SUMMARY
Distal cis-regulatory elements play essential roles in the T lineage-specific expression of cytokine genes. We have mapped interactions of three transacting factors – NF-κB, STAT4 and T-bet – with cis elements in the Ifng locus. We find that RelA is critical for optimal Ifng expression and is differentially recruited to multiple elements contingent upon T cell receptor (TCR) or interleukin-12 (IL-12) plus IL-18 signaling. RelA recruitment to at least four elements is dependent on T-bet-dependent remodeling of the Ifng locus and co-recruitment of STAT4. STAT4 and NF-κB therefore cooperate at multiple cis elements to enable NF-κB–dependent enhancement of Ifng expression. RelA recruitment to distal elements was similar in Th1 and Tc1 effector cells, although T-bet was dispensable in CD8 effectors. These results support a model of Ifng regulation in which distal cis-regulatory elements differentially recruit key transcription factors in a modular fashion to initiate gene transcription induced by distinct activation signals.
INTRODUCTION
Specification of distinct effector T cell lineages from naïve T cell precursors is controlled by transcriptional networks that are activated by cytokine signals received in concert with antigen recognition. T-bet, Gata-3, and RORγt and RORα, and specific members of the STAT family, among others, are key transcription factors that are recruited by cytokine signals to regulate T helper 1 (Th1), Th2 and Th17 cell differentiation programs (Afkarian et al., 2002; Ivanov et al., 2006; Kaplan et al., 1996; Shimoda et al., 1996; Szabo et al., 2000; Yang et al., 2008; Zheng and Flavell, 1997). The generation of functionally mature effector T cells is contingent, in turn, upon lineage-specific remodeling of cytokine gene loci that is orchestrated by the interplay of trans-acting factors with cis-regulatory elements (Agarwal and Rao, 1998; Lee et al., 2006; Wilson et al., 2009). However, a detailed understanding of the interactions between these transacting molecules and the cis-regulatory elements to which they bind to regulate transcription of effector cytokines remains to be defined.
Production of IFN-γ in response to antigen re-encounter represents an innovation of Th1 cells of the adaptive immune system. However, Th1 cells can also secrete IFN-γ in a TCR–independent fashion in response to synergistic actions of the cytokines IL-12 and IL-18, akin to cytokine-driven production of IFN-γ by NK and NKT cells (Berenson et al., 2004). Although these pathways have evolved to complement each other in the generation of a successful Th1 cell immune response, they differ in several respects. The amplitude and kinetics of IFN-γ production induced by TCR versus cytokine signaling are distinct, and the utilization of several transcription factors, including NFAT, STAT4, and T-bet, differ in their contributions to these two pathways (Yang et al., 1999; Yang et al., 2001). More widely expressed transcriptional activators, such as AP-1 and NF-κB, appear to play roles in enhancing Ifng gene transcription in response to both TCR-dependent and TCR-independent activation (Robinson et al., 1997; Sica et al., 1997). Despite extensive characterization of the signaling cascades activated by these pathways, relatively little is known regarding interactions between these factors and cis-regulatory elements of the Ifng locus that might control stimulus-specific transcription of Ifng (Wilson et al., 2009).
Several functionally distinct distal regulatory elements have been identified and characterized in the Th2 cytokine gene cluster, including multiple enhancers, a silencer element (HSIV) and a locus control region that coordinate expression of the Il4, Il5 and Il13 genes (Ansel et al., 2006; Lee et al., 2006). Analogous studies to identify distal regulatory elements that impact Ifng gene transcription are relatively nascent. Initial DNase I mapping identified three hypersensitive sites within introns of the Ifng gene (Agarwal and Rao, 1998). Despite their intrinsic enhancer activities, transgenic analysis indicated that these introns were insufficient to confer lineage-specific expression of IFN-γ (Soutto et al., 2002). Subsequent analysis using a BAC transgene that contained ~191kb flanking the human IFNG gene successfully recapitulated lineage-specific expression of human IFN-γ in murine effector T cells (Soutto et al., 2002). Further, transgenic reporter mice that incorporated ~160kb surrounding the murine Ifng gene display lineage-specific transcription of a reporter molecule, Thy1.1 (Harrington et al., 2008; Hatton et al., 2006). Collectively, these studies have strongly affirmed essential roles for distal regulatory elements in regulating lineage-specific expression of IFN-γ.
Comparative genomics has emerged as a powerful tool to identify putative distal regulatory elements (Loots et al., 2000), and has advanced characterization of the Ifng locus (Hatton et al., 2006; Schoenborn et al., 2007; Sekimata et al., 2009; Shnyreva et al., 2004). To date, nine evolutionarily conserved non-coding sequences (CNS) have been identified within ~120kb flanking the murine Ifng locus (Hatton et al., 2006; Lee et al., 2004; Schoenborn et al., 2007; Shnyreva et al., 2004). Of these, CNSs -34, -22 and -6 have drawn attention as T-bet–responsive elements that markedly impact Ifng gene transcription (Hatton et al., 2006; Lee et al., 2004; Shnyreva et al., 2004). In a previous report, we used a BAC-transgenic model to demonstrate that one of these elements, CNS-22, plays an obligatory role in driving Ifng gene transcription in both effector T cells and NK cells (Hatton et al., 2006). Two recent studies identified CTCF-dependent boundary elements that insulate the IFNG and Ifng loci from neighboring gene loci (Hadjur et al., 2009; Sekimata et al., 2009). Using chromosome conformation capture, Th1-specific, T-bet–dependent interactions between multiple Ifng CNSs and the Ifng gene itself were identified, indicating that these distal elements employ chromosomal looping to transactivate promoter-driven gene expression (Sekimata et al., 2009). Although these recent studies have assigned broad functional attributes to other Ifng CNSs, their precise functions remain unknown (Chang and Aune, 2005, 2007; Schoenborn et al., 2007).
Here, we have mapped the chromatin state of the extended Ifng locus prior to and after Th1 and Th2 cell differentiation and have carried out analyses of multiple distal regulatory elements that impact Ifng gene transcription under conditions of TCR versus cytokine induced signaling. We demonstrate that key distal cis-regulatory elements in the Ifng locus become permissive upon Th1 cell differentiation whereas repressive chromatin remodeling of this locus during Th2 cell differentiation limits accessibility to these elements. Th1 differentiation is accompanied by progressive recruitment of key transcription factors to distal elements that ultimately determine the transcriptional competence of the Ifng locus. Specifically, we show that Ifng CNSs -54, -34, -22, +40, +46 and +54 are NF-κB consensus sequence-containing elements that modulate Ifng gene transcription through differential recruitment of RelA in response to TCR versus cytokine induced signaling. Further, we have delineated specific roles for T-bet and STAT4 in positively modulating the functions of these NF-κB response elements. Taken together, our study provides new insights into the dynamics between distal cis-regulatory elements and the trans-acting factors they recruit to dictate lineage- and stimulus-specific Ifng gene transcription.
