Abstract
Type I interferons (IFNs) are known to be critical factors in the activation of host antiviral responses and are also important in protection from influenza A virus infection. Especially, the RIG-I- and IPS-1-mediated intracellular type I IFN-inducing pathway is essential in the activation of antiviral responses in cells infected by influenza A virus. Previously, it has been reported that influenza A virus NS1 is involved in the inhibition of this pathway. We show in this report that the influenza A virus utilizes another critical inhibitory mechanism in this pathway. In fact, the viral polymerase complex exhibited an inhibitory activity on IFNβ promoter activation mediated by RIG-I and IPS-1, and this activity was not competitive with the function of NS1. Co-immunoprecipitation analysis revealed that each polymerase subunit bound to IPS-1 in mammalian cells, and each subunit inhibited the activation of IFNβ promoter by IPS-1 independently. In addition, by a combinational expression of each polymerase subunit, IPS-1-induced activation of IFNβ promoter was more efficiently inhibited by the expression of PB2 or PB2-containing complex. Moreover, the expression of PB2 inhibited the transcription of the endogenous IFNβ gene induced after influenza A virus infection. These findings demonstrate that the viral polymerase plays an important role for regulating host anti-viral response through the binding to IPS-1 and inhibition of IFNβ production.
Keywords: Interferon, Pattern Recognition Receptor, Signal Transduction, Viral Polymerase, Viral Protein, RNA Polymerase, Influenza A Virus, Interferon Beta Promoter Stimulator 1, Polymerase Basic Protein 2, Type I Interferon
Introduction
Influenza A virus, which belongs to the Orthomyxoviridae, is a negative sense, single-stranded RNA virus, which causes respiratory disease in humans. Influenza A virus carries eight segmented RNA as its genome, and viral proteins are translated from viral mRNA transcripted by the RNA-dependent RNA polymerase of influenza A virus. The influenza A virus polymerase complex is a heterotrimer consisting of PA (polymerase acidic protein), PB1 (polymerase basic protein 1), and PB2, and each component is crucial in order for the virus to replicate (1).
Type I interferons (IFNs) represented by IFNα and IFNβ are known to be important factors in the activation of host defensive mechanisms against invasion of exogenous pathogens. Type I IFNs are produced by various cells and activate transcription of IFN-stimulated genes through the IFNα receptor, JAK (Janus kinase), and STAT (signal transducers and activator of transcription) (2, 3). Production of proteins from IFN-stimulated genes is also important in the acquisition of resistance to influenza A virus infection. For instance, mouse Mx1 (myxovirus resistance 1) or its human homologue MxA, which is known to be a member of the IFN-stimulated genes, displays a strong antiviral activity against influenza A virus infection by overexpression (4–6).
The induction pathway for Type I IFN is activated by cellular sensors, which recognize exogenous pathogens, such as Toll-like receptor family receptors (7, 8). Recently, the gene products of RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated gene-5) were identified as intracellular RNA sensors, which are responsible for the activation of IFN induction in cells infected with viruses (9, 10). These molecules recognize distinct viruses in each other (e.g. RIG-I recognizes Flaviviridae, Orthomyxoviridae, Paramyxoviridae, and Reoviridae, whereas MDA5 recognizes Picornaviridae) (11–13) and bind to IPS-1 (IFNβ promoter stimulator 1; also called MAVS, Cardif, or VISA). IPS-1 is known to be a downstream mitochondrial adapter protein that transmits the induction signal of type I IFN through the activation of transcription factors IRF3 (IFN regulatory factor 3), IRF7, and NF-κB (nuclear factor κB) (14–17). These molecules are assumed to be physiologically important in the inhibition of viral replication; in fact, the functions of these molecules are inhibited by a variety of viruses (18–22). In addition, mouse embryonic fibroblasts derived from IPS-1-deficient mice do not completely produce type I IFN against influenza A virus infection (23). Therefore, the intracellular induction pathway for type I IFN is also thought to be important for the host defense against influenza A virus infection through the activation of an immunoresponse, especially at the early phase of infection.
Previously, it has been reported that influenza A virus NS1 (nonstructural protein 1) binds to RIG-I and prevents IPS-1 mediated type I IFN production (24–27). On the other hand, an earlier study using a UV-irradiated virus suggested that the influenza viral RNA polymerase is also responsible in the inhibition of type I IFN production (28). A viral RNA polymerase recognizes cap structure at the 5′-end of host mRNA and snatches it as a primer for viral mRNA synthesis (29, 30). Because this function of a viral RNA polymerase causes shutoff of host protein synthesis by decreasing the mature mRNA level, the inhibitory activity of IFN production by the influenza A virus polymerase has been assumed to be dependent on this “cap-snatching” activity. However, there is no direct evidence to support this hypothesis.
In this study, we report that the influenza A virus RNA polymerase performs the function of inhibition of the intracellular induction pathway for type I IFN by binding to IPS-1 independently of its cap-snatching activity. In addition, we show that this inhibitory function is mainly dependent on the PB2 subunit of a viral polymerase, and the expression of IFNβ mRNA induced after influenza A virus infection is inhibited by the expression of the PB2 protein.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
The human embryonic kidney cell line HEK293 was maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin and grown at 37 °C with 5% CO2. Transfections were performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols.
Antibodies
The specific antibodies used in this study, rat anti-HA monoclonal antibody (3F10, Roche Applied Science), mouse anti-FLAG M2 monoclonal antibody (Sigma), mouse anti-Myc monoclonal antibody, rabbit anti-IPS-1 polyclonal antibody (Alexis Biochemicals, San Diego, CA), rabbit anti-IRF3 monoclonal antibody (D83B9; Cell Signaling Technology, Beverly, MA), and rabbit anti-phospho-IRF3 (Ser396) monoclonal antibody (4D4G; Cell signaling Technology) were purchased as commercially available products. The information for the specific antibodies raised against influenza A virus PA (clone 58/1), PB1 (clone 11/3), and PB2 (clone 143/3) is described elsewhere (31, 32).
