Abstract
In the design of nanoparticles that can target disease tissue in vivo, parameters such as targeting ligand density, type of target receptor, and nanoparticle shape can play an important role in determining the extent of accumulation. Herein, a systematic study of these parameters for the targeting of mouse xenograft tumors is performed using superparamagnetic iron oxide as a model nanoparticle system. The type of targeting peptide (recognizing cell surface versus extracellular matrix), the surface coverage of the peptide, its attachment chemistry, and the shape of the nanomaterial [elongated (nanoworm, NW) versus spherical (nanosphere, NS)] are varied. Nanoparticle circulation times and in vivo tumor-targeting efficiencies are quantified in two xenograft models of human tumors (MDA-MB-435 human carcinoma and HT1080 human fibrosarcoma). It is found that the in vivo tumor-targeting ability of the NW is superior to that of the NS, that the smaller, neutral CREKA targeting group is more effective than the larger, positively charged F3 molecule, that a maximum in tumor-targeting efficiency and blood half-life is observed with ≈60 CREKA peptides per NW for either the HT1080 or theMDA-MB-435 tumor types, and that incorporation of a 5-kDa polyethylene glycol linker improves targeting to both tumor types relative to a short linker. It is concluded that the blood half-life of a targeting molecule–nanomaterial ensemble is a key consideration when selecting the appropriate ligand and nanoparticle chemistry for tumor targeting.
Keywords: imaging, magnetic materials, nanoworms, peptides, tumor targeting
1. Introduction
Targeted nanomaterial-based imaging and drug-delivery vehicles have tremendous potential to improve the current clinical paradigms of cancer diagnosis and therapeutics.[1–15] Current clinically approved nanoparticle formulations accumulate preferentially in tumors through the enhanced permeability and retention (EPR) effect,[16,17] whereby materials in the size range of 20–200nm preferentially extravasate through transendothelial pores in tumor vessels and are retained in part because of nonfunctional tumor lymphatic vessels. The attachment of tumor-specific targeting ligands to nanomaterials has the promise to enhance tumor targeting for improved detection and therapy. However, modification of nanomaterial surfaces with targeting ligands that contain charged domains (i.e., cell-penetrating peptide sequences, nucleic acid-based agents, etc.),[18–20] or that consist of large biomolecules (i.e., immunoglobulins)[21,22] generally will decrease the circulation time of a nanomaterial in vivo because these moieties are recognized and eliminated by macrophages. With reduced circulation times, the targeting groups become less effective at homing to and penetrating their tumor targets. Long, hydrophilic polymer tethers can reduce macrophage recognition of ligands,[23] but they introduce an entropic penalty to the tumor–ligand binding process.[24] Thus, it appears that optimized nanoparticle formulations for tumor targeting exist within a broad, complex chemical space and may be surrounded by a vast landscape of substandard derivatives. A careful, systematic screening of the cooperative relationships between nanomaterial properties and tumor-targeting efficiency is needed.
Despite the critical role they play in targeting nanomaterials to tumors, relatively few studies have attempted to maximize in vivo targeting efficiency through systematic variation of surface charge, targeting group, or nanomaterial shape. Recently, Gu et al. screened targeted drug-encapsulated polymer nanoparticles with different compositions of self-assembled diblock copolymers and aptamers to find optimally formulated nanoparticles.[14] However, the tumor-targeting efficiency (0.5–2%ID g−1; ID = injected dose) was too low to reproducibly differentiate the optimized formulation from the other candidates. There have been some impressive reports of in vivo tumor targeting with various targeting ligand/nanoparticle combinations, such as folic acid-modified dendrimers (≈9%ID g−1),[3] synthetic small-molecule-modified iron oxide (IO) nanoparticles (≈4%ID g−1),[4] polyethylene glycol-coated (PEGylated) arginine–glycine–aspartic acid (RGD) peptide-modified carbon nanotubes (10–15%ID g−1),[10] and PEGylated single-chain variable-fragment (ScFv) antibody-modified gold nanoparticles (≈8%ID g−1).[13] However, the extent to which these materials represent optimized formulations for a given targeting ligand/nanomaterial combination remains unclear, providing relatively little insight into the design of more powerful nanoprobes.
In addition, the role of nanomaterial shape in tumor targeting has received relatively little attention, although there are indications that nanomaterials with elongated shapes and correspondingly increased surface area are more effective in vivo due to geometrically enhanced multivalent interaction between ligands and receptors.[10,25,26] It has become increasingly clear that the tumor-homing capability of a nanodevice that has been optimized in vitro is not predictive of in vivo results. In this work, we systematically optimize in vivo tumor targeting by varying the targeting ligand type (recognizing cell surface versus extracellular matrix), ligand surface coverage, attachment chemistry, and nanomaterial shape (elongated versus spherical). The particle circulation time and the in vivo tumor-targeting efficiency of each class of nanomaterial were quantified in two xenograft models of human tumors (MDA-MB-435 human carcinoma and HT1080 human fibrosarcoma).
