| miR-18a | 1.385109 | 0.4569157 | 0.496546 | 1.515717 | 5.61778 | 3.863745 | 0.2973018 | 20.25211 | 18.126142 |
| miR-190 | 0.659754 | 1.1647336 | 5.35171 | 25.63424 | 7.835362 | 3.294364 | 0.1907824 | ∞ | ∞ |
| miR-191 | 2.12874 | 1.9724654 | 22.94328 | 7.78124 | 1.164734 | 1.086735 | 49.52208 | 15.13692 | 5.5021673 |
| miR-192 | 0.554785 | 0.659754 | 0.986233 | 2.297397 | 0.363493 | 4.257481 | 781.44471 | 1.79005 | 0.0973956 |
| miR-194 | 1.021012 | 0.4796321 | 0.946058 | 1.42405 | 3.182146 | 6.773962 | 55.330383 | 2.828427 | 0.1111053 |
| miR-195 | 0.664343 | 3.2716082 | 5.464161 | 3.837056 | 0.528509 | 0.716978 | 1.2657566 | 9.57983 | 10.777869 |
| miR-196a | 0.301452 | 1.8150383 | 349.7063 | 21.85664 | 27.09585 | 168.897 | 2.0139111 | 5.314743 | 152.21851 |
| miR-197 | 1.071773 | 1.0210121 | 2.361985 | 3.758091 | 0.732043 | 1.474269 | 0.6643429 | 4.112455 | 8.1116758 |
| miR-198 | 2.042024 | 0.2414841 | 0.049037 | 0.198884 | 1.474269 | 1.753211 | 0.1755556 | 1.591073 | 2.7894873 |
| miR-199a+b | 16.56424 | 0.6925547 | 3.458149 | 2.732081 | 18.50701 | 0.888843 | 3.0314331 | 51.62507 | 33.824577 |
| miR-30b | 1.101905 | 1.8150383 | 5.696201 | 2.969047 | 0.578344 | 0.482968 | 8.8152409 | 18.50701 | 10.852835 |
| miR-19a+b | 1.70527 | 0.6029039 | 0.737135 | 0.913831 | 0.297302 | 1.94531 | 4.3771748 | 1.071773 | 1.3472336 |
| miR-95 | 0.065154 | 6.1050368 | 4.027822 | 2.013911 | 0.175556 | 2.042024 | 58.89201 | 40.22443 | 24.933267 |
| miR-20a | 0.959264 | 0.5904963 | 1.028114 | 2.188587 | 2.158456 | 2.989698 | 26.354913 | 18.76536 | 14.723002 |
| miR-200a | 0.486327 | 0.0079767 | 0.986233 | 3.41054 | 0.993092 | 1.866066 | 2.8878584 | 1.239708 | 2.2815274 |
| miR-200b | 0.325335 | 0.017337 | 0.052193 | 4.316913 | 0.180491 | 1.958841 | 464.64981 | 1.515717 | 5.8158901 |
| miR-200c | 0.153893 | 0.0044253 | 0.008669 | 3.07375 | 0.060371 | 0.441351 | 41.642939 | 0.835088 | 0.9265881 |
| miR-202 | 0.835088 | 0.6285067 | 0.598739 | 1.042466 | 0.204476 | 1.148698 | 12.906268 | 1.028114 | 0.6551967 |
| miR-203 | 0.895025 | 0.0915054 | 0.05672 | 6.868523 | 2.12874 | 16.67945 | 0.0429857 | 52.34573 | 33.590934 |
| miR-204 | 0.11908 | 0.417544 | 1.189207 | 1.101905 | 0.063813 | 0.208772 | 0.5823668 | 3.89062 | 2.0849315 |
| miR-205 | 0.716978 | 0.0024381 | 0.004809 | 2.828427 | 0.334482 | 0.80107 | 0.7071068 | 2.496661 | 268.72747 |
| miR-206 | 0.80107 | 0.5946036 | 2.084932 | 7.210004 | 0.139661 | 1.109569 | 0.1425955 | 0.926588 | 0.952638 |
| miR-21 | 3.732132 | 0.6198538 | 2.789487 | 1.753211 | 2.013911 | 1.958841 | 6888.6234 | 27.66519 | 256 |
| miR-210 | 2.620787 | 2.1584565 | 2.80889 | 3.160165 | 2.17347 | 1.972465 | 0.5 | 8.633826 | 16 |
| miR-214 | 2.86791 | 3.1166583 | 1.658639 | 2.265768 | 4.563055 | 0.732043 | 16 | 2.770219 | 3.09513 |