RESULTS
Long-range DNase-chip mapping of the Ifng locus in naïve and effector T cells
Recent studies have used global genome alignment tools to identify multiple conserved, non-coding sequences (CNSs) as candidate cis-regulatory elements in the Ifng locus (Frazer et al., 2004; Hatton et al., 2006; Schoenborn et al., 2007). To determine which CNSs correspond to functional regulatory elements in T cell subsets, we employed a microarray based approach (DNase-chip) (Crawford et al., 2006) to identify sites of DNase I hypersensitivity (HS) across an ~780kb region flanking the Ifng gene on mouse chromosome 10. Comparison of naïve CD4+ T cells with polarized Th1 and Th2 cells indicated that the extended Ifng locus undergoes extensive lineage-specific remodeling during the course of effector T cell differentiation (Fig. 1). Notably, lineage-specific HS sites were not identified outside of a region bounded by common HS sites 70kb upstream (HS-70kb) and 66kb downstream (HS+66) of the start site of Ifng transcription, each of which contain consensus CCCTC-binding factor (CTCF) sites and appear to represent boundary sites that delimit cis-elements that control the Ifng locus (Hadjur et al., 2009; Sekimata et al., 2009).
Figure 1. DNase I hypersensitivity map of extended Ifng locus in naïve, Th1 and Th2 cells.
DNase I HS profiles of T helper subsets aligned with a VISTA plot of syntenic regions of the Ifng and IFNG gene loci. DNase–chip analyses were performed on CD4+ T cells from OT-II transgenic mice that were naïve, differentiated under Th1 polarizing conditions (5 d), or differentiated under Th2 polarizing conditions (14 d). Prior to processing, cells were either left unstimulated (“U”), or were restimulated with anti-CD3 plus anti-CD28 (“TCR”) or rIL-12 plus rIL-18 (“IL-12+IL-18”). P, Ifng promoter; CTCF, CCCTC-binding factor site.
In addition to the CTCF-containing boundary sites, two additional HS sites were common to each of the subsets. A major hypersensitivity peak correlated with CNS-22, which was previously identified as an upstream element required for Ifng expression from a BAC transgene (Hatton et al., 2006). An additional HS site was identified within intronic sequence of the Ifng gene, as previously reported (Agarwal and Rao, 1998), although it was more prominent in Th1 cells (Fig. 1). This intronic HS site contains an additional CTCF element, which appears to play role in structural organization of the locus through cooperation with the distal CTCF boundary elements (Sekimata et al., 2009). Hypersensitivity peaks at CNS+19, CNS+40, CNS+46 and immediately upstream of CNS+30 (HS+28) were present in naïve T cells, consistent with possible roles for these sites, in addition to CNS-22, as foci for initiation of lineage-specific remodeling of the Ifng locus during effector T cell differentiation.
Th1 cell differentiation was associated with induction of multiple additional HS sites, including those at the promoter and at CNSs -54, -34, +17, +30 and +54 as well as those at non-CNS sites, HS-40, HS-37 and HS+60. In addition, the intronic HS site and those at +17, +28, and +46 become more sensitive to DNase I after Th1 cell differentiation. At this stage of Th1 cell development, although the Ifng locus is poised for gene transcription, high-level transcription is activation-dependent. After TCR activation or cytokine driven signaling, CNS-34 became hypersensitive and other major Th1 cell-specific sites at CNSs -54 and +54 also became more hypersensitive to DNase I. The stimulus-dependent changes in chromatin structure at these sites are consistent with their function as important enhancers of Ifng gene transcription.
While Th1 cell differentiation is accompanied by favorable epigenetic changes across the extended Ifng locus, the silencing of Ifng transcription that accompanies Th2 cell differentiation is reflected in acquired resistance to DNase I digestion at CNSs+19, +40 and +46, and HS+28, each of which is accessible in naïve CD4+ T cells. Several sites became hypersensitive in Th2 cells, but to a lesser extent than observed in Th1 cells. Importantly, CNSs -54, -34 and +54 were completely resistant to DNase I digestion in Th2 cells. These results identify candidate cis-regulatory elements likely to play important roles in regulating Ifng gene transcription in effector T cells and other IFN-γ–producing cells.
RelA is required for optimal acute transcription of the Ifng gene
Activation of Th1 cells or effector CD8+ T cells (Tc1) by either TCR or IL-12+IL-18 signaling induces Ifng transcription, albeit with different kinetics and recruitment of distinct signaling cascades (Sweetser et al., 1998; Yang et al., 1999; Yang et al., 2001). Activation of NF-κB occurs downstream of both TCR-dependent and -independent pathways (Robinson et al., 1997; Schoenborn et al., 2007; Schulze-Luehrmann and Ghosh, 2006). While several studies have examined the importance of NF-κB activation in Ifng gene transcription (Corn et al., 2003; Corn et al., 2005), the precise contribution of individual family members has remained unclear. Specifically, a possible role for RelA has not been critically examined, primarily due to embryonic lethality of RelA-deficient mice (Beg et al., 1995).
To address this, mice with a conditional deletion of RelA in T cells were generated through crosses of mice with a floxed Rela allele (Relafl/fl) with mice that express cre recombinase under control of the Cd4 locus (Cd4-cre) (Lee et al., 2001; Steinbrecher et al., 2008). Relafl/fl.Cd4-cre+ mice were produced at expected Mendelian ratios, and their deficiency of RelA was confirmed by both transcript analysis and by immunoblotting (Fig. S1). Consistent with previous studies (Steinbrecher et al., 2008), effector T cells that lacked RelA show increased activation of c-Rel (Fig. S1). Relafl/fl.Cd4-cre+ mice showed no differences in numbers of CD4+ and CD8+ T cells in peripheral lymphoid tissues (data not shown), and their T cells had similar proliferative responses to those of littermate controls (Fig. 2A). However, production of IFN-γ by RelA-deficient Th1 and Tc1 cells was compromised in response to both TCR-dependent and IL-12+IL-18–dependent signaling, with substantially greater decrements of IFN-γ found in RelA-deficient Th1 cells. Notably, expression of other Th1 cell genes, including Tbx21, Hlx, Runx3 and the receptors for IL-12 and IL-18 (Il12rb1, Il12rb2 and Il18r), was not compromised by loss of RelA (Fig. S1). Further, repression of reciprocally expressed gene loci of non-Th1 cell lineages, including Il4, Il17a and Foxp3 was intact in RelA-deficient Th1 cells, and there was no significant alteration in expression of Il21 or Il10 relative to WT controls (data not shown, and Fig. S1). Collectively, these results indicate that whereas RelA is dispensable for antigen-driven proliferation, it is indispensable for optimal induction of Ifng gene transcription, particularly in Th1 cells.
Figure 2. RelA is essential for acute induction of Ifng gene expression.

(A) CD4+ and CD8+ T cells from Rela fl/+.Cd4-cre− or Rela fl/fl.Cd4-cre+ mice were isolated and labeled with CFSE. CD4+ T cells were cultured under Th1 cell conditions for 5 d (upper panel), and CD8+ T cells were cultured under Tc1 condition for 3 d (lower panel). Recovered T cells were reactivated as indicated and intracellular IFN-γ was assessed by flow cytometry. Plots were gated on live CD4+ or CD8+ T cells. Numbers in each plot indicate the percentage of the gated cells in each quadrant. (B) Th1 cells were incubated for 1 hour with indicated concentrations of the RelA inhibitory peptide, (Ser 276), then activated with anti-CD3 plus anti-CD28 for 4 h and analyzed for intracellular IFN-γ. Dot plots are gated on live CD4+ T cells, and numbers indicate the percentage of viable CD4+ T cells in the indicated subgates. (C) Th1 cells were incubated for 1 hour with 150 μM of the inhibitory RelA peptide (lower panel) or control peptide (upper panel), then activated with the indicated stimuli for 4 h and analyzed for intracellular cytokines. Plots are gated on live CD4+ T cells; numbers indicate the percentage of viable CD4+ T cells in the indicated subgates. (D) Comparison of mean fluorescence intensities (MFI) of IFN-γ+ cells from (C). (E) Th1 cells were activated with IL-12 with or without IL-18, following 1 h pre-incubation with RelA inhibitory peptide or the indicated controls, and nuclear translocation of RelA was assesed by immunoblotting. Cyclophilin B was used as loading control. All data are representative of at least two independent experiments.
