Abstract
The mechanisms of HLA-DM catalyzed peptide exchange remain uncertain. We found that all stages of the interaction of DM with HLA-DR were dependent on the occupancy state of the peptide binding groove. High-affinity peptides were protected from removal by DM through two mechanisms: peptide binding induced dissociation of a long-lived complex of empty DR and DM, and high-affinity DR-peptide complexes bound DM only very slowly. Non-binding covalent DR-peptide complexes were converted to efficient DM binders upon truncation of an N-terminal peptide segment that emptied the P1 pocket and disrupted conserved hydrogen bonds to MHC. DM thus only binds to DR conformers in which a critical part of the binding site is vacant, due to spontaneous peptide motion.
Introduction
Efficient surveillance of the surface of antigen presenting cells by CD4+ T cells requires long-lived display of peptides bound to major histocompatibility complex class II (MHCII) molecules. High-affinity peptides are kinetically trapped in the peptide binding groove and dissociate at extremely slow rates (days to weeks at 37 °C)1,2. Such stable binding is enabled by a conserved hydrogen bonding network between the MHC helices and the backbone of bound peptides as well as occupancy of MHC pockets by peptide side chains3,4.
Empty MHCII molecules quickly lose their ability to rapidly bind peptide and aggregate5,6. Prior to arrival of MHCII in the late endosomal peptide loading compartment, the binding groove is protected by the CLIP segment of invariant chain7. Invariant chain cleavage in the late endosomal compartment leaves the CLIP peptide in the binding groove8,9, which is bound with a wide range of affinities by different allelic forms of MHCII10. DM plays a critical role in MHCII antigen presentation by accelerating removal of CLIP and by editing the peptide content of MHCII molecules such that display of high-affinity peptides is favored9,11–20. DM also acts as a chaperone that maintains empty MHCII in a highly peptide receptive state21–23. Mass spectrometry analysis of DM-MHCII complexes purified from antigen presenting cells showed them to be largely devoid of peptide22.
Crystal structures of both MHCII and DM have been available for many years (1993 and 1998, respectively), but it has been challenging to define the molecular mechanisms of DM-catalyzed peptide exchange4,24,25. Comprehensive mutagenesis identified large lateral surfaces of DM and DR required for their interaction; of particular interest are DR residues in the vicinity of the peptide N-terminus (DRα Phe51 and Glu40)26,27. The proximity of the DM interaction site to the peptide N-terminus was also demonstrated by covalent attachment of a peptide to a surface-accessible cysteine of DM (DMβ 46) and subsequent loading of this DM-linked peptide into the DR1 peptide binding groove. Such a complex was stable when DM was linked to the peptide C-terminus, but DM catalyzed rapid peptide dissociation when it was linked to the peptide N-terminus28.
Two major models of DM action have been proposed. The first model suggests that DM breaks some of the conserved hydrogen bonds between the peptide backbone and the MHC helices24,29, while the second model proposes more global conformational changes30. The first model was supported by functional data showing that the rate of DM-induced peptide dissociation was proportional to the intrinsic rate of peptide dissociation for all tested peptides and MHCII molecules29. These data suggested that bonds conserved in all peptide-MHCII interactions are targeted by DM, such as conserved hydrogen bonds formed by DRα Phe51, DRα Ser53 and DRβ His81 close to the peptide N-terminus24. An initial report implicated DRβ His81 as the target of DM action31, but other studies showed that mutation of this site did not reduce DM susceptibility32,33. Furthermore, individual mutation of all MHC side chains forming conserved hydrogen bonds to the peptide backbone (9 hydrogen bonds total) did not identify a mutation that reduced susceptibility to DM32. Finally, loss of hydrogen bonds to the main chain atoms of DRα Phe51 and DRα Ser53 enhanced DM susceptibility, rather than reducing it, suggesting that these hydrogen bonds are not direct targets of DM34.
The second model of DM action proposes that DM globally distorts the MHCII binding groove, rather than breaking a small number of hydrogen bonds. Analysis of a large number of DR-peptide complexes showed that the intrinsic stability of any one complex was a poor predictor of DM susceptibility and that interactions along the entire length of the groove affected DM susceptibility30. Dynamic light scattering and circular dichroism studies further indicated that DR undergoes conformational changes upon peptide binding35.
A central problem in defining the mechanism of DM-catalyzed peptide exchange is that it remains unknown which DR-peptide conformers interact with DM. We report that the interaction of DM and DR is highly dependent on the occupancy of the peptide binding groove, with high-affinity peptides destabilizing empty DR-DM complexes. Furthermore, we show that DM only binds DR-peptide conformers in which key interactions between the peptide N-terminus and the DR molecule have already been lost due to peptide motion.
Results
Peptide binding destabilizes the complex of DM and empty DR
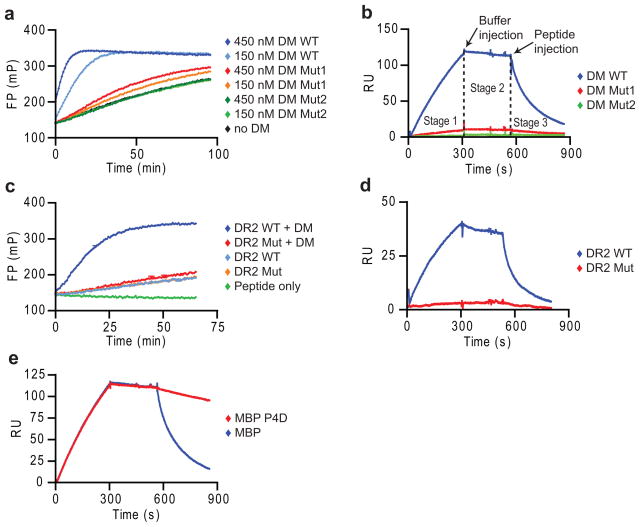
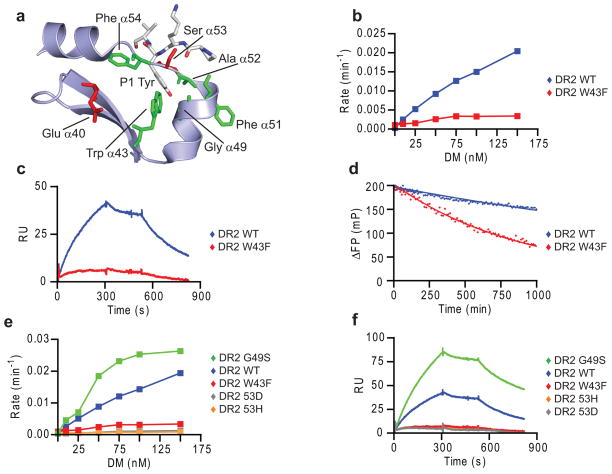
DM-catalyzed peptide exchange is a multi-step process that has been difficult to study mechanistically because most assays do not isolate individual steps but rather represent a mixture of multiple rates. We developed a real-time surface plasmon resonance (SPR, Biacore) assay to directly study the individual steps of the DM-DR interaction. DM was oriented on streptavidin chips through C-terminal site-specific biotinylation so that the large lateral surface required for DR binding was fully accessible. Soluble DR-CLIP complexes were generated with a cleavable CLIP linker. In SPR experiments, it is critical to employ suitable control proteins to ensure specificity of binding, and we therefore identified DM and DR mutants with loss of activity in a real-time peptide binding assay based on fluorescence polarization (FP) (Fig. 1a,c). Mutation of one DMα arginine residue (R98A; hereafter referred to as DM Mut1) reduced DM activity, while an additional mutation (DMα R98A and R194A; called DM Mut2) resulted in loss of activity (Fig. 1a). The SPR binding data closely correlated with these functional data, showing little DR binding to DM-Mut1 and no binding to DM Mut2 (Fig. 1b). Also, a DR2 (DRB1*1501) mutant (DRα S53D) unresponsive to DM in the FP assay (Fig. 1c) failed to bind to DM by SPR (Fig. 1d; Supplementary Figure 1). This DR2 mutant bound peptide with similar kinetics as wild-type DR2 (Fig. 1c), indicating correct folding.