Vector Construction
To construct expression plasmids, the coding regions of PB2 from various strains were amplified from the infectious clones by PCR. A series of the expression vectors for FLAG-tagged deletion mutants of PB2 were constructed by PCR from PB2 cDNA derived from an influenza virus strain A/Puerto Rico/8/34 (H1N1; PR8). PA, PB1, and NS1 coding regions were then obtained from the total RNA of PR8-infected cells by RT-PCR, and these amplified cDNAs were inserted into the cloning vectors to express epitope-tagged protein designated pcDNA5/FRT/FLAG, pcDNA5/FRT/HA, and pcDNA5/FRT/MYC, respectively. These cloning vectors were generated by an insertion of synthetic oligonucleotide, which encoded the peptide sequence for the epitope tag to pcDNA5/FRT (Invitrogen). The retrovirus vector carrying the PB2 gene was constructed by an insertion of the fragment encoding full-length PB2 derived from the expression plasmid into pMXs Puro (Cell Biolabs, San Diego, CA). The expression vectors for FLAG- and HA-tagged IPS-1, RIG-I, and MDA5 were described previously (16), and p125 luc, which carries the firefly luciferase gene under the control of an INFβ promoter, is described elsewhere (33). pcDNA 3.1(+) TLR3 carried the full-length coding region of the human TLR3 (Toll-like receptor 3) gene was constructed by RT-PCR. To construct a template plasmid carrying influenza virus matrix (M) segment cDNA used for in vitro translation, the full-length M segment genome cDNA was amplified by RT-PCR and cloned into a pCR blunt II TOPO cloning vector using a Zero blunt II TOPO PCR cloning kit (Invitrogen). pBS HCV 1B IRES, which carries the hepatitis C virus type 1B 5′-untranslated region, was kindly provided by A. Nomoto (University of Tokyo). More detailed vector information is available upon request.
Luciferase Assays
Luciferase activities were quantified with a luminometer (Mtharas LB940; Berthold, Bad Wildbad, Germany) using the Dual-Glo luciferase assay system (Promega, San Luis Obispo, CA) in accordance with the manufacturer's instructions. Single-stranded RNA used in this study to stimulate RIG-I was synthesized by in vitro transcription using a MEGAscript T7 kit (Applied Biosystems, Foster City, CA) from the vector carrying the HCV 5′-UTR and influenza virus M segment cDNA downstream of the T7 promoter. These RNA were purified by using the MEGAclear kit (Applied Biosystems) according to the manufacturer's protocols. Poly(I-C), the synthetic double-stranded RNA used to stimulate MDA5 and TLR3, was purchased from Sigma. IFNβ promoter activities were measured by firefly luciferase activity of the reporter plasmid carrying the human IFNβ promoter region, p125 luc, and activations of NF-κB were monitored by pNF-κB-luc (Stratagene, La Jolla, CA), which carries a synthetic promoter containing direct repeats of the NF-κB recognition sequence. The quantified results were normalized with the Renilla luciferase activities of the internal control vector (pGL 4.74; Promega).
Immunoprecipitation
To analyze binding of transiently expressed proteins, the expression plasmids indicated in the figures were transfected into HEK293 cells, and the cells were then harvested and lysed by radioimmunoprecipitation buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with a protease inhibitor mixture (Complete Mini; Roche Applied Science). After the cell debris was removed by centrifugation, the lysates were subjected to immunoprecipitation using anti-FLAG M2-agarose (Sigma). To analyze the binding of viral polymerase to the IPS-1 protein in virus-infected cells, the HEK293 cells were infected with the PR8 virus strain (multiplicity of infection = 2). After 8 h, the cells were harvested and were lysed with radioimmunoprecipitation buffer supplemented with the protease inhibitor mixture. The cell lysates were immunoprecipitated with anti-PB2 monoclonal antibody and protein G-Sepharose (GE Healthcare). The resins were washed five times with radioimmunoprecipitation buffer, and binding proteins were eluted by boiling for 5 min with 2× SDS-polyacrylamide gel-loading buffer (125 mm Tris-HCl (pH 6.8), 5% β-mercaptoethanol, 4% SDS, 20% glycerol, 0.01% bromphenol blue).
RT-PCR
Quantitative and semiquantitative RT-PCR for endogenously expressed IFNβ was carried out as described previously (34). Total cellular RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The isolated RNA was treated with RNase-free DNase I (Roche Applied Science). Subsequently, cDNA was synthesized from the RNA using random hexamer and reverse transcriptase (ReverTra Ace; Toyobo, Osaka, Japan). Real-time PCRs were performed using SYBR Premix Ex TaqII (Takara, Otsu, Japan) and quantified by the Mx3000P quantitative PCR System (Stratagene).
Virus Infection
HEK293 cells were transfected with 2 μg of expression vector for intact PB2. After 35 h, the cells were infected with influenza virus A/Aichi/2/1968 (H3N2; Aichi) strain (multiplicity of infection = 2), and incubated with DMEM containing 1% BSA and 0.5 μg/ml trypsin for 3 h. Subsequently, the cells were harvested and subjected to RT-PCR and Western blotting analysis.
RESULTS
Inhibition of Activation of Intracellular Induction Pathway for Type I IFN by Influenza A Virus Polymerase
To investigate the function of the influenza virus polymerase complex against the induction of type I IFN, we constructed expression vectors that encode the N-terminal epitope-tagged influenza A virus RNA polymerase subunits PA, PB1, and PB2 derived from the H1N1 subtype influenza virus strain A/Puerto Rico/8/34 (PR8). Initially, we confirmed whether these N-terminal epitope-tagged subunits are able to form a heterotrimeric complex. The expression vectors for FLAG-tagged PB2 and HA-tagged PA and PB1 were transfected into HEK293 cells, and the whole cell extracts prepared from the cells were subjected to immunoprecipitation using anti-FLAG resin. As shown in Fig. 1, the results indicate that when three subunits are expressed together, both HA-tagged PA and PB1 subunits are co-immunoprecipitated with FLAG-tagged PB2 subunit. Previous reports demonstrated that the RNA polymerase subunits of influenza A virus are able to form PA/PB1 and PB1/PB2 heterodimers as intermediates, whereas PA/PB2 is not (35, 36). In agreement with the previous observations, although the signal was relatively weak, HA-tagged PB1 was co-immunoprecipitated with FLAG-tagged PB2, whereas HA-tagged PA was not. These results suggest that these N-terminal epitope-tagged subunits are able to form the heterotrimer of the mature viral polymerases.
FIGURE 1.
The N-terminal epitope-tagged subunits of influenza A virus RNA polymerase also form the heterotrimer. Expression vectors for FLAG-tagged PB2 (1.6 μg) and HA-tagged PA and PB1 (1.2 μg each) were transfected into HEK293 cells. After 24 h, the cells were harvested and lysed, and then the whole cell extracts were subjected to the co-immunoprecipitation assay (IP) using an anti-FLAG M2-agarose.