2. Results and Discussion
Two different types of tumor-homing peptides were used as targeting ligands: F3, a 31-residue peptide, which preferentially binds to blood vessels and tumor cells in various tumors,[27,28] and CREKA, a five-residue peptide (sequence CREKA), which recognizes clotted plasma proteins in the blood vessels and stroma of tumors.[8] In addition, two types of nanomaterials that display excellent in vivo tumor targeting were studied: superparamagnetic IO nanospheres (NSs) with a mean diameter of ≈30 nm, and superparamagnetic IO nanoworms (NWs) with a long dimension of ≈70 nm and thickness of ≈30 nm. Both nanoparticle types were prepared by previously reported methods.[26,29] All samples were dextran-coated, containing varying degrees of amination on the dextran, one of two different types of dextran–peptide linkers (5 kDa PEG versus short hydrocarbon chain), and varying numbers of targeting peptides (Scheme 1 and Table 1). We have recently published a study of the in vitro tumor-cell-targeting capabilities of some of these formulations.[26]
Scheme 5.
Three parameters varied to determine optimal in vivo tumor targeting: the shape of the nanoparticle, the type of targeting ligand, and the nature of the molecular linker. Two types of surface linkers are used to attach targeting groups to IO NWs or NSs. A short hydrocarbon places the targeting peptide (either F3 or CREKA, green lines) in close proximity to the dextran-coated nanostructure. A 5-kDa PEG linker places it further away. The number of targeting groups per NW was varied to find the maximal in vivo circulation times and the optimal in vivo tumor-targeting efficiency. These same chemistries were tested on IO NSs; the NWs consist of several of these NS cores linked together in a chain. SMCC = sulfosuccinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate.
Table 1.
Characteristics of targeted and untargeted NWs and NSs.
| Sample[a] | Size [nm][b] |
Targeting peptide |
Number of peptides | |
|---|---|---|---|---|
| per NW or NS[c] |
per g Fe (×1020)[d] |
|||
| NS | 30.3 | none | ||
| NS-30-C | 34.3 | CREKA | 18 | 9.4 |
| NS-P30-C | 46.8 | CREKA | 13 | 6.8 |
| MM-500-C | 107.2 | CREKA | 350 | |
| NW | 68.7 | none | ||
| NW-42-F | 73.7 | F3 | 23 | 1.7 |
| NW-P42-F | 87.3 | F3 | 16 | 1.2 |
| NW-175-F | 76.6 | F3 | 69 | 5.1 |
| NW-P175-F | 88.2 | F3 | 48 | 3.0 |
| NW-350-F | 76.1 | F3 | 83 | 6.2 |
| NW-P350-F | 90.8 | F3 | 59 | 4.4 |
| NW-42-C | 70.9 | CREKA | 29 | 1.6 |
| NW-P42-C | 82.4 | CREKA | 23 | 1.2 |
| NW-175-C | 70.2 | CREKA | 117 | 6.3 |
| NW-P175-C | 85.0 | CREKA | 60 | 3.2 |
| NW-350-C | 72.3 | CREKA | 205 | 10.2 |
| NW-P350-C | 85.5 | CREKA | 90 | 4.9 |
The number following the letter identifier designates the number of amine groups per particle. The letter P indicates that a PEG spacer is used. The -F or -C suffix denotes an F3- or CREKA-conjugated particle, respectively. For example, NW-P175-C denotes a NW with 175 amines to which CREKA is conjugated through a PEG spacer. MM = aminated Micromod. Micromod is a commercially available IO nanoparticle preparation.
Mean hydrodynamic size based on dynamic light scattering measurements.
Number of targeting peptides per single NW or NS.
Number of targeting peptides (×1020) per gram of Fe.
We first tested various formulations of F3-conjugated NW (NW-F) for in vivo tumor targeting, since it has been reported that F3 peptide effectively transports payloads, such as nanoparticles or oligonucleotides, into tumor vasculature in vivo (Table 1).[1,30,31] Mice bearing MDA-MB-435 tumors were injected with the various formulations of NW-F. In all cases, the NW-F preparations were cleared from the blood mainly by the liver within 1 h, regardless of the number of attached peptides or the presence of a PEG layer, and no tumor homing was observed. This somewhat contrasts with previous findings with other types of nanoparticles[1,30,31] (see Supporting Information, Figure S1). It may be that IO nanoparticles are particularly prone to uptake by the mononuclear phagocytic system (MPS), and that this tendency is augmented by the large number of positively charged residues of the many F3 peptides on the NW surface. We conclude that the kinetics of NW-F binding to the intended tumor target is not large or favorable enough to compete with rapid uptake by the MPS in vivo.