Figure 1. Peptide disrupts long-lived complex between empty DR and DM.
(a,b) Specificity of SPR assay. (a) Activity of DM WT, Mut1 (DMα R98A) and Mut2 (DMα R98A/R194A) in accelerating binding of Alexa-488 labeled MBP85-99 peptide (30 nM) to DR2 (150 nM), measured by fluorescence polarization (FP). (b) Binding of DR2-CLIP complexes to DM WT and mutants by SPR. DR2-CLIP (2 μM) was run over DM WT and mutant surfaces (5 min, pH 5.35, 25 μl/min, 30°C) (stage 1), followed by buffer (stage 2) and 1 μM MBP85-99 (stage 3). Readings from streptavidin flow cell were subtracted from DM WT and mutant flow cells. (c,d) DR2 mutant without DM interaction. (c) DM-accelerated binding (+/− 25 nM DM) of labeled MBP85-99 to DR2 WT and DRα S53D mutant (150 nM) was measured as in (a). (d) DM binding of DR2 WT and mutant. WT or mutant DR2-CLIP (1 μM) were injected (5 min), followed by buffer and 10 μM CLIP87-101. (e) Dissociation of DM-DR2 complex by high-affinity peptide. DR2-CLIP complexes (2 μM) were injected (5 min), followed by buffer and MBP85-99 or MBP P4D analog (1 μM). Data are representative of two (a, c, d) or more than three (b, e) independent experiments.
In these SPR experiments, DR-CLIP complexes were injected over the DM surface (stage 1), followed by injection of buffer (stage 2) (Fig. 1b,d). Surprisingly, the dissociation of DR from DM during stage 2 was very slow, even though soluble DM and DR are thought to interact only weakly36. We reasoned that the catalytic activity of DM removed the DR-bound CLIP peptide which exited with the buffer flow, while peptide remains available for rebinding in typical tube-based experiments. We therefore postulated that peptide injection would destabilize the complex of DM and empty DR. Indeed, injection of a high-affinity peptide from myelin basic protein (amino acids 85–99, MBP85-99) induced dissociation of DR2 from DM (Fig. 1e). A single amino acid mutant of the MBP85-99 peptide with very low-affinity for DR2 (MBP P4D, aspartic acid [D] at P4 position) induced minimal dissociation at the same peptide concentration (1 μM) (Fig. 1e). Similar binding profiles were obtained at flow rates of 15 to 50μl/min, indicating that rebinding (mass transport) was negligible (Supplementary Fig. 2).
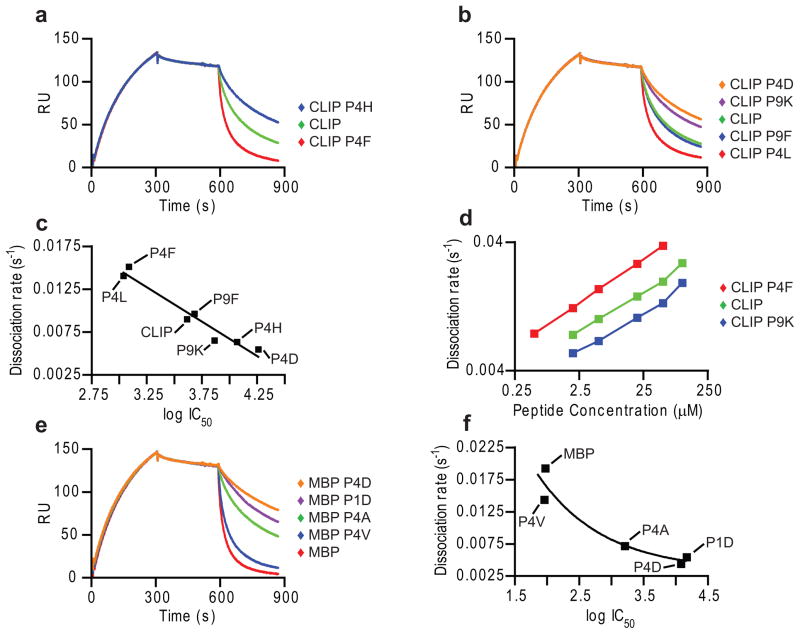
The rate of peptide-induced DM-DR dissociation (stage 3) correlated closely with the affinity of these peptides for the DR2 binding site, both for sets of CLIP (Fig. 2a-d) and MBP85-99 (Fig. 2e,f) mutants, regardless of which peptide anchors were mutated. Peptides with high affinities for DR2 induced DM-DR dissociation at faster rates, such as CLIP P4F (Fig. 2c) or MBP85-99 (Fig. 2f), than lower affinity peptides. Peptides with very low affinity, such as MBP P4D, did not induce dissociation at low concentrations (such as 1 μM) (Fig. 1e) but led to some dissociation at higher concentrations (10 μM) (Fig. 2e).
Figure 2. Rate of DM-DR complex dissociation is determined by peptide affinity.
(a,b) Dissociation of DM-DR complex by CLIP and CLIP mutants. DR2-CLIP (5 μM) was injected (5 min, pH 5.35, 15 μl/min, 30 °C), followed by buffer and CLIP or CLIP mutants (10 μM). (c) Relationship between peptide affinity and DM-DR dissociation rate. DR2 (100 nM) binding of peptides (3-fold dilutions, 10 μM to 14 nM) was examined in competition assay against Alexa-488 labeled MBP85-99 (10 nM). IC50 values were plotted against rates of peptide-induced DM-DR dissociation (a,b). (d) Titration of CLIP mutants. CLIP and two CLIP mutants were injected in stage 3 (0.5 to 100 μM). Rates of DR dissociation from DM were plotted against peptide concentration. (e) Testing of DM- DR dissociation by MBP85-99 and mutants. Indicated MBP peptides were injected in stage 3 as in (a). (f) Relationship between MBP peptide affinity and DM-DR dissociation rate. IC50 values determined in competition assay (as in c) were plotted against rate of DR dissociation from DM (e). Data are representative of two independent experiments as shown (a-f) and more than three independent experiments under similar conditions (a–c, e, f). Peptide competition assays were performed in triplicates.