| miR-215 | 0.353553 | 0.7900413 | ∞ | 2.188587 | 0.514057 | 4.890561 | 2005.8528 | 3.41054 | 0.208772 |
| miR-372 | 0.264255 | 0.6029039 | 1.385109 | 2.042024 | 0.125869 | 0.668964 | 0.2448551 | 1.70527 | 1.6586391 |
| miR-373 | 0.264255 | 1.1809927 | 3.031433 | 2.188587 | 0.153893 | 0.939523 | 0.060371 | 0.687771 | 0.2102241 |
| miR-218 | 2.114036 | 0.0112028 | 1.079228 | 5.314743 | 0.594604 | 0.747425 | 0.0824692 | 11.31371 | 16 |
| miR-219 | 0 | 1.1647336 | 2.751084 | 1.972465 | 0.134904 | 0.678302 | 0.1073207 | 1.231144 | 0.5034778 |
| miR-22 | 0.993092 | 0.4263174 | 1.347234 | 1.879045 | 3.138336 | 0.582367 | 1.8531761 | 3.863745 | 2.6026837 |
| miR-488 | 0.403321 | 0.5509526 | 1.515717 | 2.639016 | 0.102238 | 0.011518 | 0.8585654 | 0.303549 | 0.0940779 |
| miR-221 | 2.602684 | 3.6553258 | 3.340352 | 6.680703 | 9.12611 | 2.07053 | 0.3977682 | 28.05138 | 19.159659 |
| miR-222 | 1.972465 | 2.2657678 | 1.765406 | 3.758091 | 3.271608 | 3.630077 | 0.2016604 | 43.71329 | 54.1917 |
| miR-223 | 4.14106 | 0.3535534 | 2.566852 | 10.05611 | 4.723971 | 4.469149 | 79.341293 | 13.54792 | 13.737047 |
| miR-224 | 1.148698 | 0.0021373 | 0.003262 | 2.86791 | 1.36604 | 6.868523 | 14.025692 | 168.897 | 45.886568 |
| miR-23a | 0.655197 | 0.8408964 | 4.626753 | 4.169863 | 0.241484 | 1.569168 | ∞ | 101.1253 | 95.670352 |
| miR-24 | 0.702222 | 0.8293195 | 3.944931 | 3.605002 | 3.031433 | 1.840375 | 62.682899 | 13.8326 | 4.6913398 |
| miR-25 | 0.707107 | 0.7791646 | 1.815038 | 2.445281 | 0.279322 | 1.453973 | 52.709825 | 48.50293 | 12.295001 |
| miR-26a | 0.479632 | 0.8293195 | 3.434262 | 4.531536 | 0.29937 | 0.61132 | 22.161751 | 26.72281 | 23.26356 |
| miR-26b | 0.586417 | 1.0717735 | 6.498019 | 5.205367 | 0.325335 | 1.042466 | 29.040613 | 56.49299 | 43.411338 |
| miR-27a+b | 0.496546 | 0.6417129 | 3.605002 | 3.732132 | 1.494849 | 1.214195 | 19.027314 | 24.42015 | 9.3178687 |
| miR-30c | 0.450625 | 1.591073 | 5.426417 | 1.931873 | 0.539614 | 0.423373 | 37.530718 | 14.02569 | 8.168097 |
| miR-29a+b+c | 1.972465 | 0.8408964 | 1.729074 | 0.993092 | 0.566442 | 1.22264 | 704.27741 | 3.5801 | 8.3977335 |
| miR-30a-3p | 0.140632 | 1.4948492 | 5.540438 | 1.879045 | 0.065154 | 0.376312 | 15.242208 | 2.188587 | 1.1647336 |
| miR-30a-5p | 0.273573 | 2.2657678 | 5.314743 | 3.706352 | 0.389582 | 0.435275 | 2.3133764 | 1.375542 | 0.528509 |
| miR-296 | 0.539614 | 0.1103379 | 0.432269 | 1.310393 | 1.013959 | 1.505247 | 1.4539725 | 5.028053 | 1.4640857 |
Chronic pancreatitis tissue (P2). Pancreatic cancer cell lines (MIA-CaPa2, Panc-1 and BxPC-3). Surgical specimens of pancreatic cancer tissues (T2, T7, T22, T33 and T35).
the miRNA was increased in cancer tissues or cells while normal controls had no expression. The expression of all miRNAs was normalized to the U6 level in all tissue samples and cell types.