In contrast to the in vivo behavior of F3-modified NWs, when CREKA is used as the targeting peptide (NW-C), the NWs effectively home to their tumor targets (Figure 1). CREKA is a short linear peptide that is neutrally charged and most likely nonimmunogenic.[8,22] A tradeoff between the number of attached peptides and the efficiency of tumor targeting is observed for the NW-C preparations; the most effective in vivo tumor targeting is observed with ≈60 CREKA peptides per NW. This maximum correlates with a substantial decrease in blood half-life that is observed when >60 CREKA peptides are attached to a NW. The trend is observed for both HT1080 and MDA-MB-435 tumor types, although the overall targeting efficiency of NW-C is greater for HT1080 tumors. Additionally, in contrast to the NW-F preparations, significantly long circulation times (>10 h) are observed with some of the NW-C preparations.
Figure 1.
Comparison of in vivo blood circulation half-life and tumor-targeting efficiency of NWs as a function of CREKA targeting peptide density (mouse model). Blood half-life in mice without tumors is shown in the bottom plot, and the percentage of injected dose (%ID g−1) of NWs that target MDA-MB-435 and HT-1080 tumors are shown in the middle and the top plots, respectively. The effect of using a PEG linker to attach the CREKA targeting peptide (●, “NW-P-C”) is compared with a short hydrocarbon linker (○, “NW-C”). Data obtained from SQUID measurements performed on blood or tissue samples, obtained 24 h post-injection.
For both HT1080 and MDA-MB-435 tumors, greater targeting efficiency is observed for NW-C when a PEG linker is used to attach the CREKA targeting group. It is postulated that the PEG linker facilitates CREKA homing by providing a less restrictive environment (relative to the short sulfo-SMCC linker), thus improving the peptide’s ability to bind to clotted plasma proteins associated with the tumor. Additionally, the PEG linker increases the residence time of the nanostructure in the bloodstream. These results are distinct from the previous in vitro observations with PEGylated NW-F,[26] because the targeting moiety in that case (F3) was a cell-internalizing peptide that is found in this study to be less suitable for in vivo targeting of nanoparticles. The reason is that F3-coated particles are cleared by MPS-related organs, presumably because multiple copies of the highly cationic F3 cause a large increase in surface charge of the particles. Moreover, we previously showed that PEGylated F3-NWs are less effectively internalized into tumor cells than F3-NWs without PEG.[26] Thus, the PEG linker can impede the uptake of nanoparticles into cells, but it does not appear to significantly interfere with the binding of nanoparticles to receptors.
The decrease in circulation time observed for NWs containing >60 CREKA peptides is attributed to the presence of unreacted amines and damage to the dextran coating (exposing bare IO cores) that occurs during preparation of the more extensively functionalized nanoparticles. The data indicate that the blood half-life of a targeting molecule/nanoparticle ensemble must be considered when selecting the appropriate ligand to target a tumor. As also observed with NW-F, a dramatic decrease in circulation time and a corresponding decrease in targeting efficiency can occur when targeting ligands are linked to nanomaterials.
A control experiment using KAREC, a scrambled version of CREKA, was performed in mice bearing MDA-MB-435 tumors. KAREC was attached to the NW using a PEG linker, and the formulations displayed similar circulation times to the PEGylated NW-C formulations. Significantly lower tumor-targeting efficiency was observed with the scrambled peptide (Supporting Information, Figure S2).