High-affinity DR-peptide complexes bind weakly to DM
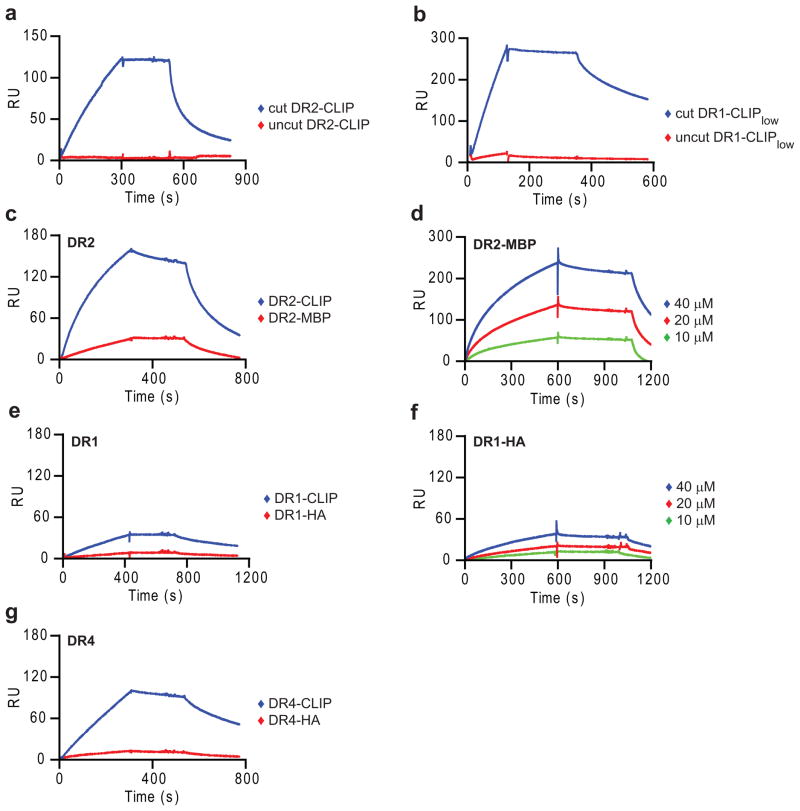
The fact that peptide binding induces dissociation of DM-DR complexes suggests that the DR-bound peptide is likely to also have a major effect on the reverse process, the binding of DR to DM. Initial experiments showed that DR molecules with covalently bound peptides did not bind to DM, both for DR2-CLIP and DR1 (DRB1*0101) with a low-affinity CLIP variant (DR1-CLIPlow,), even though binding was readily detectable upon linker cleavage (Fig. 3a,b). These results could either be explained by steric hindrance caused by the linker attached to the N-terminus of the DRβ chain or by a substantial affinity gain resulting from covalent peptide attachment (rebinding of peptide is likely to be extremely rapid). The linker is flexible and also not in the immediate proximity of the predicted DM interaction site, making the latter explanation more plausible.
Figure 3. High-affinity DR-peptide complexes interact only slowly with DM.
(a, b) Absence of detectable DM binding by DR molecules with covalent peptides. (a) DR2-CLIP complexes with or without thrombin cleavage of the peptide linker (5μM) were injected for 5 min, followed by buffer (stage 2) and 20 μM CLIP87-101 (stage 3). (b) 3C cut or uncut DR1-CLIPlow complexes (5 μM) were injected (2 min), followed by buffer and HA306-318 (50 μM). (c,e,g) Slow DM binding by DR molecules with bound high-affinity peptides. DR complexes preloaded with indicated peptides (2 μM for DR2 and DR4; 10 μM for DR1) were injected for 5-7 min, followed by buffer and 10 μM CLIP87-101 (DR2) or 50 μM HA306-318 (DR4 and DR1). (d,f) Concentration-dependent binding of high-affinity DR-peptide complexes under optimal conditions (37°C, long injection time, high DR-peptide concentrations). DR-peptide complexes preloaded with the indicated peptides were injected for 10 min, followed by buffer (stage 2) and 5 μM MBP85-99 (DR2) and 50 μM HA306-318 (DR1) (stage 3). (a-g) SPR assays were carried out at 25°C (a, b) or 37°C (c-g) (pH 5.35, 15 μl/min). Data are representative of three (a, c, e) and of two (b, d, f, g) independent experiments.
We next examined the effect of peptide affinity on DM-DR association using DR molecules loaded with single peptides; peptides were synthesized with a dinitrophenol (DNP) tag for affinity isolation of homogenous DR-peptide complexes. There was strong DM binding by the low-affinity DR2-CLIP complex, but substantially slower binding by the high-affinity DR2-MBP85-99 complex (Fig. 3c,d). This effect was also observed for DR4 preloaded either with low-affinity CLIP or high-affinity influenza hemagglutinin 306-318 peptide (HA306-318) (Fig. 3g). CLIP binds with high affinity to DR1 (ref.37) and binding of the DR1-CLIP complex to DM was also slow (Fig. 3e). Nevertheless, concentration-dependent DM binding of the high-affinity DR2-MBP85-99 and DR1-HA306- 318 complexes could be demonstrated (Fig. 3d,f), indicating that both low and high-affinity DR-peptide complexes bound to DM in this SPR assay, only with different rates. In a functional peptide binding assay, DM-catalyzed binding of a fluorescently labeled peptide occurred with an 11.8-fold slower rate when DR2-MBP85-99 rather than DR2-CLIP was used as input protein. In the SPR assay, a similar difference in the rates of DR2-MBP85-99 and DR2-CLIP binding to DM was observed (8.7-fold), showing overall agreement between the SPR and peptide binding assays (Supplementary Fig. 3). Previous studies with full length DR and DM molecules isolated from cells reported a stronger DM interaction with DR-CLIP than DR-peptide complexes21, 38; also a previous SPR experiment showed stronger binding of immobilized soluble DM to detergent solubilized full-length DR-CLIP than DR-peptide complexes39. In both sets of experiments, a complex mixture of peptides was bound to these cell-derived DR-peptide complexes. In the experiments reported here, differences in binding were much more substantial because purified low and high-affinity DR-peptide complexes were utilized.
Comparison of the DR-CLIP binding data among these three allelic forms (DR1, DR2 and DR4) showed large differences in the DM binding kinetics. DR2-CLIP bound with the fastest and DR1-CLIP with the slowest kinetics (Fig. 3). Large differences in peptide induced dissociation were also observed among these allelic forms, with DR2 being more sensitive to peptide-induced dissociation than DR1 and DR4 (Fig. 3).