Compared with the relatively normal human pancreatic ductal epithelium (HPDE).
compared with the relatively normal pancreatic tissues.
Validation of eight over-expressed miRNAs in more pancreatic cancer cell lines and surgical specimens
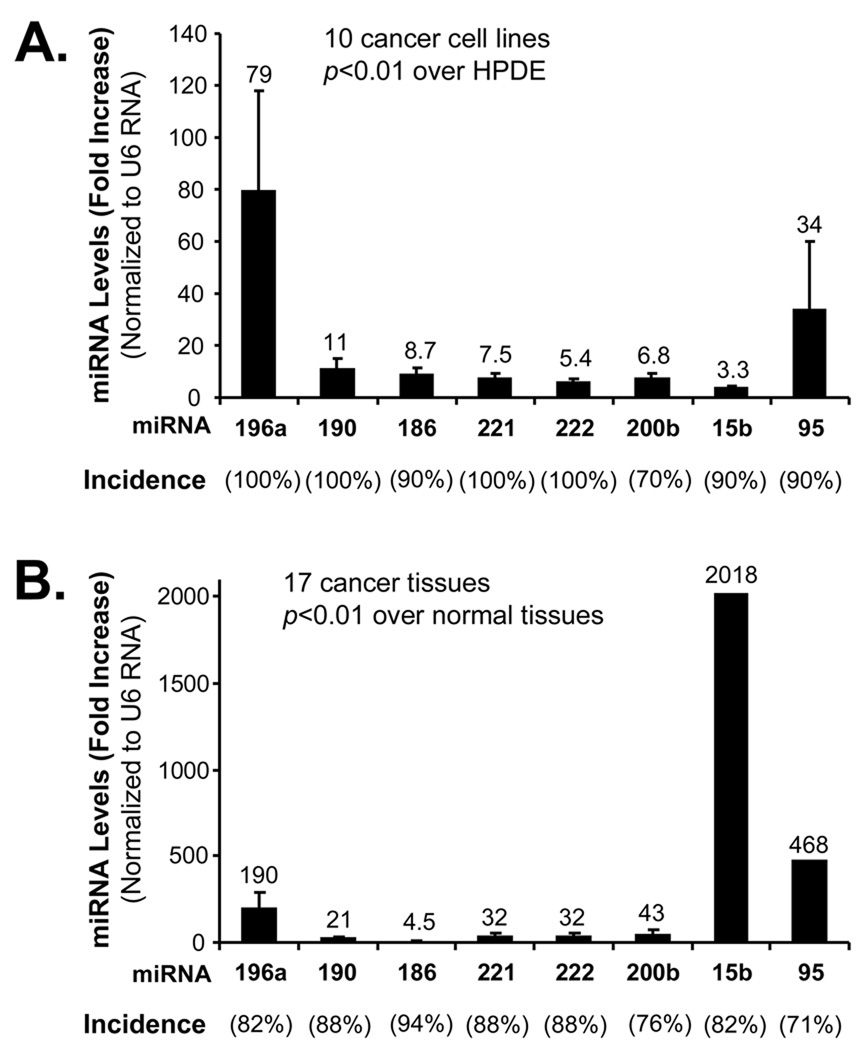
From 95 miRNAs, 8 miRNAs (miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b and miR-95) were identified to have high expression levels more than 3.3-fold both in pancreatic cancer tissue samples and cell lines compared with that in normal pancreatic tissues and HPED cells. The expression of these miRNAs was further analyzed in more samples of pancreatic cancer and normal pancreatic tissue pairs (n=17) as well as more pancreatic cancer cell lines (n=10) by real time PCR. The incidence of expression increase and average fold increase of 8 miRNAs were shown in Fig. 2 and Table 3. Compared with normal HPDE cells, the incidence of 10 pancreatic cancer cell lines exhibited elevated levels of miR-196a (100%), miR-190 (100%), miR-186(90%), miR-221(100%), miR-222 (100%), miR-200b (70%), miR-15b (90%) and miR-95 (90%) and the increase levels ranged from 3.3 to 79 fold (P < 0.01, n=10, Fig. 2A). For the pancreatic cancer tissues compared with normal pancreatic tissues, the expression increases (incidence and fold increase) of miR-196a (82% and 190), miR-190 (88% and 21), miR-186 (94% and 4.5), miR-221 (88% and 32), miR-222 (88% and 32), miR-200b (76% and 43), miR-15b (82% and 2018) and miR-95 (71% and 468) were also observed (P <0.01, n=17, Fig. 2B). These data indicate that these miRNAs may share common pathways in the pancreatic cancer pathogenesis.