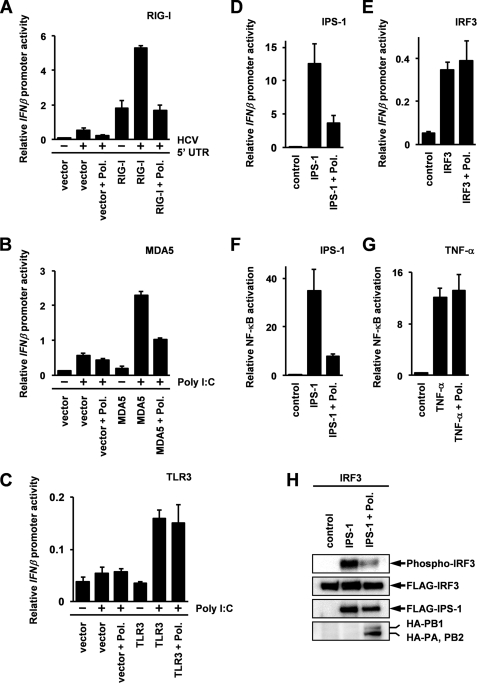
Next, we examined the effect of the expression of these subunits on the activation of the IFNβ promoter by stimulation with exogenous RNA by using a reporter gene assay. The results show that activation of IFNβ promoter mediated by MDA5 or RIG-I was significantly inhibited by the overexpression of the viral RNA polymerase complex (Fig. 2A and 2B), whereas TLR3-mediated activation of the IFN promoter was not influenced (Fig. 2C). Because it has been reported that the conserved flanking sequence lying on viral genomic RNA is required for the activation of the cap-snatching function of the influenza A virus polymerase (37–39), it was assumed that stimulation by both HCV 5′-UTR and poly(I-C) could not activate cap-snatching activity. Therefore, we concluded that the inhibitory effect of a viral polymerase on RIG-I-mediated IFNβ promoter activation was independent of its cap-snatching function. This conclusion is also supported by the finding that the expression of a viral polymerase did not inhibit TLR3-mediated IFNβ promoter activation.
FIGURE 2.
Inhibition of intracellular IFN inducing pathways by overexpression of influenza viral polymerase complex. A and B, the expression plasmids for each subunit of influenza viral RNA polymerase derived from PR8 strain (200 ng) were transfected with the p125 luc (80 ng) and pGL4.74 (20 ng) reporter constructs together with 100 ng of expression vector for RIG-I (A) or MDA5 (B) into HEK293 cells. After a 24-h post-transfection incubation period, the cells were stimulated with 800 ng of HCV 5′-UTR (A) or poly(I-C) (B) by transfection. After an additional 24-h incubation, luciferase activities were measured by luminometer. C, PA (300 ng), PB1 (150 ng), PB2 (300 ng), and TLR3 (125 ng) expression plasmids were transfected with the p125 luc (100 ng) and pGL4.74 (25 ng) reporter constructs into HEK293 cells. After 24 h, the cells were treated with poly(I-C) (50 μg/ml) for 12 h, and luciferase activities were measured. D, HEK293 cells were transfected with the p125 luc (80 ng) and pGL4.74 (20 ng) reporter constructs together with expression vectors for IPS-1 (50 ng) and each component of influenza virus RNA polymerase (200 ng). After a 24-h period of incubation, luciferase activities were measured. Total amounts of DNA in each transfection were equalized by empty vectors. E, the expression vector for the FLAG-tagged IRF3 and each subunit for the viral polymerase were transfected into HEK293 cells with p125 luc and pGL4.74 plasmids. After 24 h, the activation of IFNβ promoter was analyzed by a luciferase assay. F, the expression plasmids for FLAG-tagged IPS-1 (50 ng) and each subunit of viral polymerase (250 ng) were transfected into HEK293 cells together with the reporter construct plasmids, pNF-κB luc (50 ng) and pGL4.74 (50 ng). After 24 h, the activation of NF-κB was measured by a luciferase assay. G, the expression plasmids for the FLAG-tagged each viral polymerase subunits (250 ng) were transfected with pNF-κB (50 ng) luc and PGL 4.74 (50 ng) into HEK293 cells. After 24 h, the cells were stimulated by 25 ng/ml TNF-α. After an additional 8-h post-stimulation incubation period, activation of NF-κB was analyzed by the luciferase assay. Data represent relative IFNβ promoter activities or NF-κB activations, which were normalized with Renilla luciferase activities. Error bars indicating the S.D. values were calculated from at least three independent experiments. H, the expression plasmids for FLAG-tagged IRF3 (0.8 μg) and IPS-1 (40 ng) were transfected with expression vectors for the HA-tagged subunits of influenza viral RNA polymerase (1.0 μg) into HEK293 cells. After 24 h, the cells were harvested, and phosphorylation of IRF3 was analyzed by immunoblotting specific antibody against phospho-IRF3 (Ser396).
Although subcellular localization of RIG-I or MDA5 is different from that of TLR3, the overall induction pathway for type I IFN by RIG-I and MDA5 is mediated by the same signaling molecules as the pathway mediated by TLR3. These molecules transduce the signals to the same transcription factors, IRF3, IRF7, and NF-κB, mediated by the intermediate signaling molecules, such as TBK1 (TANK-binding kinase 1) and IKKϵ (IκB kinase ϵ) (40, 41). Therefore, our data, derived by using a reporter gene assay (Fig. 2, A–C), suggest that the targets of viral RNA polymerase are not these overlapping signaling molecules that inhibit the intracellular induction pathway for type I IFN. From these facts, we thought that there is a possibility that the viral polymerase complex targets to IPS-1 and inhibits its function, because IPS-1 is an adapter molecule that binds RIG-I and MDA5 but not TLR3.
Initially, we investigated the effect of an expression of the influenza A virus polymerase to the IFNβ promoter activated by the overexpression of IPS-1. As shown in Fig. 2D, the results show that a viral polymerase strongly inhibited IFNβ promoter activation induced by the expression of IPS-1. To confirm that the viral polymerase specifically inhibits the IPS-1-mediated signaling pathway, we evaluated the effects of the viral polymerase on the IFNβ promoter activation induced by overexpression of IRF3. The results show that activation of the IFNβ promoter induced by overexpression of IRF3 was not affected by the expression of the viral polymerase (Fig. 2E).
It is known that activation of the IFNβ promoter is regulated by IRF3, IRF7, and NF-κB (40, 41), and NF-κB is involved in the induction of a variety of inflammatory cytokines in addition to type I IFN (42). Taking this into consideration, we used a reporter plasmid that carries the luciferase gene under the control of the synthetic NF-κB binding site (pNF-κB-luc; Promega) and evaluated the effect of the viral polymerase expression on IPS-1-induced NF-κB activation. The results show that the viral polymerase significantly inhibited NF-κB activation induced by the IPS-1 overexpression (Fig. 2F) but did not inhibit the activation induced by TNF-α stimulation (Fig. 2G). TNF-α is known to be an inflammatory cytokine belonging to the TNF superfamily and is also known to be a strong NF-κB inducer that activates a signaling pathway distinct from the IPS-1-mediated signaling pathway (43).