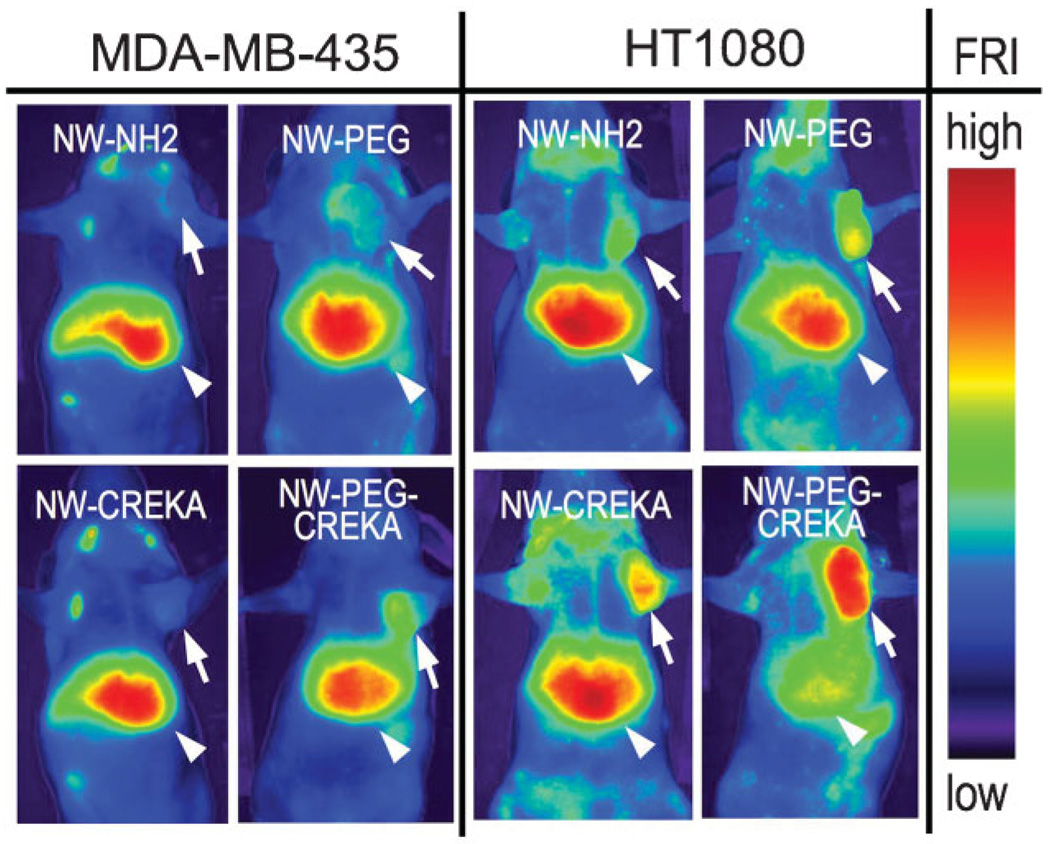
Near-infrared (NIR) fluorescence images of mice injected with NW-C confirm the tumor uptake results obtained by superconducting quantum interference device (SQUID) magnetic measurements (Figure 2 and Supporting Information, Figure S3). Significant increases in the tumor/liver fluorescence signal ratio are observed in both tumor types for PEGylated NW-C (sample NW-P175-C) compared with the other samples studied. Targeting of PEGylated NW-C could be observed in smaller tumors (size 0.2 cm, see Supporting Information, Figure S4a), which indicates that the formulation is applicable for the detection of tumors at the early stages of growth. NIR fluorescence images of organs and biodistribution results in mice bearing MDA-MB-435 tumors 24 h post-injection reveal that most of the NW-C is cleared by the liver and spleen of the mouse, similar to what is observed with other targeted nanomaterials[3–5,10,14] (Figure 3a and Supporting Information Figure S4a). PEGylated NW-C shows relatively greater uptake by the spleen, while NW-C formulations containing the short-chain linker display somewhat greater uptake by the liver.
Figure 2.
NIR fluorescence images of mice injected with various NW formulations, obtained 24 h post-injection (NW-NH2 = NW-175, NW-PEG = NW-P175, NW-CREKA = NW-175-C, NW-PEG-CREKA = NW-P175-C; see Table 1 for definitions of these abbreviations). All samples contain a Cy7 fluorescent dye covalently linked to the dextran coating of the NW, and the images are obtained by observation in the Cy7 channel. Arrows and arrowheads point to the tumors and the livers, respectively.
Figure 3.
a) SQUID quantification of biodistribution of unmodified NW, PEGylated NW-C (NW-P175-C), and NW-C (NW-175-C) in mice bearing MDA-MB-435 tumors, obtained 24 h post-injection. b) Histological images of PEGylated NW-C (NW-P175-C, green) in MDA-MB-435 and HT1080 tumors. The tumors were collected 24 h post-injection. Blood vessels are stained red and cell nuclei are blue. Scale bars: 100µm.