To determine whether impaired IFN-γ production by RelA-deficiency was due to impairment of effector cell development or impaired acute signaling, additional experiments were performed in which RelA signaling was specifically blocked after differentiation of WT Th1 cells. In the presence of a peptide that is a RelA phosphorylation decoy ((peptide Ser 276) (Derossi et al., 1994; Takada et al., 2004)), there was a dose-dependent inhibition of TCR-driven IFN-γ expression by Th1 cells (Fig. 2B), which was associated with inhibition of RelA nuclear translocation (Fig. 2C). Both TCR- and IL-12+IL-18-driven induction of IFN-γ were markedly compromised, as was IFN-γ induction by synergistic actions of IL-12+IL-18 and antigen receptor activation (Fig. 2D). Under all conditions examined, pre-incubation with the RelA inhibitory peptide led to decreases in both the frequency of IFN-γ+ cells and single-cell expression of IFN-γ (Fig. 2E), and could not be ascribed to cell toxicity (data not shown). Thus, RelA plays an important role in acute transcription of Ifng gene expression in Th1 cells whether induced by either TCR or IL-12+IL-18 signaling.
RelA is differentially recruited to cis-regulatory elements by TCR-dependent and -independent activation
Previous studies have demonstrated recruitment of NF-κB subunits to the Ifng promoter and intronic sites (Sica et al., 1997; Tato et al., 2006). However, analysis of NF-κB binding to more distal elements has not been reported, and promoter binding alone may not account for defects in Ifng transcription (Corn et al., 2003; Corn et al., 2005). We therefore evaluated whether distal elements defined by comparative genomic and DNase-chip analyses might bind NF-κB under conditions of active Ifng gene transcription. Sequence scans for consensus NF-κB binding sequences were performed and compared to CNSs and DNase HS sites (Fig. 1). Although numerous potential NF-κB consensus sites were identified across the Ifng locus, relatively few of these coincided with identified CNSs or HS sites (Fig. S2, and data not shown). Thus, in addition to NF-κB sites previously described (Sica et al., 1997; Tato et al., 2006), highly conserved NF-κB consensus sites were identified in multiple distal CNSs, including CNSs -54, -22, -6, +17, +40, +46 and +54, as well as a tandem site identified in CNS-34 (Fig. S2).
To screen for functional binding sites for RelA, we initially performed ChIP-chip analysis of Th1 cells without re-stimulation (“resting”), or after activation with IL-12+IL-18, anti-CD3 and anti-CD-28, or PMA plus ionomycin (Fig. S3, and data not shown). Binding sites identified from these analyses were restricted to previously identified CNS or HS sites (Fig. 1), so further studies were performed using conventional ChIP–RT-PCR analysis of these sites (Fig. 3). Upon TCR activation, significant RelA binding was detected at three CNSs: CNS-34, CNS-54 and CNS+40 (Fig. 3A). Comparable RelA recruitment was observed at each of these sites in response to stimulation by either anti-CD3 plus anti-CD28 or the TCR signaling surrogate, PMA plus ionomycin (Fig. 3A). In some experiments, RelA binding achieved significance at other sites (eg, CNSs +17–19 and +54), but this was less prominent and varied between experiments and activation conditions. Modest binding at the Ifng promoter was seen in some experiments, but did not achieve statistical significance.
Figure 3. Differential recruitment of NF-κB p65 to distinct Ifng CNSs.
Th1 cells that were either left unstimulated (“resting”), stimulated for 4 hours with 50ng/ml PMA plus 750 ng/ml Ionomycin or with anti-CD3 plus anti-CD28 (A), or restimulated with 10 ng/ml rIL-12 plus 25 ng/ml rIL-18 (B) were processed for ChIP using an NF-κB p65-specific antibody, and analysis of immunoprecipitated DNA was performed using real-time PCR with primer sets designed to detect DNA at the indicated CNS elements (-55, -34, -22, -5, +18–20, +29, +40, +46 and +55) and the Ifng promoter (Pro). The Il2 promoter (Il2 Pro) and 16S ribosomal protein (16Srp) promoter were used as positive and negative controls, respectively. Values were normalized against input DNA and expressed relative to resting Th1 cells, which was <0.05% of input DNA. Data represent Mean ± SEM from at least three independent experiments. Statistical significance is relative to binding at 16Srp under the same conditions (* p<0.01).
Activation of Th1 cells with IL-12+IL-18 induced a different pattern of RelA recruitment (Fig. 3B). CNS-34 retained prominent RelA binding, and there was strong recruitment to three additional distal sites that were not consistently detected following TCR-dependent signaling: CNS-22, CNS+46 and CNS+54. No significant binding was detected at CNS-54. In agreement with the ChIP-chip data (Fig. S3), RelA bound two additional CNSs (ie, CNS+30 and CNS+40), albeit substantially weaker than observed for the four other binding sites. Again, modest, variable RelA binding was detected at the Ifng promoter, but did not achieve statistical significance, despite a significant peak of binding at the promoter in the ChIP-chip analyses. No RelA recruitment was detected outside of mapped DNase I HS sites (see Fig. 1). Collectively, these results establish that RelA is differentially recruited to distinct binding sites to drive Ifng gene transcription in response to distinct stimuli. Based on the high degree of cross-species conservation of NF-κB binding sites of the six distal cis-elements that bind RelA most prominently in Th1 cells (i.e., CNSs -54, -34, -22, +40, +46 and +54), it would appear that coordinated, inducible recruitment of RelA to multiple distal elements in the Ifng locus is an evolutionarily conserved mechanism to drive Ifng gene transcription.
RelA recruitment induced by IL-12 plus IL-18 is STAT4-dependent
Given the differential recruitment of RelA to distal CNS elements by TCR- versus cytokine-driven signaling, we speculated that other transcription factors that are differentially activated by these pathways might regulate usage of distinct NF-κB response elements in the Ifng locus. Because IL-12-induced STAT4 activation is critical to cytokine-driven, but not TCR-driven activation of Ifng transcription, it was plausible that STAT4 might cooperate with IL-18-induced NF-κB under conditions of cytokine-driven activation. This possibility was supported by the proximity of evolutionarily conserved NF-κB and STAT binding sites, particularly in CNSs -22, -34 and +46 (Fig. S2). We therefore evaluated whether the observed binding of RelA to Ifng CNSs was dependent on coordinate IL-12-induced STAT4 activation.