Fig. 2.
The expression of 8 miRNAs in more pancreatic cancer cell lines and surgical specimens. Expression of 8 miRNAs (miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b and miR-95) selected from the 95 miRNAs was determined in 10 pancreatic cancer cell lines and 17 pairs of pancreatic cancer tissues and their adjacent normal pancreatic tissues. A. The expression of 8 miRNAs were significantly increased in pancreatic cancer cell lines compared with HPDE cells (n=10, P < 0.01). B. The expression of 8 miRNAs were significantly increased in pancreatic cancer tissues compared with their adjacent normal pancreatic cancer tissues (n=17, P <0.01).
Table 3.
| Sample | miR-196a | miR-190 | miR-186 | miR-221 | miR-222 | miR-200b | miR-15b | miR-95 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | P2 | 0.301452 | 0.659754 | 0.97942 | 2.602684 | 1.972465 | 0.325335 | 0.90125 | 0.065154 |
| 2 | Panc-1 | 349.7063 | 5.35171 | 7.727491 | 3.340352 | 1.765406 | 0.052193 | 5.388934 | 4.027822 |
| 3 | MIA | 1.815038 | 1.164734 | 2.907945 | 3.655326 | 2.265768 | 0.017337 | 1.558329 | 6.105037 |
| 4 | BxPC-3 | 21.85664 | 25.63424 | 9.38268 | 6.680703 | 3.758091 | 4.316913 | 4 | 2.013911 |
| 5 | ASPC-1 | 234.753 | ∞ | 21.18542 | 19.63016 | 13.54792 | 21.55574 | 11.75335 | 273.4247 |
| 6 | Capan1 | 13.68952 | ∞ | 4.228072 | 2.505329 | 1.802501 | 8.724062 | 0.737135 | 11.08088 |
| 7 | Capan2 | 7.43844 | ∞ | 1.918528 | 7.361501 | 7.210004 | 11.3924 | 1.840375 | 9.849155 |
| 8 | Panc3.27 | 9.849155 | ∞ | 1.328686 | 2.799172 | 1.898684 | 1.765406 | 1.068065 | 0.747425 |
| 9 | Hs766T | 36.12686 | ∞ | 27.28432 | 14.17228 | 10.81529 | 0.031577 | 2.838247 | 16.39291 |
| 10 | PL45 | 2.666597 | ∞ | 0.70466 | 2.694467 | 2.099433 | 1.252664 | 1.630145 | 4.084049 |
| 11 | HPAFII | 117.784 | ∞ | 10.30305 | 11.71269 | 8.969329 | 19.15966 | 1.879045 | 8.724062 |
| 12 | T7 | 168.897 | 3.294364 | 1.109569 | 2.07053 | 3.630077 | 1.958841 | 1.547565 | 2.042024 |
| 13 | T13 | 648.0674 | 21.63057 | 6.680703 | 17.75311 | 21.70567 | 9.000468 | 15.03236 | 7.412704 |
| 14 | T18 | 0.353553 | 0.503478 | 0.018517 | 25.45717 | 0.63728 | 10.30305 | 261.3791 | 61.60604 |
| 15 | T19 | 1.094294 | 4.773343 | 1.500039 | 1.69937 | 2.313376 | 3.680751 | 9.986644 | 9.094536 |
| 16 | T22 | 2.013911 | 0.190782 | ∞ | 0.397768 | 0.20166 | 464.6498 | 8.633826 | 58.89201 |
| 17 | T29 | 1.125058 | 4.890561 | 2.020903 | 3.271608 | 2.531513 | 1.01748 | 1.261377 | 1.049717 |
| 19 | T31 | 101.8287 | 8.574188 | 1.287882 | 1.986185 | 2.938337 | 0.784584 | 4.789915 | 1.248331 |
| 20 | T32 | 7.727491 | 6.083915 | 1.168777 | 1.419123 | 1.404445 | 1.132884 | 1.314943 | 0.787308 |
| 21 | T33 | 5.314743 | ∞ | 3.09513 | 28.05138 | 43.71329 | 1.515717 | 29.04061 | 40.22443 |
| 22 | T34 | 0.809442 | 6.940309 | 7.412704 | 21.03908 | 6.84476 | 6.750526 | 3.193194 | 31.45026 |
| 23 | T35 | 152.2185 | ∞ | 1.464086 | 19.15966 | 54.1917 | 5.81589 | 28.44297 | 24.93327 |
| 24 | T36 | 11.31371 | 174.8532 | 8.196455 | 32.1111 | 22.08508 | 1.086735 | 62.03455 | 19.7667 |
| 25 | T37 | 140.0696 | 32.67239 | 18.37917 | 76.63864 | 325.1587 | 61.1805 | 68.35619 | 21.78103 |
| 26 | T38 | 1557.482 | ∞ | 11.91742 | 292.0355 | 40.64483 | 150.6441 | 33806.19 | 7669.942 |
| 27 | T39 | 2.938337 | 1.433955 | 1.172835 | 0.760489 | 1.132884 | 1.85961 | 0.750019 | 0.78187 |