Histological analysis revealed that most of the PEGylated NW-C localizes with large blood vessels in the MDA-MB-435 tumor, whereas it extravasates into the tumor tissue along the smaller vessels in the HT1080 tumor (Figure 3b). In addition, NWs in the MDA-MB-435 tumor co-localize with fibrin(ogen) in the blood vessels, indicative of the self-amplifying homing that has been observed previously (Supporting Information, left panel in Figure S4b).[8] NWs in the HT1080 tumor localize with fibrin(ogen) in blood vessels as well as in tumor stroma (Supporting Information, right panels in Figure S4b). These results suggest that HT1080 tumors, like other tumors,[32,33] contain clotted plasma proteins that provide initial binding sites for the CREKA peptide, and that the nanoparticles induce additional clotting within the tumor. Thus, the larger uptake of NW-C observed in the HT1080 tumor relative to the MDA-MB-435 tumor is attributed to passive transport across a highly vascularized and porous microstructure,[34] active peptide-mediated binding, and self-amplifying homing due to clotting induced by the CREKA-coated particles.
Lastly, we compared the targeting efficiency of NW-C with CREKA-conjugated nanospheres (NS-C, blood half-lives: 10.6 h for NS-30-C and 17.9 h for NS-P30-C, Table 1). We have previously reported that the elongated NWs more effectively attach to tumor cells in vitro while exhibiting comparable blood circulation times relative to spherical NSs. The superior in vitro targeting efficiency was attributed to multivalent interactions between the elongated NWs and receptors on the tumor cell surface.[26] We find a similar improvement in tumor targeting by NWs in vivo. The optimized NW-C formulation (NW-P175-C) displays significantly higher levels of uptake in MDA-MB-435 tumors relative to NS-C (NS-P30-C; Figure 4). We also compared the targeting efficacy of NW-C with CREKA-conjugated commercial IO nanoparticles (MM-500-C, blood half-life: ≈30 min). This latter formulation was used in the first study demonstrating CREKA-mediated nanoparticle targeting to tumors.[8] In that study, CREKA-conjugated IO nanoparticles accumulated in tumors, but only after preinjection with Ni-liposomes designed to inhibit MPS uptake. This inability of the nanoparticles to evade the MPS by themselves highlighted a significant limitation to the practical application of nanoparticle therapies that is overcome in the present study.
Figure 4.
SQUID quantification of in vivo tumor-targeting efficiency of NW, NS, and Micromod samples, with and without CREKA targeting peptide, in mice bearing MDA-MB-435 tumors, 24 h post-injection.
3. Conclusions
We have demonstrated that careful, systematic evaluation of nanomaterials across a diverse material and surface chemical space, which incorporates varying nanoparticle structural (i.e., spherical versus elongated) and surface characteristics (i.e., targeting ligand type, ligand surface density, and linker chemistry), can produce dramatic increases in tumor-targeting potency. We suggest a few “design rules” for the future development and evaluation of targeted nanomaterial diagnostics and therapeutics:
Because tumor vascular volumes represent a small fraction of the complete host vasculature, parent nanomaterial formulations should be optimized to exhibit long-term stability and long blood circulation times prior to incorporation of targeting ligands.
Ligand affinity should not be considered as the only determinant of effective nanoparticle targeting. Highly cationic ligands and large biomolecules may produce dramatic alterations in blood circulation times, which prevent access to receptors and prohibit effective targeting.
Due to the dramatic effect circulation time has on targeting efficiency, it should be emphasized that multiple, circulation- matched controls (including unmodified materials and scrambled ligand controls) should be included in the assessment of targeting in order to decipher EPR-mediated accumulation from targeted accumulation.
Finally, our finding that the targeting efficiency of one-dimensional NWs is enhanced compared to that of their spherical counterparts suggests that shape is an important determinant for effective imaging and drug delivery with nanomaterials. The elongated shape prolongs in vivo circulation and provides a more effective scaffold to generate multivalent interactions between homing moieties and their in vivo targets.
This study suggests that a systematic approach to the engineering of nanomaterials that includes a strong in vivo component is essential in transitioning nanodevices into clinical applications.