DM binding requires partial peptide dissociation
The results with covalently linked peptides (Fig. 3a,b) raised the question whether DR molecules with fully bound peptides do not bind to DM and whether partial release of the bound peptide is actually required to generate a DM-sensitive conformer. To investigate this hypothesis, we generated DR1 molecules to which peptides could be covalently attached in one of the pockets so that peptide termini could be truncated without peptide loss. Mutant HA peptides were either covalently linked in the P6 pocket (HA6 peptides) or the P3 site (HA3 peptides) by mutation of DRα Val65 or Gly58 to cysteine; both residues are located on the DRα chain helix. These complexes were made using a DR1 molecule with a very low-affinity peptide as input protein; redox conditions were chosen that enabled efficient formation of a disulfide bond between the peptide and the DRα helix. A large set of such complexes was generated (Table 1) which were affinity purified using DNP affinity tags on the peptides to ensure the absence of empty or other peptide-loaded DR species (Supplementary Fig. 4). Full occupancy was confirmed by monitoring the ratio of 280 nm (DR) and 350 nm (DNP group on peptide) measurements. Also, DR1 molecules with linked HA6 P2-P11 peptide could be immunoprecipitated by conformation-sensitive antibodies to DRα and DRβ (Supplementary Fig. 5).
Table 1.
HA-P6Cys and HA-P3Cys peptide variants.
| Name | Sequence | Change/Mutation |
|---|---|---|
| Cysteine residue at P6 | ||
| HA6 | PKYVKQNCLKLAT* | |
| HA6 P1A | PKAVKQNCLKLAT | P1 Tyr to Ala |
| HA6 P1G | PKGVKQNCLKLAT | P1 Tyr to Gly |
| HA6 P-1-P11 | KYVKQNCLKLAT | P-2 truncated |
| HA6 P1-P11 | YVKQNCLKLAT | P-2 and P-1 truncated |
| HA6 P1me-P11 | Me-YVKQNCLKLAT | P-2 and P-1 truncated; P1 Tyr N-methylated |
| HA6 P1Gme-P11† | Me-GVKQNCLKLAT | P-2 and P-1 truncated; P1 Tyr to Gly N-methylated |
| HA6 P2-P11 | VKQNCLKLAT | P-2, P-1 and P1 truncated |
| HA6 (N-DNP) | PKYVKQNCLKLAT | N-terminal DNP-label |
| HA6 P-2-P6 | PKYVKQNC | P7, P8, P9, P10 and P11 truncated; N-term. DNP-label |
| Cysteine residue at P3 | ||
| HA3 | PKYVCQNTLKLAT | |
| HA3 P1G | PKGVCQNTLKLAT | P1 Tyr to Gly |
| HA3 P1me-P11 | Me-YVCQNTLKLAT | P-2 and P-1 truncated; P1 Tyr N-methylated |
| HA3 P1Gme-P11 | Me-GVCQNTLKLAT | P-2 and P-1 truncated; P1 Tyr to Gly N-methylated |
| HA3 P2-P11 | VCQNTLKLAT | P-2, P-1 and P1 truncated |
| HA3 P3-P11 | CQNTLKLAT | P-2, P-1, P1 and P2 truncated |
All peptides except HA6 (N-DNP) and HA6 P-2-P6 had a DNP group attached through a C-terminal lysine.
P1 anchor residues are underlined.
HA6 peptides have Thr to Cys mutation at position 6.
HA3 peptides have Lys to Cys mutation at position 3.
DR/peptide complexes with HA peptides shown in bold font bind rapidly to DM.
Covalent attachment of the high-affinity HA306-318 peptide in the P6 pocket (HA6) (Fig. 4a) yielded a complex that failed to bind to DM by SPR (Fig. 4a,d). The full length HA peptide, which spanned from the P-2 to the P11 residue, was then systematically truncated from the N-terminus, which resulted in loss of conserved hydrogen bonds between the peptide backbone and the DR helices. Removal of the P-2 residue (peptide HA6 P-1–P11) resulted in loss of hydrogen bonds to Phe α51 and Ser α53 and further truncation of P-1 (peptide HA6 P1–P11) also eliminated a hydrogen bond to His β81 (Fig. 4a). Deletion of these two N-terminal peptide residues was not sufficient for DM binding (Fig. 4d). However, strong DM binding was observed with further truncation of the P1 tyrosine (peptide HA6 P2–P11), a critical anchor residue for DR1 binding (Fig. 4b-d)40. In contrast, truncation of five C-terminal residues (HA6 P-2–P6) resulted in little DM binding; the small amount of apparent binding was possibly due to an overall increase in protein mobility due to loss of five peptide residues (Fig. 4e).
Figure 4. DM binds with fast kinetics to DR-peptide complexes without engaged peptide N-terminus.
(a, b) The hydrogen bonding network between DR1 and full length, covalently linked HA306-318 peptide (a) is compared to a linked HA mutant peptide lacking three N-terminal residues (b). Peptide residues are numbered from P-2 to P11. DRα Val65 was mutated to cysteine to enable disulfide bond formation to HA peptides with a cysteine at the P6 position (HA6 peptides). (c) Space filling model of the empty DR1 groove showing residues contacted by two N-terminal peptide residues (red) and the P1 anchor (green). (d, f) DM binding was enabled by loss of two N-terminal residues and the P1 anchor. DR1 complexes with linked HA6 peptides (5 μM) were injected for 2 minutes (pH 5.35, 15 μl/min 25 °C), followed by injection of buffer. (e) C-terminal truncation of HA6 peptide did not result in substantial DM binding. Complexes linked at the P6 position were tested as in (d). Data are representative of at least three (d, f) and of two (e) independent experiments.
To determine whether this effect was solely due to removal of the P1 anchor residue, full-length peptides with alanine or glycine substitutions at P1 were tested. Mutation of the P1 anchor to alanine (HA6 P1A) or glycine (HA6 P1G) resulted in only low amounts of DM binding (Fig. 4f). In contrast, substantial binding was enabled by simultaneous loss of the P1 side chain (P1 glycine) and the two N-terminal residues (P1Gme–P11) (Fig. 4f). Substantial DM binding therefore requires release of both the P1 anchor and the peptide residues N-terminal to it.
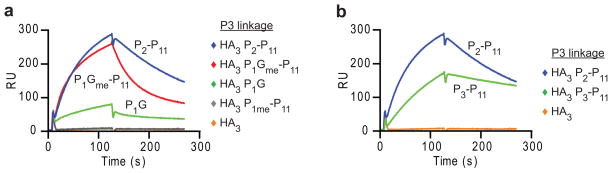
Loss of the P1 anchor could enable dissociation of a longer peptide segment toward the P6 position, which could be required for DM binding. To address this possibility, we moved the covalent attachment site from the P6 pocket to the P3 site by mutation of DRα Gly58 to cysteine (Table 1). Again, removal of P-2 and P-1 did not permit DM binding (HA3 P1me–P11) (Fig. 5a) but further removal of the P1 side chain (HA3 P1Gme–P11) or the P1 residue (HA3 P2–P11) resulted in rapid binding of these two complexes to DM (Fig. 5a). Interestingly, additional truncation of the P2 residue (HA3 P3 P11) appeared to slow DM binding (Fig. 5b), indicating that disengagement of the P1 anchor and two N-terminal peptide residues was sufficient for DM binding. DRβ Asn82 forms bidentate hydrogen bonds to the peptide backbone at the P2 position and mutation to alanine increases the intrinsic dissociation rate of the HA peptide >3,000-fold, indicating that it plays a dominant role in stabilizing peptides in the binding groove32, 41. Loss of the P2 peptide residue may thus reduce the activity of such a DR-peptide complex due to loss of these critical hydrogen bonds. These results show that DM only binds to DR-peptide complexes with a disengaged peptide N-terminus, but not to complexes in which the peptide is fully bound.