Finally, we examined whether IPS-1-mediated activation of IRF3 is inhibited by the expression of the viral RNA polymerase. It has been reported that the Ser396 residue of IRF-3 is phosphorylated by the infection of RNA viruses and the transfection of double-stranded RNA (44) and is also phosphorylated by the overexpression of IPS-1 (16). We evaluated the activation of IRF3 by monitoring the phosphorylation level of IRF3 at Ser396, using its specific antibody. As shown in Fig. 2H, The results show that the phosphorylation of Ser396 in IRF3 was increased by the IPS-1 overexpression and was significantly inhibited by the expression of the viral RNA polymerase. These results suggest that the influenza A virus RNA polymerase has an activity to inhibit internal IFN induction pathway through the inhibition of IPS-1 function.
Inhibition of IPS-1-mediated Induction of Type I IFN through Binding to IPS-1 by Influenza A Virus Polymerase
To reveal the molecular mechanism for the inhibition of the IPS-1-mediated signaling pathway, we initially examined a possibility that viral RNA polymerase inhibits the IPS-1 function through the binding to the IPS-1. To investigate the binding of a viral polymerase with IPS-1, we performed a co-immunoprecipitation assay. IPS-1 with PA, PB1, or PB2 was expressed in HEK293 cells, and the binding of IPS-1 with PA, PB1, or PB2 was analyzed. Each component of the influenza viral polymerase complex was co-immunoprecipitated with IPS-1 (Fig. 3, A and B). These results indicated that all subunits of a viral RNA polymerase have the ability to bind with IPS-1.
FIGURE 3.
Binding of each component of influenza virus RNA polymerase to IPS-1. A, HEK293 cells were transfected with expression vectors for HA-tagged PA, PB1, or PB2 (3.5 μg) together with 0.5 μg of FLAG-tagged IPS-1. B, FLAG-tagged PA, PB1, PB2, or M (3.5 μg) expression vectors were transfected with HA-tagged IPS-1 expression constructs (0.5 μg) into HEK293 cells. C, the expression plasmids for Myc-tagged PB2 (1.5 μg), HA-tagged PA (1.5 μg) and PB1 (0.6 μg) were transfected into HEK293 cells with 0.4 μg of expression plasmid for FLAG-tagged IPS-1. Whole cell extracts from the transfected cells were immunoprecipitated (IP) by using FLAG M2-agarose and subjected to Western blotting analysis. D, HEK293 cells were infected with influenza A virus PR8 strain (multiplicity of infection = 2). After 8 h, the cells were harvested and lysed, and then the lysates were subjected to immunoprecipitation using a mouse monoclonal antibody against PB2. The antibody that recognizes Ebola ZGP was used for the control. IPS-1 and each subunits of the viral polymerase were detected by using the specific antibodies.
Next, we investigated whether all components of viral polymerase bind to IPS-1 simultaneously. As shown in Fig. 3, A and B, PA and PB2 were detected to have the same mobility by Western blotting, so it was difficult to distinguish between the two proteins. To resolve this problem, we constructed a Myc-tagged PB2 expression vector. Expression vectors for HA-tagged PA, PB1, Myc-tagged PB2, and FLAG-tagged IPS-1 were transfected into HEK293 cells, and the cell lysates were immunoprecipitated by anti-FLAG resin. The results show that all components of the viral polymerase were co-immunoprecipitated with IPS-1 at the same time (Fig. 3C). A viral polymerase forms a stable complex (1, 36, 45), and we confirmed that the N-terminal epitope-tagged polymerase subunits used in this study are also able to form a complex (Fig. 1). From these results, it is likely that a viral polymerase binds to IPS-1 as a complex.
Finally, we investigated whether the viral polymerase binds to IPS-1 in virus-infected cells. The PB2 proteins in the lysates of the influenza virus-infected cells were immunoprecipitated with anti-PB2 monoclonal antibody, and co-immunoprecipitated proteins were analyzed by Western blotting. As shown in Fig. 3D, the results indicate that PA and PB1 subunits were largely co-immunoprecipitated with PB2, and IPS-1 was also co-immunoprecipitated together with these proteins. The data suggested that the PB2 subunit binds to IPS-1 in the virus-infected cells and probably makes a complex.
Dependence on PB2 in Inhibition of IPS-1-mediated Signaling Pathway for Induction of IFNβ by Influenza A Virus Polymerase
To investigate which subunit of viral polymerase is responsible for the inhibition of IPS-1 function, we examined the inhibitory activity of each polymerase subunit by using a reporter gene assay. Each polymerase subunit and NS1 were independently expressed in HEK293 cells together with RIG-I, and the cells were stimulated with single-stranded RNA derived from the influenza virus matrix (M) segment. The results indicated that IFNβ promoter activation by single-stranded RNA was equally inhibited by each subunit of the viral polymerase (Fig. 4A). However, the expression level of PB1 was higher than that of PA and PB2 (Fig. 4B). In addition, the expression level of NS1 was higher than that of PA, PB1, and PB2. However, the inhibitory activity of NS1 on IFNβ promoter activation was detected as stronger than that of each polymerase. Therefore, it may be assumed that the inhibitory effect of PB2 on IFNβ promoter activation was very strong.
FIGURE 4.
PB2 as a major component for inhibition of IPS-1-mediated IFNβ and NF-κB promoter activation by influenza viral polymerase. A, the expression plasmids for HA-tagged NS1, PA, PB1, and PB2 (600 ng) were transfected with the p125 luc (80 ng) and pGL4.74 (20 ng) reporter constructs together with 100 ng of expression vector for FLAG-tagged RIG-I into HEK293 cells. After a 24-h post-transfection incubation period, the cells were stimulated by the transfection of single-stranded RNA (800 ng) derived from the matrix (M) segment of influenza virus PR8 strain. The luciferase activities were measured after a 24-h poststimulation incubation period. B, Western blotting analysis of whole cell extracts from the same transfectants. C, the expression plasmids for HA-tagged PA (280 ng), PB1 (140 ng), and PB2 (280 ng) were transfected with the expression construct for FLAG-tagged IPS-1 (20 ng) together with p125 luc (64 ng) and pGL 4.74 (16 ng) into HEK293 cells. After 24 h, the luciferase activities were measured, and part of the transfectants were harvested and subjected to Western blotting analysis. D, the vectors to express FLAG-tagged NS1, PA, PB1, and PB2 (800 ng) were transfected with the pNF-κB luc (80 ng) and pGL4.74 (20 ng) reporter plasmids together with 50 ng of expression vector for FLAG-tagged IPS-1 into HEK293 cells. After 24 h, the luciferase activities were measured. Total amounts of DNA in each transfection were equalized by empty vectors. Data represent relative IFNβ promoter activities, which were normalized with Renilla luciferase activities. Error bars indicating the S.D. values were calculated from at least three independent experiments. ssRNA, single-stranded RNA.