| 28 | T40 | 401.7071 | 21.48116 | 3.797368 | 7.210004 | 8.969329 | 9.679953 | 0.604997 | 0.982821 |
| 29 | T2 | 27.0958 | 7.8356 | 2.362 | 9.1261 | 3.2716 | 0.1805 | 0.395 | 0.1756 |
Chronic pancreatitis tissue (row 1). Pancreatic cancer cell lines (rows 2–11). Surgical specimens of pancreatic cancer tissues (rows 12–29).
the miRNA was increased in cancer tissues or cells while normal controls had no expression. The expression of all miRNAs was normalized to the U6 level in all tissue samples and cell types.
Compared with the relatively normal human pancreatic ductal epithelium (HPDE).
compared with the relatively normal pancreatic tissues.
Discussion
In the current study, a unique 95 miRNA expression profile was observed in human pancreatic cancer tissues and cell lines, and eight miRNAs (miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b and miR-95) were significantly increased in the most of pancreatic cancer tissues and cell lines compared with normal pancreatic tissues and cells. Many of these miRNAs have not been reported in pancreatic cancer. This study provides new opportunities for studying novel molecular pathways of pancreatic cancer pathogenesis and for developing new strategies for pancreatic cancer diagnosis and treatment.
The mechanism of action of a specific miRNA is usually involved in its nucleotide complementary pairing to the 3' UTR of its specific targeting mRNAs, primarily functioning as a negative regulator by repressing target mRNA translation. miRNAs may directly regulate tissue or organ development and cell differentiation as well as maintain normal functions of many organ systems [27]. The alterations in miRNA expression may play an important role in many diseases including pancreatic cancer formation. Using the QuantiMir™ RT Kit, we tagged and converted mature miRNAs into detectable and quantifiable cDNAs. We used a highly sensitive real time PCR analysis to profile 95 cancer-related miRNAs. This method is more reliable and accurate for detection of miRNA expression and has much less technical noise, but greater reproducibility than traditional cDNA microarray or northern blot analysis. All 95 miRNAs chosen for the array have functional implications with regard to their potential roles in cancer, cell development and apoptosis. Our expression profiling data indicate a large number of miRNAs that are aberrantly expressed in pancreatic cancer tissues and cell lines compared with normal pancreatic tissues and cells. From these profiling data, we observed a diversity nature of miRNA expression among individual pancreatic cancer tissues or cells, which may support the concept of personalized medicine in care of these patients. However, we also observed the expression pattern of many miRNAs was reserved in the most pancreatic cancer tissues and cell lines studied in the current study. For example, 8 miRNAs (miR-196a, miR-190, miR-221, miR-222, miR-200b, miR-15b and miR-95) were consistently increased in the majority of pancreatic cancer tissues and cell lines. These data indicate that pancreatic cancer may share some common pathways for cancer pathogenesis by regulation of miRNAs. Many of these miRNAs have not been reported before in pancreatic cancer and their biological functions are largely unknown in pancreatic cancer pathogenesis.