Figure 5. Truncated peptides covalently linked through the P3 peptide position also bind DM.
(a, b) DRα Gly58 was mutated to a cysteine to permit disulfide bond formation with HA peptides carrying a cysteine at the P3 position (HA3 peptides). DM binding of DR molecules disulfide-linked to the indicated HA3 peptides (5 μM) was examined as in Fig. 4. Data are representative of three (a) and of two (b) independent experiments.
Important role for DRα Trp43 in DM binding
DRα Trp43 directly interacts with the P1 tyrosine and is partially accessible on the lateral surface of the DRα40-54 segment (Fig. 6a) implicated in DM binding. Conservative mutation to phenylalanine greatly reduced susceptibility to DM in a peptide binding assay (Fig. 6b), and little DM binding by this mutant was observed by SPR (Fig. 6c). This mutation also accelerated spontaneous release of labeled MBP85-99 peptide from DR2 (Fig. 6d), a property that would actually be expected to increase DM binding based on the data shown above. We also directly compared a panel of four DR mutants for DM-catalyzed peptide exchange and DM binding (Fig. 6e,f). Three mutants had greatly reduced activity (αW43F, αS53D, αS53H) in both assays while one mutant (αG49S) showed enhanced activity. These data show that DR binding to DM as well as DM-catalyzed peptide exchange require the same DR residues and hence involve the same binding site on DR.
Figure 6. DRα Trp43 is important for interaction with DM.
(a) DRα Trp43 is surface accessible and interacts with the P1 tyrosine of HA306-318. Hydrophobic residues in the DRα40-54 segment interacting with P1 anchor (Trp43, Ala52 and Phe54) are indicated, as well as residues implicated in DM interaction (Glu40, Phe51 and Ser53). HA306-318 P-2 to P2 segment is shown as a stick model. (b-d) Conservative DRα W43F mutation reduced DM susceptibility (b), DM binding (c) and intrinsic stability of DR2-peptide complexes (d). (b) Dissociation rate of Alexa-488 labeled MBP85-99 from DR2 WT or W43F was measured by FP over a range of DM concentrations. (c) DR2 WT or W43F with bound CLIP (1 μM) were injected over DM WT and DM Mut2 flow cells (pH 5.35, 30 °C, 15 μl/min) followed by buffer and 5 μM CLIP87-101. (d) Dissociation of Alexa-488 labeled MBP85-99 from preloaded DR2 WT or W43F complexes (100 nM) was measured by FP in presence of 10 μM unlabeled MBP85- 99 competitor peptide (pH 5.4, 25°C). (e,f) Comparison of a panel of four DRα chain mutants in FP (e) and SPR (f) assays, using conditions described in b and c, respectively. Data are representative of two independent experiments.
High energy barrier for DM binding
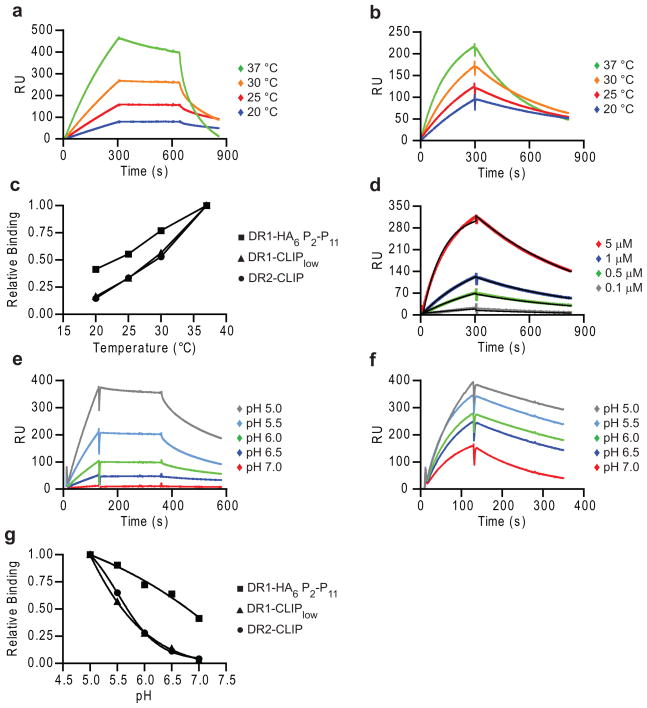
Peptide exchange in the presence or absence of DM is strongly dependent on both temperature and pH11, 12, 42. DR2-CLIP and DR1-CLIPlow (Fig. 7a,c, Supplementary Fig. 6) showed a steep temperature dependence in DM binding that is unusual for protein-protein interactions. However, binding of DM to DR1 with a covalent peptide lacking three N-terminal peptide residues (DR1-HA6 P2–P11) was substantially less temperature dependent (Fig. 7b,c), demonstrating that dissociation of the peptide N-terminus represents a significant component of the energy barrier. With higher affinity peptides this energy barrier is expected to be even higher. Based on the DM association and dissociation kinetics of the covalent DR1-HA6 P2–P11 complex, a Kd of 1 μM was calculated (Fig. 7d), which is consistent with the Michaelis constant (KM) of ~750 nM for the enzymatic reaction executed by DM39. Interestingly, both on- and off-rates increased to a similar extent with temperature, and the affinity is therefore maintained over this temperature range (Supplementary Fig. 7a-c).
Figure 7. Large energy barrier for DR-peptide binding to DM.
(a,b) Temperature profiles for DM binding by DR with full length or linked truncated peptide (HA6 P2-P11). (a,b) DR1-CLIPlow (1 μM, a) and DR1-HA6 P2-P11 (1 μM, b) complexes were run at indicated temperatures (15 μl/min, pH 5.35), followed by buffer and 50 μM HA306-318. (c) Relative binding at time 250-270 sec of the association phase in (a,b and Supplementary Fig. 6a) was plotted against corresponding temperature. (d) DM affinity for DR1 with covalently linked HA6 P2-P11 peptide. DR1-HA6 P2-P11 complexes were injected at indicated concentrations as in (a). Data were fitted with a 1:1 Langmuir binding model using BIAevaluation software. (e-g) Comparison of pH profiles for binding by DR molecules with full-length peptides (e) and covalent truncated peptide (f). (e,f) DR1-CLIPlow complexes (5μM, e) and DR1-HA6 P2-P11 complexes (5 μM, f) were injected as in (a) for 2 min at the indicated pH (25 °C). (g) Relative binding at time 91-110 seconds of the association phase in (e,f and DR2-CLIP) was plotted against pH, with the binding at pH 5.0 defined as 1.0. Data are representative of two (a, e) and of three (b, d, f) independent experiments.