4. Experimental Section
NW and NS preparation
Magnetic IO NWs with a longitudinal size of ≈70 nm were synthesized as previously described.[26] Magnetic IO NSs with a size of ≈25 nm were synthesized using the published preparation of crosslinked dextran-coated IO nanoparticles.[35] Micromod nanoparticles (MM, spherical dextran-coated IO nanoparticles with free amines) with a size of ≈100 nm were obtained from Micromod Partikeltechnologie GmbH (Rostock, Germany). NWs or NSs with different numbers of free amines were prepared for peptide conjugation by reacting them with different concentrations of aqueous ammonia at room temperature for 48 h. The amine number per NW or NS was measured with the SPDP assay.[29] The amine number per NW was calculated by assuming that the molecular weight of a NW is 7 times higher than that of a NS, based on the mean value of aggregated IO cores for one NW observed in transmission electron microscopy (TEM) images and supported by dynamic light scattering (DLS) data. The sizes, shapes, and surface charges of NWs, NSs, or MM were characterized by TEM and DLS measurements as previously reported.[26]
Targeting peptide conjugation
One of two targeting peptides was used with the NW, NS, or MM samples: KDEPQRRSARLSAKPAPPKPEPKPKKAPAKK (F3), which preferentially binds to blood vessels and tumor cells in various tumors,[27,28] and CREKA, which recognizes clotted plasma proteins in the blood vessels and stroma of tumors.[8] The fluorescein (FITC)-conjugated peptides were synthesized by Fmoc chemistry in a solid-phase synthesizer, and purified by preparative HPLC. The sequences and compositions were confirmed by mass spectrometry. For the F3 peptide, an extra cysteine residue was added to the N terminus to allow conjugation to the aminated dextran coating of the NWs or NSs. For NIR fluorescence imaging, NWs or NSs were first labeled with either Cy5.5 or Cy7 fluorescent molecules. The remaining free amines were used for conjugation with the targeting peptides. Briefly, aminated NWs (500 µg Fe) or NSs (900 µg Fe) were reacted in phosphate-buffered saline (PBS) with Cy5.5-NHS ester (8 µg) or Cy7-NHS ester (6 µg, GE Healthcare Bio-Sciences) in DMSO (Sigma) for 1 h to display the same fluorescence intensity per iron atom for both NWs and NSs (one Cy5.5/Cy7 dye per single IO core). Next, Cy5.5/Cy7-labeled NWs or NSs (500µg Fe) were reacted with sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC; 200 µg, Pierce Chemicals) or NHS-PEG(5 kDa)-MAL (2 mg, Nektar) in PBS for 1 h and then purified using an ACA54 desalting column (Pall, USA). Targeting peptide with a free terminal cysteine (200 µg) was then added to the NW or NS sample (500 µg Fe) in PBS. After incubation for 2 h with mild shaking at room temperature, the sample was purified on the desalting column and then resuspended in PBS. The number of Cy5.5/Cy7 dye or FITC-peptide molecules per single NW or NS was determined from the absorbance spectrum. Peptide conjugation to the particles through PEG chains resulted in fewer peptides per particle (Table 1).
Blood half-life determination
All animal work was performed in accordance with the institutional animal protocol guidelines in place at the Burnham Institute for Medical Research, and it was reviewed and approved by the Institute’s Animal Research Committee. NWs, NSs, or MM in PBS (100 µL) were intravenously injected into nude BALB/c mice (3 mg Fe kg−1, n = 3–4 for each formulation). Heparinized capillary tubes (Fisher) were used to draw blood (15µL for fluorescence, or 70 µL for magnetization) from the periorbital plexus at several different times after intravenous injection. The extracted blood samples were immediately mixed with ethylenediaminetetraacetate (10mM in PBS) to prevent coagulation. For Cy7-labeled formulations, the blood samples were imaged in a 96-well plate in the Cy7 channel (760 nm excitation/800 nm emission) with a NIR fluorescence scanner (LI-COR Biosciences, NE, USA). The images were analyzed by using the ImageJ (NIH) or Osirix (Apple) programs. For nonlabeled samples, the blood samples were immediately freeze-dried in gelatin capsules, and then analyzed for magnetization using a SQUID magnetometer.[36] The blood half-lives of NWs, NSs, and MM were calculated by fitting the fluorescence or magnetization data to a single-exponential equation using a one-compartment open pharmacokinetic model.[18]
Biodistribution measurement
For the mouse biodistribution studies, unmodified NWs, NW-C, and PEGylated NW-C in PBS (100 µL) were intravenously injected into MDA-MB-435 tumor-bearing nude BALB/c mice at a dose of 3 mg Fe kg−1 body mass (n = 3–4 for both the PBS controls and the NW samples). The animals were sacrificed 24 h after injection by cardiac perfusion with PBS under anesthesia, and the brain, kidney, liver, lung, spleen, and tumor were collected. For magnetic measurements, the organs were immediately weighed, freeze-dried in gelatin capsules, and then analyzed for magnetization using SQUID. Percentages of injected dose of the NWs per wet weight of each organ were further corrected by subtracting the magnetization of the PBS-injected organs (controls) from the magnetization of the particle-injected organs. For fluorescence imaging, the organs were imaged in the Cy7 channel with a NIR fluorescence scanner. All the NIR images for animals or organs were taken at the same exposure time.