Although IL-18 induced NF-κB activation is independent of IL-12 (Yang et al., 1999), and the target NF-κB sites in Th1 cells are accessible (Fig. 1), IL-18 signaling alone failed to induce significant RelA recruitment compared to that induced by combined activation with IL-18+IL-12 (Fig. 4A). Thus, although there were modest increases in binding detected at several sites after stimulation with IL-18 alone (eg, CNS+46 and the Ifng promoter), these were not significant, and were substantially reduced relative to binding induced by IL-18+IL-12. Accordingly, RelA recruitment by IL-18+IL-12 was significantly impaired in Th1-polarized CD4+ T cells from Stat4−/− mice (Fig. S4). These results are consistent with a role for IL-12-induced activation of STAT4 for recruitment of IL-18-induced RelA to the Ifng locus.
Figure 4. Co-recruitment of STAT4 and RelA at multiple cis elements that drive Ifng gene transcription.
Th1 cells were either left unstimulated or stimulated as indicated for 4 h and ChIP was performed for the indicated CNS sites using antibodies specific for NF-κB p65 (A) or STAT4 (B). Relative RelA binding (A) was calculated as described in Fig. 3. Data are representative of at least three independent experiments (* p<0.01 for Th1 samples reactivated with IL-12+IL-18 versus IL-18 alone). (B) Recruitment of STAT4 was calculated as percentage of input DNA. Data represent mean ± SEM from at least three independent experiments. (# p<0.05 versus STAT4 recruitment in resting Th1 samples; ** p<0.01 versus recruitment of STAT4 to 16Srp in resting Th1 cells).
While signaling downstream of the IL-12 and IL-18 receptors may converge and synergize at multiple levels, the proximity of STAT and NF-κB binding sites in several of the CNSs led us to evaluate whether STAT4 and RelA might be coordinately recruited to these sites. We therefore surveyed the extended Ifng locus for STAT4 binding sites by ChIP-chip analysis of Th1 cells (Fig. S3), followed by confirmatory ChIP–RT-PCR analyses (Fig. 4B).
Upon activation of Th1 cells with IL-12+IL-18, we observed robust recruitment of STAT4 to both CNS-22 and CNS+46 (Fig. 4B), with lesser, but significant binding to CNSs -34, +30, +40 and +54, and the promoter. In contrast to STAT4–dependent recruitment of RelA, STAT4 recruitment to the Ifng locus in response to IL-12+IL-18 was not strictly RelA-independent (Fig. S4). Thus, with the exception of CNS-34 and CNS+54, STAT4 binding was independent of NF-κB, as comparable binding was observed in the absence of IL-18 signaling (Fig. S4A) and in RelA-deficient T cells (Fig. S4B). No significant STAT4 binding was detected at CNS-34 or CNS+54 in response to IL-12 signaling alone, despite the requirement for IL-12 for optimal RelA binding to these sites. CNS-54 lacks a STAT consensus sequence (data not shown), suggesting that localization of STAT4 to this site might be indirect and perhaps independent of direct STAT4-DNA interactions. Notably, we detected significant binding of STAT4 to CNS+46 in resting Th1 cells, prior to stimulation by cytokines (Fig. 4B), suggesting that STAT4 might be poised on this site even in the absence of on-going IL-12 signaling. These results establish that RelA binding to target elements in the Ifng locus is substantially enhanced by concurrent STAT4 binding, and suggest that NF-κB complexes that contain RelA might require STAT4 as a chaperone for binding to the majority of sites in the Ifng locus.
RelA recruitment to the Ifng locus is T-bet-dependent in Th1 but not Tc1 cells
The T-box family members, T-bet and Eomes, play redundant and non-redundant roles in effector T cell differentiation and cell fate (Intlekofer et al., 2008; Intlekofer et al., 2005; Pearce et al., 2003). Whereas production of IFN-γ by cytotoxic T cells is independent of T-bet, T-bet is required for optimal production of IFN-γ in Th1 cells (Szabo et al., 2000). T-bet has been shown to regulate Ifng gene transcription in concert with several other transcription factors, including NFAT, Runx3, and Hlx (Djuretic et al., 2007; Lee et al., 2004; Mullen et al., 2001), and was more recently shown to cooperate with the architectural factor, CTCF, to restructure the Ifng locus for active transcription (Sekimata et al., 2009). In addition to its role in locus remodeling, T-bet also functions as an enhancer through its interactions with several CNS elements (Chang and Aune, 2005; Hatton et al., 2006; Shnyreva et al., 2004). In view of sequence scans that identify evolutionarily conserved composite T-box and NF-κB consensus sites in these elements (Fig. S2), and our findings that RelA interacts with these sites and additional CNS elements identified herein, we examined whether recruitment of RelA to CNSs in the Ifng locus is T-bet–dependent.
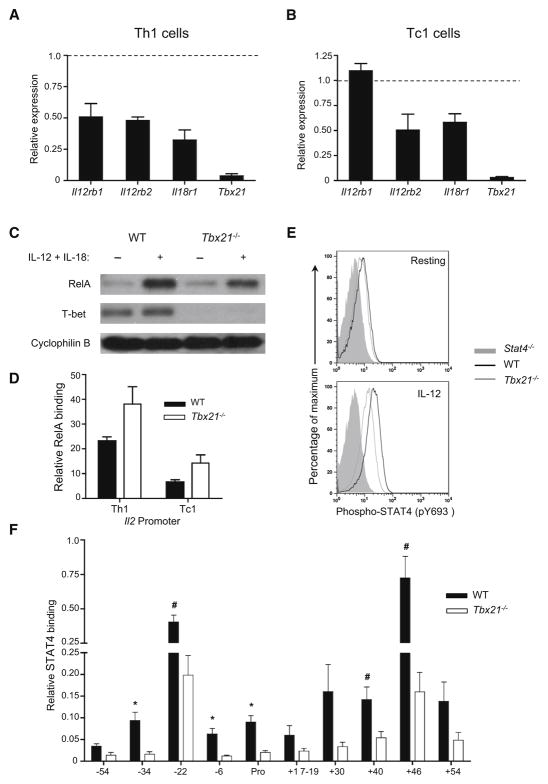
Th1 and Tc1 were derived by polarization of CD4+ or CD8+ T precursors from WT or Tbx21−/− (T-bet–deficient) mice, and RelA ChIP–RT-PCR analyses were performed after stimulation with PMA+ionomycin or IL-12+IL-18 (Fig. 5). Notably, the pattern of RelA recruitment to CNSs in Tc1 cells mirrored that of Th1 cells under both conditions of activation (Fig. 5B,D). Like Th1 cells, Tc1 cells also demonstrated differential utilization of CNSs contingent upon the stimulus. However, consistent with the disparate requirement for T-bet in Th1 and Tc1 cells, inducible recruitment of RelA was selectively impaired in T-bet–deficient Th1 cells (Fig. 5B,D); RelA recruitment to distal CNSs in Tc1 cells was independent of a requirement for T-bet, irrespective of the mode of activation. Thus, T-bet’s role in the recruitment of RelA to the Ifng locus is CD4+ T cell-specific (see also, Fig. S6).
Figure 5. RelA recruitment is dependent on T-bet in Th1 cells but not in Tc1 cells.