Bloomston et al. reported that the high expression of miR-196a-2 was found to predict poor survival in pancreatic cancer patients [16]. miR-196a involves organ development by negatively regulating Hoxb8 [28]. miR-190 was found to be upregulated in human hepatocellular carcinomas [7]. The miR-200 family has been shown to regulate epithelial to mesenchymal transition (EMT) by targeting ZEB1 and SIP1. However, miR-200b was markedly downregulated in cells that had undergone EMT in response to transforming growth factor (TGF)-beta or to ectopic expression of the protein tyrosine phosphatase Pez [29]. Over-expression of miR-15b sensitized human gastric cancer cells to anticancer drugs by targeting BCL2 [30]. Inhibition of miR-95 decreased cell growth in HeLa cells [31]. miR-221 was reported to be overexpressed in glioblastoma [32] and in thyroid cancer [33]. miR-221 and miR-222 are clustered on the X chromosome, and both of them are predicted to regulate cell cycle by targeting on kit [33] and p27Kip1 [8]. Our data showed that miR-222 was increased in pancreatic cancers at the level similar to miR-221. Based on the miRNA profiling and their functional studies, miRNA/RNAi-based therapeutics could be attractive strategies for pancreatic cancer treatment.
In summary, pancreatic cancer may have a unique miRNA expression pattern at each individual basis. However, common pathways for pancreatic cancer pathogenesis may exist. Our study suggests that the expression of 8 miRNAs (miR-196a, miR-190, miR-221, miR-222, miR-200b, miR-15b and miR-95) was signficantly increased in the majority of pancreatic cancer tissues and cell lines. Further investigations are required for determination of their molecular functions and mechanisms as well as characterization of these miRNAs as prognostic and/or diagnostic markers in pancreatic cancer. Since miRNAs may regulate multiple oncogenic pathways, they may serve as potential targets for cancer therapy. For examples, antagomirs and chemically modified antisense nucleotides for miRNAs can be used to silence specific endogenous miRNA in vivo [34]. This may provide a novel strategy to treat pancreatic cancer.
Acknowledgements
This study was partially supported by the Michael E. DeBakey Department of Surgery at the Baylor College of Medicine and the Michael E. DeBakey VA Medical Center, Houston, Texas, USA.
The symposium was supported by a grant from the National Institutes of Health (R13 CA132572 to Changyi Chen).
Footnotes
This work was presented at the Molecular Surgeon Symposium on Personalized Genomic Medicine and Surgery at the Baylor College of Medicine, Houston, Texas, USA, on April 12, 2008.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–977. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 4.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Hornstein E, Mansfield JH, Yekta S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 7.Datta J, Kutay H, Nasser MW, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 9.Jeffrey SS. Cancer biomarker profiling with microRNAs. Nat Biotechnol. 2008;26:400–4001. doi: 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Saif MW. Pancreatic cancer: highlights from the 42nd annual meeting of the American Society of Clinical Oncology, 2006. JOP. 2006;7:337–348. [PubMed] [Google Scholar]
- 13.Jemal A, Tiwari RC, Murray T, et al. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 14.Sun M, Estrov Z, Ji Y, et al. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 15.Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 17.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 18.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuda N, Ishiyama S, Li Y, et al. Synthetic microRNA designed to target glioma-associated antigen 1 transcription factor inhibits division and induces late apoptosis in pancreatic tumor cells. Clin Cancer Res. 2006;12:6557–6564. doi: 10.1158/1078-0432.CCR-06-0588. [DOI] [PubMed] [Google Scholar]
- 20.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa T, Duguid WP, Rosenberg L, et al. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 22.Ouyang H, Mou L, Luk C, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157:1623–3123. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Bharadwaj U, Zhang R, et al. Mesothelin is a malignant factor and therapeutic vaccine target for pancreatic cancer. Mol Cancer Ther. 2008;7:286–296. doi: 10.1158/1535-7163.MCT-07-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Zhang Y, Liu Z, et al. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci U S A. 2007;104:18636–18641. doi: 10.1073/pnas.0709307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Yang H, Chai H, et al. Pancreatic carcinoma cells express neuropilins and vascular endothelial growth factor, but not vascular endothelial growth factor receptors. Cancer. 2004;101:2341–2350. doi: 10.1002/cncr.20634. [DOI] [PubMed] [Google Scholar]
- 26.Li M, Zhai Q, Bharadwaj U, et al. Cyclophilin A is overexpressed in human pancreatic cancer cells and stimulates cell proliferation through CD147. Cancer. 2006;106:2284–2294. doi: 10.1002/cncr.21862. [DOI] [PubMed] [Google Scholar]
- 27.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Mansfield JH, Harfe BD, Nissen R, et al. MicroRNA-responsive 'sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. 2004. [DOI] [PubMed] [Google Scholar]
- 29.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 30.Xia L, Zhang D, Du R, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 31.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 33.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]