DM has a sharp pH optimum in the acidic range found in late endosomes11,12. As expected, the binding of DR-CLIP to DM was increased by lowering the pH from 5.5 to 4.6 (Supplementary Fig. 8a). However, peptide binding experiments with DR2 molecules rendered empty by photocleavage of bound peptide showed that peptide binding was faster at higher pH (Supplementary Fig. 8b)23. Consistent with this result, peptide-induced dissociation of the complex was slower at pH 4.6 (Supplementary Fig. 8a). DR1 with a covalent truncated peptide (DR1-HA6 P2–P11) showed 40% of maximal DM binding at pH 7.0, while complexes with a full-length peptide (DR1-CLIPlow and DR2-CLIP) did not bind to DM at this pH (Fig. 7e-g). Furthermore, raising the pH during stage 2, where empty DR dissociates from DM, had little to no effect on dissociation rates (data not shown). The pH-facilitated release of the peptide N-terminus therefore explains, in part, the low pH optimum of DR-peptide binding to DM.
Model of DM-DR interaction
These results significantly change our model of DM action (Supplementary Fig. 9). We show that DR molecules with fully engaged peptides fail to interact with DM until the N-terminal part of the peptide dissociates through constant motion within the DR-peptide complex. DM captures this short-lived transition state and peptide quickly departs following DM binding. The empty DM-DR complex retains the ability to quickly bind a new peptide over extended periods of time. If an interacting peptide has a low affinity, DM may catalyze its removal (editing) while binding of a high-affinity peptide is more likely to induce dissociation of the DM-DR complex. The resulting high-affinity DR-peptide complex has a low likelihood of rebinding DM. This model is consistent with a large body of prior work in the field, including the identification of empty DR-DM complexes in cells and the demonstration of an editing function of DM that drives selection of high-affinity peptides14–22, 43.
Discussion
Two mechanisms therefore protect high-affinity peptides from rapid removal by DM: dissociation of the DM-DR complex by such peptides, and slow rebinding of high-affinity DR-peptide complexes to DM. These mechanisms explain how editing by DM produces DR-peptide complexes with high intrinsic stability for long-lived display on the cell surface. Forced linkage of DM and DR through leucine zipper dimerization domains attached to DMβ and DRβ chains resulted in rapid dissociation of both low and high-affinity peptides (t1/2 of 10 seconds and 2 minutes for CLIP and HA306-318 peptides bound to DR1), illustrating the importance of rapid DM-DR dissociation for keeping high-affinity peptides in the binding groove27.
DR molecules with full-length peptides covalently linked at the P3 or P6 positions did not bind to DM. However, efficient binding resulted after removal of two N-terminal residues (P-1 and P-2) and truncation of the P1 side chain which serves as the primary anchor for many DR-bound peptides40. Removal of the P1 peptide side chain alone only resulted in weak DM binding. The two peptide residues N-terminal to the P1 anchor (P-2 and P-1) form conserved hydrogen bonds to DRα Phe51, DRα Ser53 and DRβ His813; in addition, the side chains of DRα Phe51 and Ser53 are critical for DM action. These results show that DM interacts only with DR molecules when the P1 pocket is vacant and conserved N-terminal hydrogen bonds to HLA-DR are disrupted; loss of either set of interactions is not sufficient for substantial DM binding. Consistent with these results, it was previously shown that filling of the P1 pocket by mutation of DRβ 86 (Gly to Tyr) significantly reduced DM susceptibility44 and that loss of hydrogen bonds between the peptide N-terminus and DRα Phe51 and Ser53 increased susceptibility to DM six to nine- fold34. DM thus only interacts with DR conformers in which a critical part of the binding site is already empty.
The DRα 40-54 segment not only forms three hydrogen bonds with the backbone of the N-terminal peptide segment (DRα 51 and 53) but also makes substantial hydrophobic contacts that pack against the P1 pocket3. Prior mutagenesis studies had shown that DRα Phe51 and Glu40 are important for DM susceptibility26, and our studies showed that the side chains of DRα Ser53 and Trp43 are also critical. DRα Trp43 is of particular interest because it interacts with the P1 anchor and is also partially solvent accessible in an area implicated in the DM interaction. Even a conservative substitution from tryptophan to phenylalanine resulted in greatly reduced DM binding, even though this mutation moderately increased the off-rate of DR-bound peptide (a change predicted to increase DM binding). DRα Phe51 and Ser53 (hydrogen bonds to peptide) and DRα Phe54 and Ala52 (hydrophobic contacts to P1 anchor) are located on an extended strand at the N-terminal side of the DRα chain helix which may readily adopt a new conformation when the close interactions with the peptide N-terminus and the P1 anchor are lost. This specific area of the DRα chain shows higher flexibility than the DR helices, as evidenced by higher B-factors in crystal structures of MHCII3, 4. Molecular dynamics simulations of the peptide-occupied state have further shown that the DRα 50–59 segment shows greater mobility than other segments that flank the peptide binding groove45.
All proteins are in constant motion and local motions may lead to transient loss of interactions in part of the binding groove. We propose that the DR1 complex with the N-terminally truncated HA peptide mimics a transient conformation in which part of the peptide has temporarily left the binding site. Structurally, the hydrogen bonds formed by DRα 51, 53 and DRβ 81 are surface exposed while other conserved hydrogen bonds to the peptide backbone are sheltered within the helical walls of the binding groove24. In the absence of DM, the most likely outcome is rebinding of this peptide segment in the groove. DM appears to have a higher affinity for fully empty compared to partially peptide-filled DR molecules because the dissociation rate of empty DR-DM complexes (stage 2) is significantly slower (~8.5-fold) compared to covalent DR-peptide complexes lacking the N-terminal peptide segment. When DM senses the loss of interactions surrounding the peptide N-terminus, its higher affinity for the empty state could shift the equilibrium from a partially peptide-filled to a fully empty form.
DR-peptide binding to DM shows a temperature dependence that is highly unusual for protein interactions. The temperature dependence was less steep for truncated DR-peptide complexes, suggesting that a substantial part of the input energy for DM binding is required for dissociation of the N-terminal peptide segment. DM can catalyze peptide exchange for a wide variety of DR-peptide complexes, but the enhancement of peptide dissociation by DM varies substantially among DR-peptide complexes. In one study, the enhancement of peptide dissociation by DM for high-affinity DR-peptide complexes ranged from 1.4 to 227-fold30. The SPR data show slow DM binding by high-affinity DR-peptide complexes, strongly suggesting that the initial binding event is a rate-limiting step. How frequently such DR-peptide complexes assume a conformation compatible with DM binding is likely to be influenced by a number of factors, including the tightness with which the N-terminal peptide segment is bound and how the binding energy is distributed along the groove. Some high-affinity peptides may become almost resistant to the action of DM because the DM-interacting conformation is accessed only infrequently.