(A),(C) Cytokine production by Th1 and Tc1 cells generated from WT or Tbx21−/− mice was evaluated by intracellular cytokine staining. Flow cyometry plots are gated on live CD4+ (A) or CD8+ (B) T cells. Percentage values shown indicate frequencies of cells positive for the indicated cytokines. (B),(D) ChIP was performed for NF-κB p65 and enrichment was calculated as described in Fig. 3 following restimulation with PMA and ionomycin (top panel) or IL-12 plus IL-18 (bottom panel). Data are representative of at least three independent experiments. T cells from Tbx21−/− or WT mice were polarized under Th1 (B) or Tc1 (D) conditions and restimulated prior to ChIP. * P < 0.01, # P<0.05 versus Tbx21−/− samples.
Since RelA recruitment in T-bet–deficient Th1 cells was impaired in response to both TCR- and cytokine-dependent signals, we speculated that T-bet might play a Th1-specific role in the epigenetic remodeling of key distal elements that regulate Ifng gene transcription. Previous studies demonstrated that Th1 cell differentiation is associated with increased acetlyation of histone H4 and methylation of lysine 4 on histone H3 (H3K4), and decreased trimethylation of lysine 27 on histone H3 (H3K27) across the Ifng locus (Schoenborn et al., 2007; Wei et al., 2009). Consistent with previous findings (Chang and Aune, 2005), we observed that T-bet is dispensable for the changes in acetylation of histone H4 at sites across the Ifng locus, both in Th1 and Tc1 cells (Fig. S5). In contrast, methylation of H3K4, which is also associated with a permissive chromatin state, was different between Th1 and Tc1 cells. There was Th1 cell-specific impairment of H3K4 methylation at CNS-34 in the absence of T-bet (Figs. 6A). Additionally, Tbx21−/− Th1 cells showed significantly decreasd H3K4 methylation at CNS-6 and the Ifng promoter compared to WT controls (Fig. 6A, upper panel). Interestingly, there was increased H3K4 methylation at CNS-22 in the absence of T-bet. In contrast, T-bet was dispensable for H3K4 methylation at all sites examined in Tc1 cells, including CNS-34 (Figs. 6B, upper panel); no significant decreases in H3K4 methylation were evident at any of the Ifng CNSs examined in Tc1 cells, and, in general, methylation of H3K4 at most of the CNSs was substantially higher in Tc1 cells (Fig. S5).
Figure 6. Remodeling of the Ifng locus in Th1 cells is T-bet–dependent.
ChIP was performed on the indicated T cell effectors with antibodies specific for mono,di,tri-methylated H3K4 (top panels) or trimethylated H3K27 (bottom panels). CD4+ T cells (A) or CD8+ T cells (B) from wildtype or Tbx21−/− mice were activated under Th1- or Tc1 polarizing conditions respectively, then subject to ChIP. * P < 0.01, # P<0.05 versus Tbx21−/− Th1 samples. Values were normalized against the 16S ribosomal protein promoter, which was assigned a value of 1. Data are representative of at least three independent experiments.
Deficits in H3K4 methylation observed in Tbx21−/− Th1 cells, which were limited to the promoter and upstream CNS elements, were associated with more extensive increases in repressive H3K27 methylation, which extended broadly both upstream and downstream of the Ifng gene (Fig. 6A, lower panel). Thus, in comparison to wild-type controls, Tbx21−/− Th1 cells had significant increases in H3K27 trimethylation at CNSs -54, -34, -6, +30, +40 and +46, and at the promoter. In contrast, no significant changes in repressive H3K27 methylation were observed in Tc1 cells. These findings suggest that T-bet is specifically required to remodel cis-regulatory elements across the extended Ifng locus in Th1 cells, with removal of repressive histone modifications as a basis for remodeling downstream regulatory elements that are required for both TCR-dependent and -independent binding of RelA-containing NF-κB complexes.
Activation of STAT4 and RelA in Tbx21−/− T cells
In view of the critical role for T-bet in regulating expression of key receptors upstream of NF-κB and STAT4 signaling, we determined whether decreased expression of inducible components of the IL-12 and IL-18 receptors in Tbx21−/− Th1-polarized cells might explain the defective chromatin re-structuring of the Ifng locus in this population. Although transcript levels of Il12rb2 and Il18r1 were decreased in both Th1 and Tc1 cells derived from T-bet–deficient mice, there remained substantial expression of both receptors relative to WT naïve T cells (Figs. 7A,B, and data not shown). Accordingly, whereas nuclear translocation of RelA in response to IL-12+IL-18 stimulation in Tbx21−/− Th1-polarized cells was reduced, there remained considerable nuclear accumulation of RelA downstream of the IL-18 receptor (Fig. 7C). In response to TCR signaling in contrast to the Ifng locus, recruitment of RelA to the Il2 promoter was substantially enhanced (Fig. 7D). Similarly, despite decreased expression of IL-12Rβ2 in Tbx-21−/− Th1-polarized cells, substantial IL-12-induced signaling was retained, as indicated by the significant induction of STAT4 phosphorylation (Fig. 7E). Nevertheless, while there was retention of some STAT4 binding to selected CNS elements in Tbx21−/− Th1 cells (i.e., CNS-22 and CNS+46), this was significantly suppressed at all STAT4 binding sites identified in the Ifng locus (Fig. 7F, and see Fig. 4, above). Thus, the defects in RelA recruitment that result from T-bet deficiency appear to be due more to diminished recruitment of STAT4 to the poorly remodeled Ifng locus than a deficit of activated RelA.
Figure 7. Impaired IL-12 and IL-18 signaling do not account for diminished RelA and STAT4 recruitment to the Ifng locus in T-bet–deficient Th1 cells.
(A),(B) mRNA transcripts of the indicated genes were quantitated in Th1- and Tc1-polarized cells generated from WT or Tbx21−/− mice. Expression was normalized against β2-microglobulin, and relative expression compared to wildtype controls (dashed lines; assigned value of “1.00”). Data represent mean ± SEM from three independent experiments. (C) Th1-polarized cells derived from WT or Tbx21−/− mice were restimulated with IL-12 + IL-18 for 4 h and nuclear translocation of p65 evaluated by immunoblotting, using cyclophilin B as a loading control. Blots were re-probed for T-bet as a control for T-bet expression. (D) RelA ChIP was performed on indicated T cell populations and relative recruitment at the Il2 promoter was assessed as in Fig 3. Data represent mean ± SEM from at least three independent experiments. (E) Th1-polarized cells from WT, Tbx21−/− or Stat4−/− mice were left untreated (resting) or reactivated (IL-12) and phosphorylation of tyrosine residue 693 (pY693) of STAT4 was evaluated by flow cytometry. Histograms are gated on viable CD4+ T cells. (F) STAT4 ChIP was performed following IL-12+IL-18 activation of Tbx21−/− and WT Th1-polarized cells, as in Fig. 4. Data are represented as fractions of input DNA and represent mean ± SEM from at least three independent experiments (*p<0.01, #P<0.05; stimulated WT versus stimulated Tbx21−/−).
DISCUSSION
In this study, we have extended the functional map of the Ifng locus through delineation of interactions between evolutionarily conserved cis-regulatory elements and key transacting factors that dictate lineage- and stimulus-specific transcription of Ifng in Th1 and Tc1 cells. We identify RelA as a central Rel-NF-κB family member that controls Ifng transcription through interactions with six major CNSs in the Ifng locus (CNSs -54, -34, -22, +40, +46 and +54), and show that these sites are differentially utilized in response to TCR- or cytokine-driven mechanisms. Recruitment of RelA to key cis elements induced by cytokine-driven Ifng transcription was found to be coincident with, and dependent upon, coordinate recruitment of IL-12–induced STAT4, whereas STAT4 binding to the same sites was largely independent of IL-18–induced NF-κB. Thus, acute STAT4 binding to accessible sites in the Ifng locus appears to enhance Ifng transcription, at least in part, by chaperoning the recruitment of RelA-containing NF-κB complexes.