Next, we adjusted the amount of expression vectors for the transfection to equalize the protein expression level of each subunit, and then we tested their inhibitory effect on IPS-1-induced INFβ promoter activation. The results show that the relative inhibitory activity of PB2 on IFNβ promoter activation depended on the PB1/PB2 heterodimer and the viral polymerase complex (Fig. 4C). To confirm this strong inhibitory activity of PB2 against the IPS-1 function, we investigated the inhibitory activities of each viral polymerase component on IPS-1-mediated activation of NF-κB by using a reporter gene assay. The results show that NF-κB activation by overexpression of IPS-1 was also most strongly inhibited by PB2 (Fig. 4D). The amounts of transfected expression plasmids for viral polymerase subunits and NS1 were the same in this case. In addition, because NS1 binds to PI3K and activates the NF-κB pathway (46–48), NS1 did not show strong inhibitory activity on IPS-1-mediated NF-κB activation. Taken together, it was demonstrated that the PB2 protein acts mainly to inhibit the function of IPS-1 in the activation of the IFNβ promoter and NF-κB.
The Binding Activity of PB2 to IPS-1 Is Mainly Dependent on the N-terminal Region of PB2
Because our results using a reporter gene assay revealed that the PB2 subunit plays a pivotal role in the inhibition of activation of IFNβ promoter mediated by IPS-1, we next analyzed the binding region of PB2 to IPS-1 by a co-immunoprecipitation assay using deletion mutants of PB2 for further analysis of the inhibitory mechanism. As shown in Fig. 5A, we constructed the expression vectors for a series of FLAG-tagged deletion mutants of PB2 for the analysis. These expression vectors were transfected with that of HA-tagged IPS-1 into HEK293 cells, and then the PB2 mutants were immunoprecipitated using anti-FLAG resin from the whole cell extracts of the cells. The results show that although no deletion mutant of PB2 indicated a complete loss of binding activity to the HA-tagged IPS-1, PB2 ΔN242 and PB2 ΔN482 exhibited significantly weak binding activity to the IPS-1 in comparison with that of wild-type PB2 (Fig. 5B). Both of these mutants lack the N-terminal 242-amino acid region of PB2. On the other hand, PB2 ΔC257 which expresses the N-terminal 256-amino acid region of PB2 exhibits binding activity to the IPS-1 as well as that of wild-type PB2. Therefore, the N-terminal 242-amino acid region of PB2 is thought to be mainly responsible for the binding to IPS-1. Comparable results were obtained by the reporter gene assay using these mutants. As shown in Fig. 5C, the deletion mutants, PB2 ΔN242 and PB2 ΔN482, exhibited significantly weak inhibitory activity against IFNβ promoter activation induced by overexpression of IPS-1 in comparison with that of wild-type PB2.
FIGURE 5.
Cap-binding region of PB2 is not required for inhibition of the activation of IFNβ promoter mediated by IPS-1. A, schematic diagram of PB2 deletion mutants. The numbers indicate the amino acid sequence position of PB2, and the minimal cap-binding domain of PB2 is indicated in the bold bar (61). B, HEK293 cells were transfected with the expression vectors for the series of FLAG-tagged deletion mutants of PB2 (3.5 μg) together with the expression vector plasmid for HA-tagged IPS-1 (0.5 μg). At 24 h post-transfection, the cells were harvested and subjected to an immunoprecipitation assay (IP) as described under “Experimental Procedures.” C, the expression vectors for the HA-tagged IPS-1 (100 ng) and the series of FLAG-tagged deletion mutants of PB2 (400 ng) were transfected with p125 luc (80 ng) and pGL 4.74 (20 ng) into HEK293 cells. After 24 h, luciferase activities in the cells were quantified by the luminometer. Error bars indicating the S.D. values were calculated from at least three independent experiments. D, expression vectors for HA-tagged IPS-1 (0.4 μg) and FLAG-tagged PB2 (3.6 μg) were transfected into HEK293 cells using FuGENE HD transfection reagent (Roche Applied Science). The cells were harvested at 24 h post-transfection and lysed with radioimmunoprecipitation buffer. The lysates were then separated into two aliquots. One aliquot was treated with RNase A at 4 °C for 2 h, whereas the other aliquot was left untreated. Both aliquots were then subjected to the immunoprecipitation.
Most of the other viral molecules that are known to be functional for the inhibition of IPS-1-mediated type I IFN production exhibit the inhibitory function through the binding to the viral RNA and inhibit the function of RNA recognition molecules (e.g. RIG-I and MDA5) by competition (18–22). Taking this into consideration, we investigated whether the IPS-1 binding activity of PB2 is dependent on the RNA binding activity of PB2. As shown in Fig. 5D, the IPS-1 binding activity of the PB2 subunit was not affected by RNase treatment. These results suggest that PB2 binds to IPS-1 in an RNA-binding independent manner, and it is likely that PB2 inhibits the function of IPS-1 through the protein-protein interaction.
Inhibition of IFN Induction by Influenza A Virus Polymerase Is Independent of NS1 Function
It is known that NS1 inhibits the induction of IFN by binding to RIG-I, which is located upstream of IPS-1 and acts as a sensor molecule for viral RNA. To investigate whether the function of viral polymerase in the inhibition of IFNβ promoter activation is competitive with that of NS1, we transfected NS1 and each polymerase component at the same time and measured the inhibitory activity by using a reporter gene assay. The results show that inhibition of IPS-1-induced IFNβ transcription by the viral polymerase is enhanced by the expression of NS1 (Fig. 6A). Therefore, it is indicated that the inhibitory function of the viral polymerase is independent of the NS1 function. Although we transfected fewer expression vectors for NS1 than for each polymerase component in this case, the protein expression levels of NS1 in the cells simultaneously transfected were detected, as were the protein expression levels of polymerase subunits, by Western blotting (Fig. 6B). Based on these data, we have concluded that the relative inhibitory activity normalized with the protein expression level did not significantly differ between NS1 and the viral polymerase.