Finally, we observed that allelic DR variants are differentially susceptible to DM action (fast DM binding for DR2-CLIP, intermediate for DR4-CLIP, slowest for DR1-CLIP). These differences in the on-rates were, in part, explained by differences in CLIP affinity: DR1 has a high affinity for CLIP, while DR4 binds CLIP with very low affinity37,46. However, additional factors appear to contribute because DR4-CLIP complexes bound DM with slower kinetics than DR2-CLIP complexes. Also, peptide-induced dissociation of the DM-DR complex appears to be substantially more efficient for DR2 than for DR1 or DR4. The structural mechanisms contributing to these differences in DM binding kinetics may be complex and will need to be carefully dissected. These results also raise the possibility that MHCII polymorphisms, including those associated with autoimmune diseases, not only change the specificity of MHCII binding pockets but also broadly affect the peptide repertoire by modifying the kinetics of DM-catalyzed peptide editing.
Methods
DM expression
Soluble DM proteins carried C-terminal BirA (GLNDIFEAQKIEWHE) and protein C tags on DMα and DMβ extracellular domains (EC), respectively. Two N-linked glycosylation sites (DMα N165D, DMβ N92D) were removed to reduce heterogeneity, leaving glycosylation site DMα N1547. Sf9 cells were infected with recombinant Baculoviruses at an MOI of >5 and proteins were purified after three days via affinity chromatography using an anti-Protein C matrix (Roche Applied Science); aggregates/impurities were subsequently removed with a Superose 6 gel filtration column (GE Healthcare Bio-Sciences Corp.).
Site-specific biotinylation of the BirA tag was performed at a protein concentration of 2-3mg/ml at a molar ratio of 20:1 DR to BirA. The reaction proceeded for 2 h at 30 °C in the presence of biotin (100 μM), ATP (10 mM) and protease inhibitors. Excess biotin was removed by dialysis and biotinylation was confirmed by gel shift with streptavidin on native polyacrylamide gels.
DR expression
Peptides were synthesized by Peptide 2.0 and JPT Peptide Technologies. Soluble DR–CLIP complexes were produced using stable CHO cell transfectants in hollow fiber bioreactors, as previously described48. The CLIP peptide (PVSKMRMATPLLMQA) was covalently attached via a thrombin-cleavable linker at the N-terminus of the respective DRβ chain (DRB1*0101, DRB1*0401, DRB1*1501).
For covalent attachment of peptides, soluble DR1 proteins were generated with cysteines at either DRα 58 (G58C) or 65 (V65C). A low-affinity mutant of CLIP (SKARMATGALAQA, DR1-CLIPlow; mutated anchor residues underlined) was covalently attached to the N-terminus of the DR1β chain (DRB1*0101) to enable efficient exchange after linker cleavage with human rhinovirus 3C protease (provided by J. Fraser, University of Auckland, Auckland, New Zealand). This protease site enabled cleavage under low temperature conditions (4 °C) that minimized dissociation of CLIPlow prior to peptide loading. These DR variants were produced using a Baculovirus system. Soluble DR proteins were purified via affinity chromatography with mAb L243 (American Type Culture Collection); aggregates and impurities were removed using a Superose 6 gel filtration column. Depending on the construct, covalently linked CLIP or low-affinity CLIP were cleaved either with thrombin (Novagen, EMD Chemicals) or 3C protease. Thrombin cleavage was performed using 20 units thrombin per mg of protein at room temperature for 1.5 h. 3C cleavage was done using 50 μg 3C protease per mg of protein under reducing conditions (2 mM GSH) at 4 °C for 2 h. Protein purity and completeness of cleavage were assessed by SDS-PAGE.
Peptide loading and production of disulfide-linked DR-peptide complexes
The key aspects of covalent linkage of very low-affinity peptides to DR1 were as follows: 1. Use of a very low-affinity DR1-peptide complex as input protein (DR1-CLIPlow); 2. Accelerated dissociation of this low-affinity peptide from DR1 by the small molecule J10-1; 3. Use of redox conditions that enabled efficient formation of a disulfide bond between the DRα helix and incoming peptide. Most peptides with a DNP group were labeled via a Lys side chain at the peptide C-terminus; HA306-318 C-terminal truncation variants were labeled via the Lys side chain at the P-1 position. DNP-labeled peptides were exchanged onto 5μM cut DR-CLIP complexes at a 10- to 20-fold molar excess of peptide in citrate-phosphate buffer (pH 5.3) in the presence or absence of a small molecule that accelerates peptide loading (J10-1)49 overnight at 25 °C, 30 °C or 37 °C, depending on the DR and peptide. Unbound peptide was removed using a Superose 12 gel filtration column (GE Healthcare Bio-Sciences Corp.) and loaded complex was separated from other DR species via affinity chromatography using an anti-DNP-1 column (Biotrend Chemikalien GmbH). DNP and protein absorbance were monitored at 350 nm and 280 nm, respectively49.
DNP-labeled HA peptides with a cysteine at the P3 or P6 position were reduced in 500 μM GSH and added to 3C-cleaved DR1-Cys protein; the reaction was then incubated for 2 h in the presence of 50 μM J10-1 and 750μM GSH. The reaction was then shifted for 2 h to oxidizing conditions (950 μM GSSG) to promote disulfide bond formation. Unbound peptide and unloaded complex were removed by gel filtration and DNP affinity chromatography, as described above. Disulfide linkage of HA-Cys peptides to DR1- P6Cys and DR1-P3Cys proteins was confirmed based on decreased DRα motility by SDS-PAGE.
Surface Plasmon Resonance
Streptavidin chips (GE Healthcare Bio-Sciences Corp.) were primed and normalized before immobilization of ligand. Biotinylated DM proteins (ligand) were diluted to 0.5μg/ml in HBS-EP buffer (GE Healthcare Bio-Sciences Corp.) and 500 RU of protein (usually DM Mut2 and DM WT) were immobilized in two consecutive flow cells on a SA chip at a flow rate of 2-10 μl/min at 25 °C unless otherwise indicated. SPR experiments were usually performed at 25 °C (range of 20 °C to 37 °C). When an experiment was carried out at a different temperature, the chip was again normalized after change of temperature was completed. DR-peptide complexes were purified by gel filtration chromatography prior to SPR experiments to remove any aggregates. DR-peptide complexes (analyte) were diluted into the running buffer just before injection and injected at the indicated concentrations and conditions (stage 1). This was followed by buffer (stage 2) and injection of peptide (stage 3). Regeneration of the chip was carried out via injection of 50–100 μM of high-affinity peptide for the respective DR allele. Experiments were carried out in degassed 50 mM citrate-phosphate buffer (pH 5.35) with 150 mM NaCl and 0.06% C12E9 detergent (Calbiochem, EMD Chemicals) unless otherwise indicated. Binding in the reference flow cell (in most cases DM Mut2) was subtracted from binding in the DM WT flow cell.
Peptide binding assay
FP assays were carried out as previously described47. Briefly, DR-peptide complexes were incubated with Alexa488-labeled MBP85-99 or HA306-318 peptide in a 40 μl volume in black polystyrene 384-well flat bottom plates (Corning) in the indicated conditions and binding of labeled peptide was measured using a Victor3 V plate reader (PerkinElmer). Measurements were made in triplicates. MBP85-99 was labeled at the P5 position (P5 Lys to Cys mutation) with a maleimide derivative of Alexa-488 (MolecularProbes) 47.