In previous studies, it was established that NF-κB signaling is critical to Th1 cell development and function (Corn et al., 2003). The canonical, or classical pathway of NF-κB activation, which is activated by downstream of the TCR and IL-18 signaling, has been primarily linked to homo- or heterodimers of RelA, c-Rel and NF-κB p50 (Vallabhapurapu and Karin, 2009). While it has been speculated that the functions of c-Rel and RelA are at least partially redundant, studies of c-Rel–deficient mice have identified several instances where RelA cannot compensate for absence of c-Rel (Hayden et al., 2006). In particular, c-Rel plays an obligatory role in the induction of IL-2 and antigen-driven proliferation of T cells (Liou et al., 1999, Kontgen et al., 1995), establishing a requirement for c-Rel in the differentiation of naïve precursors, but also complicating analysis of its function in effector T cells by limiting their development (refs. (Hilliard et al., 2002; Kontgen et al., 1995; Liou et al., 1999; Mason et al., 2004), and data not shown). Herein, we find that T cells deficient for RelA had minimal impairment of proliferation, but instead demonstrated profound defects in Ifng transcription that could not be rescued by c-Rel, whether driven by the TCR-dependent or TCR-independent pathway. Thus, c-Rel and RelA have distinct, non-redundant roles in effector T cell biology.
In studies to identify NF-κB family members that complex with RelA in the Ifng locus, we found that binding of NF-κB p50 (NF-κB1), the most common partner for RelA (Vallabhapurapu and Karin, 2009), was primarily limited to CNS-22 — both in unactivated or activated Th1 cells (Fig. S6). Accordingly, RelA-p50 heterodimers are not the major CNS-binding NF-κB complex in the Ifng locus, in agreement with a previous study of Nfkb1−/− mice, in which Th1-polarized cells had unimpaired Tbx21 and Ifng expression (Das et al., 2001). Although we were unable to identify binding of other NF-κB family members to the Ifng locus, this could reflect the unavailability of good antibodies for ChIP analysis of these factors (data not shown). Because RelA does not typically partner with NF-κB p52 (NF-κB2), we infer that with the exception of CNS-22, RelA might bind as a homodimer to most, if not all, of the other RelA sites in the Ifng locus, although a contribution of complexes with c-Rel and RelB cannot be absolutely excluded (Corn et al., 2005). Alternatively, RelA has been reported to heterodimerize with a phophorylated variant of T-bet (Hwang et al., 2005). Given previous findings that T-bet constitutively occupies several of the CNS elements identified as RelA binding sites herein (Hatton et al., 2006; Sekimata et al., 2009), RelA recruitment via this mechanism is also a possibility, and will require further study.
The unique occupancy of CNS-22 by NF-κB p50 in both naïve and Th1 cells is notable, in that selective deletion of CNS-22 in our previous study abrogated Ifng reporter expression in a BAC transgenic reporter model (Hatton et al., 2006). This suggests a particularly important role for this element in regulating Ifng transcription and raises the possibility that NF-κB p50 plays a central role in coordinating the function of this element, whether to repress it prior to effector T cell differentiation as a homodimer, or to activate it in the context of RelA recruitment and generation of p50-RelA heterodimers at this site.
Because TCR stimulation does not activate STAT4, RelA recruitment downstream of TCR signaling must occur by STAT4-independent mechanisms. In contrast, IL-12–induced STAT4 activation clearly impacts recruitment of RelA activated by IL-18; at each of the sites of RelA binding following IL-12+IL-18 co-signaling, STAT4 was also bound and was required for RelA binding. Indeed, ChIP-chip analysis revealed that there were few, if any, sites of that bound STAT4 without RelA. While this suggested a possible physical interaction between STAT4 and RelA–containing hetero- or homodimers, an interaction between STAT4 and RelA was not found in the absence of DNA in co-precipitation studies (data not shown). Nevertheless, the coordination of these two factors at multiple sites in the Ifng locus indicates that cooperative binding is essential for optimal Ifng expression and provides a basis for the requirement for coordinate IL-12 and IL-18 receptor signaling. This extends the range of cooperative interactions identified between the STAT and NF-κB transcription factor families downstream of immunomodulatory cytokine receptors (Shen and Stavnezer, 1998; Tran et al., 2010), and suggests that cooperative binding of STAT and NF-κB family members at conserved non-coding sequences might be a common feature of cytokine responsive genes.
An alternative mechanism by which IL-12 and IL-18 cooperate to induce Ifng is via a mechanism that requires de novo synthesis of GADD45β, which acts through MEKK4 to sustain activation of p38 MAPK (Yang et al., 1999; Yang et al., 2001). Taken together with the findings herein, this suggests that sustained activation of the p38 MAPK pathway downstream of IL-12+IL-18-induced expression of Gadd45b might cooperate with more transient recruitment of STAT4 and RelA complexes to generate the pattern of prolonged expression that characterizes Ifng induction by IL-12 and IL-18 (Berenson et al., 2006; Lu et al., 2004). It will be of interest to determine whether factors downstream of the p38 MAPK pathway are recruited to the same CNS elements defined for STAT4 and RelA herein, where they might cooperate with and/or stabilize these complexes, or alternatively, might act at independent cis-regulatory sites to enhance Ifng transcription.
High-level transcription of Ifng is contingent upon epigenetic modifications that remodel the Ifng locus during Th1 differentiation (Mukasa et al., 2010; Wilson et al., 2009). STAT4 and T-bet cooperate in this process (Thieu et al., 2008), although details of the mechanisms by which this occurs, and the sites within the Ifng locus where they might directly interact are only beginning to emerge. A recent study found that both T-bet and STAT4 were necessary for removal of Sin3A-associated HDAC complexes that repress Ifng gene transcription (Chang et al., 2008). Other studies have identified conserved proximal and distal cis-regulatory elements to which T-bet binds (Hatton et al., 2006; Lee et al., 2004; Sekimata et al., 2009; Shnyreva et al., 2004), and with this report, we identify IL-12-dependent STAT4 binding at six CNS elements, and confirm its binding to the promoter (Chang and Aune, 2005; Thieu et al., 2008). With the exception of CNS-54, which binds T-bet in the absence of STAT4, each of the CNS sites that binds T-bet also binds STAT4, suggesting cooperativity between these factors following acute activation of STAT4 by IL-12. Remarkably, however, STAT4 was constitutively bound to CNS-22 and CNS+46, independently of acute Th1 cell activation, and the same sites were unique in their binding of STAT4 in Th1-polarized Tbx21−/− cells. This indicates that STAT4 recruitment to these sites during Th1 differentiation is sustained without a requirement for acute reactivation, and suggests that STAT4 might initially bind these sites without requirement for co-factors, such as T-bet. It is notable that both of these sites are DNase I hypersensitive in naïve T cells, supporting the possibility that CNS-22 and CNS+46 are critical in orchestrating early chromatin remodeling of the Ifng locus, perhaps as sites for recruitment of STAT4 as a “pioneer” factor that initiates this process.