FIGURE 6.
Inhibition of IFN induction by influenza A virus polymerase is not competitive with the NS1 function. A, the expression plasmids for HA-tagged NS1 (100 ng), PA (200 ng), PB1 (100 ng), and PB2 (200 ng) were transfected with the expression construct for FLAG-tagged IPS-1 (20 ng) together with p125 luc (64 ng) and pGL 4.74 (16 ng) into HEK293 cells. After 24 h, the luciferase activities were measured. B, Western blotting analysis of whole cell extracts from the same transfectants. C, the expression constructs for IPS-1 (20 ng) and PB2 derived from various influenza virus strains indicated in the figure (500 ng) were transfected into HEK293 cells with p125 luc (64 ng) and pGL 4.74 (16 ng). After 24 h, the luciferase activities were measured, and part of the transfectants were harvested and subjected to Western blotting analysis. The total amounts of DNA in each transfection were equalized by empty vectors. The data represent relative IFNβ promoter activities, which were normalized with Renilla luciferase activities. Error bars indicating the S.D. values were calculated from at least three independent experiments.
Inhibition of IFNβ Promoter Activation by Suppression of IPS-1 Function by PB2 of Various Strains of Influenza A Virus
In the experiments described thus far, we used viral components derived from the PR8 strain, which belongs to the H1N1 subtype influenza virus. To confirm whether the RNA polymerase subunits of other strains of influenza A virus perform a similar function, we tested the inhibitory effect of influenza A virus PB2 derived from other strains on IFNβ promoter activation. Expression vectors for PB2 proteins from H1N1 influenza virus, A/WSN/33 (WSN), and H5N1 highly pathogenic avian influenza virus, A/Hong Kong/483/97 (HK483) and A/Hong Kong/486/97 (HK486), were used for this assay, and the results show that PB2 derived from these strains also inhibited IPS-1-induced IFNβ promoter activation (Fig. 6C). From these data, it can be assumed that the function of PB2 in the inhibition of IFNβ promoter activation is common to various influenza A viruses.
Modulation of Cellular IFNβ Gene Activation Induced by Influenza A Virus by Expression of PB2
To investigate whether the protein expression level of PB2 affects the inhibition of IPS-1-mediated IFNβ promoter activation, we monitored IPS-1-induced IFNβ promoter activities after the expression of various amounts of PB2. The results show that IPS-1-induced IFNβ promoter activation was inhibited by the expression of PB2 in a dose-dependent manner (Fig. 7A). This finding suggests that the expression level of PB2 regulated the type I interferon production induced by influenza A virus infection.
FIGURE 7.
Inhibition of transcriptional activation of cellular IFNβ gene in influenza virus-infected cells by overexpression of PB2. A, various amounts of PB2 expression vector indicated in the figure were transfected with 25 ng of IPS-1 expression plasmid into HEK293 cells together with p125 luc (80 ng) and pGL4.74 (20 ng). After a 24-h post-transfection incubation period, the luciferase activities were measured by a luminometer. B, HEK293 cells were transfected with expression vectors for IPS-1 (0.1 μg), PA (1.5 μg), PB1 (0.7 μg) and PB2 (1.5 μg). After 24 h, the cells were harvested, and the total RNA was isolated from the cells. The expression levels of the endogenous INFβ gene were monitored by semiquantitative RT-PCR. The glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene was used for internal control. C and D, the PB2-expressed cells were infected with influenza A virus Aichi strain (multiplicity of infection = 2). At 3 h postinfection, the expression levels of IFNβ mRNA (C) and nucleoprotein (NP) RNA (D) were quantified by real-time RT-PCR. The data are presented as relative amounts of IFNβ mRNA, which were normalized with the expression level of G3PDH mRNA. E, Western blotting analysis of whole cell extracts from the same transfectants using an anti-PB2 monoclonal antibody. The mouse anti-actin monoclonal antibody was used as a loading control. N.D., not detected.
Next, to confirm that influenza A virus PB2 actually inhibits an endogenously expressed IFNβ gene expression mediated by IPS-1, we investigated the cellular IFNβ mRNA expression level by using RT-PCR. As shown in Fig. 5B, expression of influenza A virus PB2 remarkably repressed the endogenous IFNβ expression induced by IPS-1.
Finally, to determine whether the expression level of PB2 actually affects IFNβ gene expression in the cells infected by influenza A virus, we investigated the effects of PB2 overexpression on the endogenous IFNβ mRNA level after influenza A virus infection by RT-PCR. The results show that the amount of endogenous INFβ mRNA expression induced by viral infection was suppressed by overexpression of PB2 (Fig. 7, C and D), and the expression level of the nucleoprotein (NP) gene was not significantly changed by the overexpression of PB2 (Fig. 7E). These data suggested that the expression level of PB2 in virus-infected cells is involved in the inhibition of type I IFN response.
DISCUSSION
We show in this report that the influenza A virus RNA polymerase inhibits production of type I IFNs through binding to IPS-1 and suppression of its function. Interestingly, our findings reveal that influenza A virus inhibits the intracellular type I IFN-inducing pathway by additional components that are distinct from NS1, and this indicates the physiological importance of this function in the protection of host cells against influenza A virus infection. The influenza A virus NS1 protein is expressed more abundantly in the virus-infected cells than is the viral polymerase (49, 50). NS1 is known to be a multifunctional protein and is important for the regulation of viral replications and inhibition of host anti-viral response (51). Despite its importance, the mutant virus that lacks the NS1 coding region is able to replicate in interferon-deficient systems (52), and the NS1-deficient virus-infected cells significantly increase the production of type I IFNs. However, as suggested by the previous report using a UV-irradiated virus, type I IFN production was increased by gene disruption by UV irradiation for NS1-deficient virus (28). These findings suggest that the importance of the viral polymerase function in the inhibition of type I IFN production cannot be ignored. Because it is known that the type I IFN production pathway is enhanced in an autocrine manner (33, 53–55), inhibitory activity of the interferon production pathway at the early phase of viral infection might be strongly reflected in the intensity of cellular antiviral responses at the late phase. Therefore, at the early phase of viral infection, at which time an insufficient amount of viral components exist in infected cells, viral polymerase activity to repress type I IFN response might play an important role in escape from host antiviral responses.