Magnetic measurements for blood half-life and biodistribution:[36]
SQUID magnetometry provides a direct measure of the total number of magnetic IO nanoparticles in a sample, as it measures the magnetization of a sample rather than the total iron content or the fluorescence intensity from a molecular tag. The SQUID measurements are thus more relevant to magnetic resonance imaging applications, because the magnetization data correlate with T2. The SQUID technique has the additional advantage that it can be performed on whole organs or blood, and little sample workup is needed. The blood samples and organs collected from the mice injected intravenously with the NWs, NSs, or MM were frozen and lyophilized to dryness in gelatin capsules. The capsules were inserted into the middle of transparent plastic drinking straws. The measurements were performed at 150 K with a Quantum Design (CA, USA) MPMS2 SQUID magnetometer. The samples were exposed to direct-current magnetic fields in stepwise increments up to 1 Tesla. Corrections were made for the diamagnetic contribution of the capsule and straw.
In vivo tumor homing
MDA-MB-435 human carcinoma cells or HT1080 human fibrosarcoma cells (1 × 106) were injected into the mammary fat pad or subcutaneously injected into nude BALB/c mice, respectively. Tumors were used when they reached ≈0.5 cm in size. Some 0.2- or 1-cm tumors were used to compare the dependence of tumor size on NW homing. Cy7-labeled or nonlabeled NWs or NSs were intravenously injected into mice (n = 3–8 for each formulation) with a dose of 3 mg Fe kg−1 body mass. For real-time observation of tumor/liver uptake, animals were imaged under anesthesia in the Cy7 channel by using the BonSai fluorescence-imaging system (Siemens, PA, USA) 24 h after injection. To quantify the amount of NW or NS homing, collected tumors 24 h post-injection were weighed, freeze-dried in gelatin capsules, and then analyzed for magnetization using SQUID.[36] For histologic analysis, frozen sections of tumors were prepared. The sections were fixed with 4% paraformaldehyde and stained with 4’,6-diamidino-2-phenylindole (DAPI) for observation of NWs or NSs only. Rat antimouse CD-31 (1:50, BD PharMingen) and biotinylated mouse fibrin(ogen) antiserum (1:50, Nordic) were used for immunochemical staining of tumor tissue sections. The corresponding secondary antibodies were added and incubated for 1 h at room temperature: Alexa Fluor 594 goat antirat or rabbit immunoglobulin G (1:1000, Molecular Probes), streptavidin Alexa Fluor 594 (1:1000; Molecular Probes). The slides were washed three times with PBS and mounted in Vectashield Mounting Medium with DAPI. At least three images from representative microscopic fields were analyzed for each tumor sample.
Supplementary Material
Acknowledgements
This project has been funded in part with Federal funds from the National Cancer Institute of the National Institutes of Health (Contract Nos. R01CA124427-02, U54CA119349, and U01 HL 080718). M.J.S., E.R., and S.N.B. are members of the Moores UCSD Cancer Center and the UCSD NanoTUMOR Center under which this research was conducted and partially supported by NIH Grant U54 CA 119335. J.P. thanks the Korea Science and Engineering Foundation (KOSEF) for a Graduate Study Abroad Scholarship. The authors thank Todd Sponholtz and Dr. Ralph Weissleder for use of the NIR fluorescence-imaging system.
Footnotes
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
Contributor Information
Ji-Ho Park, Materials Science and Engineering Program, Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman, La Jolla, CA 92093 (USA).
Geoffrey von Maltzahn, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA).
Lianglin Zhang, Cancer Research Center, Burnham Institute for Medical Research, 10901 N. Torrey Pines Rd., La Jolla, CA 92037 (USA); Vascular Mapping Center, Burnham Institute for Medical Research (at UCSB) Bio II, Rm. #3119, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA).
Austin M. Derfus, Department of Bioengineering, University of California, San Diego, 9500 Gilman, La Jolla, CA 92093 (USA)
Dmitri Simberg, Cancer Research Center, Burnham Institute for Medical Research, 10901 N. Torrey Pines Rd., La Jolla, CA 92037 (USA); Vascular Mapping Center, Burnham Institute for Medical Research (at UCSB) Bio II, Rm. #3119, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA).
Todd J. Harris, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA)
Erkki Ruoslahti, Cancer Research Center, Burnham Institute for Medical Research, 10901 N. Torrey Pines Rd., La Jolla, CA 92037 (USA); Vascular Mapping Center, Burnham Institute for Medical Research (at UCSB) Bio II, Rm. #3119, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA).