Two recent reports identified orthologous CTCF-containing boundary elements that appear to define limits of the Ifng and IFNG loci (Hadjur et al., 2009; Sekimata et al., 2009), and Sekimata et al. defined an important role for T-bet in acting with these boundary elements and an intronic CTCF site to restructure the Ifng locus so as to approximate distal cis-regulatory elements with the Ifng promoter region (Sekimata et al., 2009). Interestingly, in addition to its reported binding of T-bet in Th1 cells, we find that the downstream CTCF site (HS +66) also binds STAT4 and RelA upon IL-12+IL-18 signaling, suggesting that this site might act as both boundary element and distal enhancer. Here, we provide additional evidence in support of a central role for T-bet in the generation of a transcriptionally competent Ifng locus in CD4+, but not CD8+, effector T cells. In the absence of T-bet, Th1-polarized CD4+ T cells show impairment of H3K4 methylation at CNSs -34, -6 and the Ifng promoter, while exhibiting increases in H3K27 trimethylation at these same sites, in addition to increased repressive marks at CNSs -54, +30, +40, and +46. Notably, with the exception of CNS+40, which is also hypersensitive in activated Th2 cells, each of these sites demonstrates Th1-specific hypersensitivity, suggesting that T-bet regulates their H3K4 methylation and H3K27 demethylation. This is consistent with the recent identification of conserved regions within T-box domains that mediate interactions with H3K4 methyltransferase and H3K27-demethylase (Lewis et al., 2007; Miller et al., 2008). Whether T-bet is sufficient to orchestrate these changes independently of other Th1 lineage factors or effects these histone modifications via direct or indirect mechanisms is unclear, and will require further study. Our finding that T-bet is dispensable for the establishment of favorable H3K4 and H3K27 methylation marks at these sites in CD8+ effector T cells suggests a similar function for the T-box family member Eomes in remodeling the Ifng locus in this lineage (Intlekofer et al., 2008; Pearce et al., 2003).
In conclusion, we find that coordinate and differential recruitment of transcription factors to multiple distal cis-acting elements plays an essential role in Ifng transcription initiated by TCR- or cytokine-induced signaling, in essence defining a modular pattern of utilization of distal transcriptional “units” composed of complexes of trans-acting factors and the cis-acting elements to which they are recruited. Our findings suggest that CNSs -22, +40 and +46, which are accessible prior to Th1 differentiation, might act as multifunctional nodes that participate both in the initiation of early remodeling of the Ifng locus and in acute regulation of Ifng transcription, whereas other distal elements, including CNSs -54, -34, -6 and +54, may be functionally restricted to acute regulation of Ifng transcription subsequent to differentiation-driven locus remodeling. The multiplicity of sites to which STAT and RelA are coordinately recruited in the Ifng locus is remarkable, and suggests that there are advantages to having the option of recruiting graded multiples of distal STAT4/NF-κB units to the proximal promoter, perhaps as a mechanism for rheostat-like control of the rate of Ifng transcription by mass action, as has been found for repetitive cis-elements in simple promoters. Careful deletion analyses using transgenic and gene targeting approaches will be necessary to address these issues going forward.
EXPERIMENTAL PROCEDURES
Mice, antibodies and reagents
C57BL/6, BALB/c, DO11.RAG2−/− TCR transgenic mice and STAT4−/− BALB/c mice were purchased from Jackson laboratory and/or were bred at the University of Alabama at Birmingham. Tbx21−/− BALB/c mice were a gift from Dr. Dan Bullard (UAB). All mice were maintained in accordance with Institutional Animal Care and Use Committee regulations. Antibodies specific for p50 (sc-1150), p65 (sc-372), c-Rel (sc-71) and STAT4 (sc-486) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies specific for acetylated histone H4 (06–598), mono-, di-, or trimethyl H3K4 (04–791; detects all of the three possible methylation states) and trimethylated H3K27 (17–622) were purchased from Millipore (Billerica, MA). Cyclophilin B (ab16045) antibody was purchased from Abcam (Cambridge, MA). Anti-T-bet antibody has been described previously (Hatton et al., 2006). Peptides (IMG-2001, RelA peptide and control) used for inhibiting RelA were purchased from IMGENEX (San Diego, CA). All primers were custom-synthesized by IDT (Coralville, IA).
Isolation of CD4+/CD8+ T cells and generation of Th1/Tc1 effectors
CD4+ T cells were isolated by positive selection using anti-CD4 magnetic DYNA beads (Invitrogen), as per manufacturer’s instructions. For separation of CD8+ T cells, following CD4+ isolation, CD4 negative fraction was labeled with PE labeled anti-CD8 antibody followed by selection with anti-PE microbeads (Miltenyi Biotech; Bergisch Gladbach, Germany) and then fractionated over magnetic separation columns (Miltenyi Biotech). Both Th1 and Tc1 cells were generated by culturing CD4+ and CD8+ T cells with 2.5 μg/ml anti-CD3 antibody and 10 ng/ml recombinant IL-12 (R&D Systems: Minneapolis, MN) and irradiated feeder cells (3000 rads) at a ratio of 1:5. Th1 cultures were additionally supplemented with 10 μg/ml anti-IL-4. In case of OT-II and DO11.10 strains, CD4+ T cells were isolated in a similar manner, but the cells were activated with 5 μg/ml OVA peptide 323–339 instead of anti-CD3. Th1 and Tc1 cultures were split on days 3 and 2 of culture and supplemented with 50 U/ml of recombinant IL-2 (R&D Systems).
Reactivation of Th1 and Tc1 cells
Th1 and Tc1 cultures were restimulated on days 5 and 3 of culture respectively. Restimulation was carried out with 50 ng/ml PMA and 750 ng/ml Ionomycin or 10 ng/ml rIL-12 and 25 ng/ml rIL-18. For TCR restimulation, anti-CD3 antibody was diluted to 10 μg/ml in phosphate buffered saline (PBS, 150mM NaCl, 0.02M Phosphate) and coated overnight at 4°C. The following day the plates were was washed twice with PBS and the media was supplemented with 5 μg/ml of anti-CD28 antibody (eBioscience; San Diego, CA). All conditions involved reactivation of cell for 4 hours, except Tc1 cells, which were restimulated with PMA and Ionomycin for 1hour.
Intracellular cytokine staining
Cells were reactivated as described above for 4 hours in the presence of GolgiStop (BD Biosciences; San Jose, CA). Cells were stained with fluorescent-labeled antibodies against CD4, CD8, IL-4 and IFN-γ using the Cytofix/Cytperm kit (BD Biosciences). Intracellular staining for T-bet was done similarly using FoxP3 staining kit (eBioscience). In both cases, dead cells were excluded by staining with LIVE/DEAD® fixable stain kits (Invitrogen; Carlsbad, CA). Phospho-STAT4 stains were prepared using Phosflow kit (BD Biosciences) as per manufacturer’s protocol. Samples were acquired on an LSRII flow cytometer and analyzed using FlowJo software (Treestar Inc.; Ashland, OR).
Chromatin Immunoprecipitation
All ChIP experiments were carried out using ChIP assay kit (Millipore) as previously described (Mukasa et al., 2010). Relative recruitment assessed by real-time PCR is expressed as 2ΔΔCt or as a percentage of input DNA. Inputs were appropriately diluted to facilitate normalization against input DNA. Primer sequences are listed in the supplement.