As shown in Fig. 4, the PB2 subunit efficiently repressed IPS-1-mediated IFNβ promoter activation when it was expressed in isolation. And the results of combinational expression of each polymerase subunit revealed that activation of the IFNβ promoter induced by overexpression of IPS-1 was strongly repressed by expression of the PB2-containing complex. Therefore, we have concluded that PB2 performs a central role in this inhibitory function. Interestingly, our results show that the activation of IFNβ promoter induced by IPS-1 was not inhibited by the combinational expression of PA and PB1, and the inhibitory activity was apparently weakened compared with its expression in isolation (Fig. 4C). A previous report (56) demonstrated that PA and PB1 are mainly localized in cytoplasm when these subunits were expressed alone and are translocated into the nucleus when these subunits formed a heterodimer. These findings suggest that the cytoplasmic localization of PA and PB1 subunits is important for the inhibition of the IPS-1 function. On the other hand, PB2 is mainly observed in the nucleus even when it is expressed alone. However, it has been reported that PB2 carries a mitochondrial targeting sequence at the N terminus and also localizes in mitochondria (57). Because IPS-1 is known to be localized in the mitochondrial outer membrane, it is suggested that the PB2-containing complexes efficiently inhibit the IPS-1 function in this area. From these findings, mitochondrial/cytoplasmic localization of polymerase is thought to be required for the inhibition of IPS-1 function, and the mitochondrial targeting sequence of PB2 is thought to be involved in the inhibitory function of the viral polymerase through the regulation of the subcellular localization.
It was reported that PB2 is a determinant in the difference in pathogenicity to mammalians between influenza virus HK483 and influenza virus HK483, both of which belong to the H5N1 subtype highly pathogenic avian influenza virus (58, 59). However, no significant difference was observed in the inhibitory activity of PB2 derived from these virus strains. Although the inhibitory activity of PB2 on the IPS-1 function does not directly reflect the difference in the pathogenicity between these virus strains, the general expression level of the PB2 protein is influenced by its viral replication efficiency, and highly pathogenic influenza viruses are thought to replicate more efficiently than do low pathogenic viruses in the infected cells. As shown in Fig. 7A, PB2 repressed IPS-1-mediated IFNβ promoter activation in a dose-dependent manner. In addition, the data shown in Fig. 7, C–E, suggest that the expression level of PB2 actually affects the amount of type I IFN production in cells infected by influenza A virus. Therefore, these results might explain part of the reason for the previously reported results, which showed that the production of type I IFN was significantly repressed by highly pathogenic influenza virus infection, such as in the case of the 1918 Spanish flu (60).
Molecular biological analysis revealed that the inhibitory action of NS1 on the type I IFN-inducing pathway is dependent on its binding activity to double-stranded RNA (24). On the other hand, the inhibitory mechanism of viral polymerase seemed not to be dependent on its RNA binding activity, at least in the case of PB2, because PB2 could bind to IPS-1 and repress IPS-1-mediated INFβ promoter activation, even when its cap-binding region was deleted (61) (Fig. 5, A–C). Moreover, PB2 binding activity to IPS-1 was not affected by a treatment with RNase A (Fig. 5D). Therefore, it may be suggested that repression of the molecular functions of IPS-1 could be a very important function of the viral polymerase. There is a possibility that IPS-1 is involved in other unknown signaling pathways, as are other adapter molecules, such as MyD88 (myeloid differentiation protein-88), which works not only for the Toll-like receptor-mediated signaling pathway but also for the interleukin-1 receptor signal (7, 8, 62, 63). Recently, our work has shown that IPS-1 is also responsible in the inducement of anoikis, which is known to be an apoptosis induced by a loss of adhesion (64). It may be suggested that a function of IPS-1 is to control cell death that has been induced by various stresses, such as viral infection. Further investigation is required for understanding of the biological importance of the interaction between IPS-1 and a viral polymerase for the pathogenicity of influenza A virus.
Footnotes
This work was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and by a grant from the Japan Science and Technology Agency.
REFERENCES
- 1.Neumann G., Brownlee G. G., Fodor E., Kawaoka Y. (2004) Curr. Top. Microbiol. Immunol. 283, 121–143 [DOI] [PubMed] [Google Scholar]
- 2.Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 3.Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 4.Arnheiter H., Skuntz S., Noteborn M., Chang S., Meier E. (1990) Cell 62, 51–61 [DOI] [PubMed] [Google Scholar]
- 5.Garber E. A., Chute H. T., Condra J. H., Gotlib L., Colonno R. J., Smith R. G. (1991) Virology 180, 754–762 [DOI] [PubMed] [Google Scholar]
- 6.Kolb E., Laine E., Strehler D., Staeheli P. (1992) J. Virol. 66, 1709–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirk P., Bazan J. F. (2005) Immunity 23, 347–350 [DOI] [PubMed] [Google Scholar]
- 8.Kumar H., Kawai T., Akira S. (2009) Biochem. Biophys. Res. Commun. 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 9.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 10.Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., Randall R. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meylan E., Tschopp J., Karin M. (2006) Nature 442, 39–44 [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi O., Akira S. (2008) Curr. Opin. Immunol. 20, 17–22 [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M., Fujita T. (2009) Immunol. Rev. 227, 54–65 [DOI] [PubMed] [Google Scholar]