Supplementary Material
Acknowledgments
We thank A. Chakraborty (Massachusetts Institute of Technology) for helpful discussions, H. Ploegh and G. Grotenbreg (Massachusetts Institute of Technology) for providing the photolabile peptide, J. Fraser (University of Auckland) for human rhinovirus 3C protease and J. Pyrdol for help with purification of DR-peptide complexes. This work was supported by NIH grants to K.W.W. (RO1 AI057493, NS044914).
Footnotes
Author Contributions
A.-K.A., M.J.C. and K.W.W. conceived the study, designed experiments and wrote the paper, A.-K.A. generated DR-peptide complexes and performed most SPR experiments, M.-S.E.D.S. performed a set of SPR experiments with high-affinity DR-peptide complexes, E.J.S. provided advice for SPR experiments, M.J.C. performed most FP peptide binding assays, K.D.F. performed mathematical modeling of Biacore data, D.A.S. performed functional assays on DR-peptide complexes and N.P.S. generated CHO cell lines producing DR-CLIP complexes.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Lanzavecchia A, Reid PA, Watts C. Irreversible association of peptides with class II MHC molecules in living cells. Nature. 1992;357:249–252. doi: 10.1038/357249a0. [DOI] [PubMed] [Google Scholar]
- 2.Jensen PE. Long-lived complexes between peptide and class II major histocompatibility complex are formed at low pH with no requirement for pH neutralization. J Exp Med. 1992;176:793–798. doi: 10.1084/jem.176.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern LJ, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown JH, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 5.Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 6.Rabinowitz JD, et al. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 7.Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- 8.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 9.Morris P, et al. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- 10.Stebbins CC, Loss GE, Jr, Elias CG, Chervonsky A, Sant AJ. The requirement for DM in class II-restricted antigen presentation and SDS-stable dimer formation is allele and species dependent. J Exp Med. 1995;181:223–234. doi: 10.1084/jem.181.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan VS, et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 12.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 13.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 14.Nanda NK, Sant AJ. DM determines the cryptic and immunodominant fate of T cell epitopes. J Exp Med. 2000;192:781–788. doi: 10.1084/jem.192.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz JF, Stebbins C, Appella E, Sant AJ. Invariant chain and DM edit self-peptide presentation by major histocompatibility complex (MHC) class II molecules. J Exp Med. 1996;184:1747–1753. doi: 10.1084/jem.184.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lich JD, Jayne JA, Zhou D, Elliott JF, Blum JS. Editing of an immunodominant epitope of glutamate decarboxylase by HLA-DM. J Immunol. 2003;171:853–859. doi: 10.4049/jimmunol.171.2.853. [DOI] [PubMed] [Google Scholar]
- 17.Lovitch SB, Petzold SJ, Unanue ER. Cutting edge: H-2DM is responsible for the large differences in presentation among peptides selected by I-Ak during antigen processing. J Immunol. 2003;171:2183–2186. doi: 10.4049/jimmunol.171.5.2183. [DOI] [PubMed] [Google Scholar]
- 18.Pathak SS, Lich JD, Blum JS. Cutting edge: editing of recycling class II:peptide complexes by HLA-DM. J Immunol. 2001;167:632–635. doi: 10.4049/jimmunol.167.2.632. [DOI] [PubMed] [Google Scholar]
- 19.Lazarski CA, et al. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 23.Grotenbreg GM, et al. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosyak L, Zaller DM, Wiley DC. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- 25.Fremont DH, Crawford F, Marrack P, Hendrickson WA, Kappler J. Crystal structure of mouse H2-M. Immunity. 1998;9:385–393. doi: 10.1016/s1074-7613(00)80621-4. [DOI] [PubMed] [Google Scholar]
- 26.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 27.Pashine A, et al. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 28.Stratikos E, Mosyak L, Zaller DM, Wiley DC. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-DM catalysis. J Exp Med. 2002;196:173–183. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 30.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 31.Narayan K, et al. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrante A, Gorski J. Cutting edge: HLA-DM-mediated peptide exchange functions normally on MHC class II-peptide complexes that have been weakened by elimination of a conserved hydrogen bond. J Immunol. 2010;184:1153–1158. doi: 10.4049/jimmunol.0902878. [DOI] [PubMed] [Google Scholar]
- 34.Stratikos E, Wiley DC, Stern LJ. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II alpha-chain. J Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- 35.Zarutskie JA, et al. A conformational change in the human major histocompatibility complex protein HLA-DR1 induced by peptide binding. Biochemistry. 1999;38:5878–5887. doi: 10.1021/bi983048m. [DOI] [PubMed] [Google Scholar]
- 36.Weber DA, Dao CT, Jun J, Wigal JL, Jensen PE. Transmembrane domain-mediated colocalization of HLA-DM and HLA-DR is required for optimal HLA-DM catalytic activity. J Immunol. 2001;167:5167–5174. doi: 10.4049/jimmunol.167.9.5167. [DOI] [PubMed] [Google Scholar]
- 37.Chicz RM, et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358:764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 38.Kropshofer H, et al. Editing of the HLA-DR-peptide repertoire by HLA-DM. Embo J. 1996;15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- 39.Vogt AB, Kropshofer H, Moldenhauer G, Hammerling GJ. Kinetic analysis of peptide loading onto HLA-DR molecules mediated by HLA-DM. Proc Natl Acad Sci U S A. 1996;93:9724–9729. doi: 10.1073/pnas.93.18.9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jardetzky TS, et al. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. EMBO J. 1990;9:1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McFarland BJ, Katz JF, Beeson C, Sant AJ. Energetic asymmetry among hydrogen bonds in MHC class II*peptide complexes. Proc Natl Acad Sci U S A. 2001;98:9231–9236. doi: 10.1073/pnas.151131498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reay PA, Wettstein DA, Davis MM. pH dependence and exchange of high and low responder peptides binding to a class II MHC molecule. EMBO J. 1992;11:2829–2839. doi: 10.1002/j.1460-2075.1992.tb05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanderson F, Thomas C, Neefjes J, Trowsdale J. Association between HLA-DM and HLA-DR in vivo. Immunity. 1996;4:87–96. doi: 10.1016/s1074-7613(00)80301-5. [DOI] [PubMed] [Google Scholar]
- 44.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS One. 2008;3:e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patil NS, et al. Rheumatoid arthritis (RA)-associated HLA-DR alleles form less stable complexes with class II-associated invariant chain peptide than non-RA-associated HLA-DR alleles. J Immunol. 2001;167:7157–7168. doi: 10.4049/jimmunol.167.12.7157. [DOI] [PubMed] [Google Scholar]
- 47.Nicholson MJ, et al. Small molecules that enhance the catalytic efficiency of HLA-DM. J Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day CL, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Call MJ, et al. In vivo enhancement of peptide display by MHC class II molecules with small molecule catalysts of peptide exchange. J Immunol. 2009;182:6342–6352. doi: 10.4049/jimmunol.0803464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.