DNase-chip
Samples were prepared as previously described (Crawford et al., 2006). Briefly, nuclei were isolated from 5×107 cells and subject to digestion with DNase I (Roche; Basel, Switzerland) concentrations ranging from 0–12 Units. Digestion was stopped using 100mM EDTA and nuclei were embedded in equal volumes of 1% InCert Agarose (Lonza; Basel, Switzerland). The plugs were incubated overnight at 37°C in LIDS buffer (1% lithium dodecyl sulfate, 10 mM Tris-HCl (pH 7.5), 100 mM EDTA) and then washed with 50mM EDTA. Following digestion with T4 DNA polymerase (New England Biolabs; Ipswich, MA), the blunted fragments were extracted, labeled with biotin-labeled linkers, captured and amplified using ligation mediated PCR and hybridized to microarrays. Data obtained from these experiments were visualized using IGB browser (Affymetrix; Santa Clara, CA).
Statistical analyses
Statistical significance was evaluated using two-tailed unpaired t-test. Unless specifically indicated, all p-values <0.05 are considered significant.
Highlights.
NF-κB family member RelA regulates Ifng gene expression in effector T cells.
RelA cooperates with STAT-4 at multiple sites in the Ifng locus.
T-bet plays an obligatory role in long-range remodeling of the Ifng locus.
Cis-regulatory sites are modularly recruited in response to distinct stimuli.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Laurie Harrington and members of the Weaver laboratory for their helpful comments and suggestions. We gratefully acknowledge Dr. Thomas Gilmore for advice and reagents for study of NF-κB, and Benjamin Weaver for development of DNAse-chip and ChIP-chip data analysis scripts. We also thank J. Oliver, C. Song, M. Blake, B.J. Parsons and S. Sinclair for expert technical assistance, Gloria Gaskins for editorial assistance, and the UAB Digestive Diseases Research Developmental Center (DDRDC) and Epitope Recognition and Immunoreagent Core Facility for generation and genotyping of gene-targeted mice, and antibody preparations, respectively. This work was supported by grants from the NIH (AI35783, C.T.W.; AI77574, C.T.W. and R.D.H.; AI35098, A.S.B.; and K22HG003169, G.E.C.) and the Crohn’s and Colitis Foundation of America (C.T.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Berenson LS, Gavrieli M, Farrar JD, Murphy TL, Murphy KM. Distinct characteristics of murine STAT4 activation in response to IL-12 and IFN-α. J Immunol. 2006;177:5195–5203. doi: 10.4049/jimmunol.177.8.5195. [DOI] [PubMed] [Google Scholar]
- Berenson LS, Ota N, Murphy KM. Issues in T-helper 1 development--resolved and unresolved. Immunol Rev. 2004;202:157–174. doi: 10.1111/j.0105-2896.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci U S A. 2005;102:17095–17100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-γ during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- Chang S, Collins PL, Aune TM. T-bet dependent removal of Sin3A-histone deacetylase complexes at the Ifng locus drives Th1 differentiation. J Immunol. 2008;181:8372–8381. doi: 10.4049/jimmunol.181.12.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corn RA, Aronica MA, Zhang F, Tong Y, Stanley SA, Kim SR, Stephenson L, Enerson B, McCarthy S, Mora A, Boothby M. T cell-intrinsic requirement for NF-κB induction in postdifferentiation IFN-γ production and clonal expansion in a Th1 response. J Immunol. 2003;171:1816–1824. doi: 10.4049/jimmunol.171.4.1816. [DOI] [PubMed] [Google Scholar]
- Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175:2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-κB in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452:356–360. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S. NF-κB and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- Hilliard BA, Mason N, Xu LB, Sun J, Lamhamedi-Cherradi SE, Liou HC, Hunter C, Chen YH. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J Clin Invest. 2002;110:843–850. doi: 10.1172/JCI15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Hong JH, Glimcher LH. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J Exp Med. 2005;202:1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-γ (IFN-γ) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet’s ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC, Jin Z, Tumang J, Andjelic S, Smith KA, Liou ML. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int Immunol. 1999;11:361–371. doi: 10.1093/intimm/11.3.361. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Lu B, Ferrandino AF, Flavell RA. Gadd45beta is important for perpetuating cognate and inflammatory signals in T cells. Nat Immunol. 2004;5:38–44. doi: 10.1038/ni1020. [DOI] [PubMed] [Google Scholar]
- Mason NJ, Liou HC, Hunter CA. T cell-intrinsic expression of c-Rel regulates Th1 cell responses essential for resistance to Toxoplasma gondii. J Immunol. 2004;172:3704–3711. doi: 10.4049/jimmunol.172.6.3704. [DOI] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Robinson D, Shibuya K, Mui A, Zonin F, Murphy E, Sana T, Hartley SB, Menon S, Kastelein R, Bazan F, O’Garra A. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-gamma production and activates IRAK and NFkappaB. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-γ. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor κB. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-γ locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CH, Stavnezer J. Interaction of stat6 and NF-κB: direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-γ expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. Interaction of NF-κB and NFAT with the interferon-γ promoter. J Biol Chem. 1997;272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- Soutto M, Zhou W, Aune TM. Cutting edge: distal regulatory elements are required to achieve selective expression of IFN-γ in Th1/Tc1 effector cells. J Immunol. 2002;169:6664–6667. doi: 10.4049/jimmunol.169.12.6664. [DOI] [PubMed] [Google Scholar]
- Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J Immunol. 2008;180:2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- Sweetser MT, Hoey T, Sun YL, Weaver WM, Price GA, Wilson CB. The roles of nuclear factor of activated T cells and ying-yang 1 in activation-induced expression of the interferon-γ promoter in T cells. J Biol Chem. 1998;273:34775–34783. doi: 10.1074/jbc.273.52.34775. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Takada Y, Singh S, Aggarwal BB. Identification of a p65 peptide that selectively inhibits NF-kappa B activation induced by various inflammatory stimuli and its role in down-regulation of NF-κB-mediated gene expression and up-regulation of apoptosis. J Biol Chem. 2004;279:15096–15104. doi: 10.1074/jbc.M311192200. [DOI] [PubMed] [Google Scholar]
- Tato CM, Mason N, Artis D, Shapira S, Caamano JC, Bream JH, Liou HC, Hunter CA. Opposing roles of NF-κB family members in the regulation of NK cell proliferation and production of IFN-γ. Int Immunol. 2006;18:505–513. doi: 10.1093/intimm/dxh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. Signal Transducer and Activator of Transcription 4 Is Required for the Transcription Factor T-bet to Promote T Helper 1 Cell-Fate Determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, Nagaoka H. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- Yang J, Murphy TL, Ouyang W, Murphy KM. Induction of interferon-γ production in Th1 CD4+ T cells: evidence for two distinct pathways for promoter activation. Eur J Immunol. 1999;29:548–555. doi: 10.1002/(SICI)1521-4141(199902)29:02<548::AID-IMMU548>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhu H, Murphy TL, Ouyang W, Murphy KM. IL-18-stimulated GADD45 beta required in cytokine-induced, but not TCR-induced, IFN-gamma production. Nat Immunol. 2001;2:157–164. doi: 10.1038/84264. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.