- 14.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 15.Xu L. G., Wang Y. Y., Han KJ., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 16.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 17.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 18.Foy E., Li K., Sumpter R., Jr., Loo Y. M., Johnson C. L., Wang C., Fish P. M., Yoneyama M., Fujita T., Lemon S. M., Gale M., Jr. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2986–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cárdenas W. B., Loo Y. M., Gale M., Jr., Hartman A. L., Kimberlin C. R., Martínez-Sobrido L., Saphire E. O., Basler C. F. (2006) J. Virol. 80, 5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond M. S. (2009) PLoS Pathog. 5, e1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barral P. M., Morrison J. M., Drahos J., Gupta P., Sarkar D., Fisher P. B., Racaniello V. R. (2007) J. Virol. 81, 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papon L., Oteiza A., Imaizumi T., Kato H., Brocchi E., Lawson T. G., Akira S., Mechti N. (2009) Virology 393, 311–318 [DOI] [PubMed] [Google Scholar]
- 23.Kumar H., Kawai T., Kato H., Sato S., Takahashi K., Coban C., Yamamoto M., Uematsu S., Ishii K. J., Takeuchi O., Akira S. (2006) J. Exp. Med. 203, 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 25.Mibayashi M., Martínez-Sobrido L., Loo Y. M., Cárdenas W. B., Gale M., Jr., García-Sastre A. (2007) J. Virol. 81, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Z., Chen L. M., Zeng H., Gomez J. A., Plowden J., Fujita T., Katz J. M., Donis R. O., Sambhara S. (2007) Am. J. Respir. Cell Mol. Biol. 36, 263–269 [DOI] [PubMed] [Google Scholar]
- 27.Opitz B., Rejaibi A., Dauber B., Eckhard J., Vinzing M., Schmeck B., Hippenstiel S., Suttorp N., Wolff T. (2007) Cell Microbiol. 9, 930–938 [DOI] [PubMed] [Google Scholar]
- 28.Marcus P. I., Rojek J. M., Sekellick M. J. (2005) J. Virol. 79, 2880–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagen M., Chung T. D., Butcher J. A., Krystal M. (1994) J. Virol. 68, 1509–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M. L., Ramirez B. C., Krug R. M. (1998) EMBO J. 17, 5844–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatta M., Asano Y., Masunaga K., Ito T., Okazaki K., Toyoda T., Kawaoka Y., Ishihama A., Kida H. (2000) Arch. Virol. 145, 895–903 [DOI] [PubMed] [Google Scholar]
- 32.Hatta M., Asano Y., Masunaga K., Ito T., Okazaki K., Toyoda T., Kawaoka Y., Ishihama A., Kida H. (2000) Arch. Virol. 145, 1947–1961 [DOI] [PubMed] [Google Scholar]
- 33.Yoneyama M., Suhara W., Fukuhara Y., Sato M., Ozato K., Fujita T. (1996) J. Biochem. 120, 160–169 [DOI] [PubMed] [Google Scholar]
- 34.Opitz B., Vinzing M., van Laak V., Schmeck B., Heine G., Günther S., Preissner R., Slevogt H., N'Guessan P. D., Eitel J., Goldmann T., Flieger A., Suttorp N., Hippenstiel S. (2006) J. Biol. Chem. 281, 36173–36179 [DOI] [PubMed] [Google Scholar]
- 35.González S., Zürcher T., Ortín J. (1996) Nucleic Acids Res. 24, 4456–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng T., Sharps J., Fodor E., Brownlee G. G. (2005) J. Virol. 79, 8669–8674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leahy M. B., Pritlove D. C., Poon L. L., Brownlee G. G. (2001) J. Virol. 75, 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honda A., Endo A., Mizumoto K., Ishihama A. (2001) J. Biol. Chem. 276, 31179–31185 [DOI] [PubMed] [Google Scholar]
- 39.Leahy M. B., Dobbyn H. C., Brownlee G. G. (2001) J. Virol. 75, 7042–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowie A. G., Unterholzner L. (2008) Nat. Rev. Immunol. 8, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi O., Akira S. (2009) Immunol. Rev. 227, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pahl H. L. (1999) Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 43.Servant M. J., Grandvaux N., tenOever B. R., Duguay D., Lin R., Hiscott J. (2003) J. Biol. Chem. 278, 9441–9447 [DOI] [PubMed] [Google Scholar]
- 44.Bianchi K., Meier P. (2009) Mol. Cell. 36, 736–742 [DOI] [PubMed] [Google Scholar]
- 45.Hwang J. S., Yamada K., Honda A., Nakade K., Ishihama A. (2000) J. Virol. 74, 4074–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hale B. G., Jackson D., Chen Y. H., Lamb R. A., Randall R. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14194–14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ehrhardt C., Wolff T., Pleschka S., Planz O., Beermann W., Bode J. G., Schmolke M., Ludwig S. (2007) J. Virol. 81, 3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauli E. K., Schmolke M., Wolff T., Viemann D., Roth J., Bode J. G., Ludwig S. (2008) PLoS Pathog. 4, e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarowitz S. G., Compans R. W., Choppin P. W. (1971) Virology 46, 830–843 [DOI] [PubMed] [Google Scholar]
- 50.Meier-Ewert H., Compans R. W. (1974) J. Virol. 14, 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hale B. G., Randall R. E., Ortín J., Jackson D. (2008) J. Gen. Virol. 89, 2359–2376 [DOI] [PubMed] [Google Scholar]
- 52.García-Sastre A., Egorov A., Matassov D., Brandt S., Levy D. E., Durbin J. E., Palese P., Muster T. (1998) Virology 252, 324–330 [DOI] [PubMed] [Google Scholar]
- 53.Djeu J. Y., Stocks N., Zoon K., Stanton G. J., Timonen T., Herberman R. B. (1982) J. Exp. Med. 156, 1222–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy D. E., Marié I., Smith E., Prakash A. (2002) J. Interferon Cytokine Res. 22, 87–93 [DOI] [PubMed] [Google Scholar]
- 55.Grandvaux N., tenOever B. R., Servant M. J., Hiscott J. (2002) Curr. Opin. Infect. Dis. 15, 259–267 [DOI] [PubMed] [Google Scholar]
- 56.Huet S., Avilov S. V., Ferbitz L., Daigle N., Cusack S., Ellenberg J. (2010) J. Virol. 84, 1254–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carr S. M., Carnero E., García-Sastre A., Brownlee G. G., Fodor E. (2006) Virology 344, 492–508 [DOI] [PubMed] [Google Scholar]
- 58.Hatta M., Gao P., Halfmann P., Kawaoka Y. (2001) Science 293, 1840–1842 [DOI] [PubMed] [Google Scholar]
- 59.Shinya K., Hamm S., Hatta M., Ito H., Ito T., Kawaoka Y. (2004) Virology 320, 258–266 [DOI] [PubMed] [Google Scholar]
- 60.Kobasa D., Jones S. M., Shinya K., Kash J. C., Copps J., Ebihara H., Hatta Y., Kim J. H., Halfmann P., Hatta M., Feldmann F., Alimonti J. B., Fernando L., Li Y., Katze M. G., Feldmann H., Kawaoka Y. (2007) Nature 445, 319–323 [DOI] [PubMed] [Google Scholar]
- 61.Guilligay D., Tarendeau F., Resa-Infante P., Coloma R., Crepin T., Sehr P., Lewis J., Ruigrok R. W., Ortin J., Hart D. J., Cusack S. (2008) Nat. Struct. Mol. Biol. 15, 500–506 [DOI] [PubMed] [Google Scholar]
- 62.Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway CA., Jr. (1998) Mol. Cell. 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 63.Takeuchi O., Akira S. (2002) Curr. Top. Microbiol. Immunol. 270, 155–167 [DOI] [PubMed] [Google Scholar]
- 64.Li H. M., Fujikura D., Harada T., Uehara J., Kawai T., Akira S., Reed J. C., Iwai A., Miyazaki T. (2009) Cell Death Differ. 16, 1615–1621 [DOI] [PubMed] [Google Scholar]