Sangeeta N. Bhatia, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139 (USA)
Michael J. Sailor, Materials Science and Engineering Program, Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman, La Jolla, CA 92093 (USA)
References
- 1.Akerman ME, Chan WCW, Laakkonen P, Bhatia SN, Ruoslahti E. Proc. Natl. Acad. Sci. USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 3.Kukowska-Latallo JF, Candido KA, Cao Z, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR., Jr Cancer Res. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 4.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat. Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 5.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 6.Farokhzad OC, Cheng JJ, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc. Natl. Acad. Sci. USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai WB, Shin DW, Chen K, Gheysens O, Cao QZ, Wang SX, Gambhir SS, Chen XY. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 8.Simberg D, Duza T, Park JH, Essler M, Pilch J, Zhang LL, Derfus AM, Yang M, Hoffman RM, Bhatia S, Sailor MJ, Ruoslahti E. Proc. Natl. Acad. Sci. USA. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Proc. Natl. Acad. Sci. USA. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Cai WB, He LN, Nakayama N, Chen K, Sun XM, Chen XY, Dai HJ. Nat. Nanotechnol. 2007;2:47–52. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 11.Cheng J, Teply BA, Sheri I, Sung J, Luther G, Gu FX, Levy-Nissenbaum E, Radovic-Moreno AF, Langer R, Farokhzad OC. Biomaterials. 2007;28:869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Huh YM, Jun Y, Seo J, Jang J, Song HT, Kim S, Cho EJ, Yoon HG, Suh JS, Cheon J. Nat. Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 13.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 14.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. USA. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen TM. Nat. Rev. Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 16.Jain RK. Annu. Rev. Biomed. Eng. 1999;1:241–263. doi: 10.1146/annurev.bioeng.1.1.241. [DOI] [PubMed] [Google Scholar]
- 17.Maeda H, Wua J, Sawaa T, Matsumurab Y, Horic K. J. Controlled Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 18.Wunderbaldinger P, Josephson L, Weissleder R. Bioconjugate Chem. 2002;13:264–268. doi: 10.1021/bc015563u. [DOI] [PubMed] [Google Scholar]
- 19.Zorko M, Langel U. Adv. Drug Delivery Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Thomas M, Klibanov AM. Appl. Microbiol. Biotechnol. 2003;62:27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 21.Goldsmith SJ. Semin. Nucl. Med. 1997;27:85–93. doi: 10.1016/s0001-2998(97)80041-4. [DOI] [PubMed] [Google Scholar]
- 22.Becker A, Hessenius C, Licha K, Ebert B, Sukowski U, Semmler W, Wiedenmann B, Grotzinger C. Nat. Biotechnol. 2001;19:327–331. doi: 10.1038/86707. [DOI] [PubMed] [Google Scholar]
- 23.Duncan R. Nat. Rev. Cancer. 2006;6:688–701. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 24.Mammen M, Choi SK, Whitesides GM. Angew. Chem. 1998;110:2908–2953. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. Angew. Chem. Int. Ed.1998, 37, 2755–2794. [DOI] [PubMed] [Google Scholar]
- 25.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat. Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J-H, von Maltzahn G, Zhang L, Schwartz MP, Ruoslahti E, Bhatia SN, Sailor MJ. Adv. Mater. 2008;20:1630–1635. doi: 10.1002/adma.200800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porkka K, Laakkonen P, Hoffman JA, Bernasconi M, Ruoslahti E. Proc. Natl. Acad. Sci. USA. 2002;99:7444–7449. doi: 10.1073/pnas.062189599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christian S, Pilch J, Akerman ME, Porkka K, Laakkonen P, Ruoslahti E. J. Cell Biol. 2003;163:871–878. doi: 10.1083/jcb.200304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josephson L, Tung CH, Moore A, Weissleder R. Bioconjugate Chem. 1999;10:186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 30.Reddy GR, Bhojani MS, McConville P, Moody J, Moffat BA, Hall DE, Kim G, Koo YEL, Woolliscroft MJ, Sugai JV, Johnson TD, Philbert MA, Kopelman R, Rehemtulla A, Ross BD. Clin. Cancer Res. 2006;12:6677–6686. doi: 10.1158/1078-0432.CCR-06-0946. [DOI] [PubMed] [Google Scholar]
- 31.Henke E, Perk J, Vider J, de Candia P, Chin Y, Solit DB, Ponomarev V, Cartegni L, Manova K, Rosen N, Benezra R. Nat. Biotechnol. 2008;26:91–100. doi: 10.1038/nbt1366. [DOI] [PubMed] [Google Scholar]
- 32.Dvorak HF, Senger DR, Dvorak AM, Harvey VS, McDonagh J. Science. 1985;227:1059–1061. doi: 10.1126/science.3975602. [DOI] [PubMed] [Google Scholar]
- 33.Pilch J, Brown DM, Komatsu M, Jarvinen TAH, Yang M, Peters D, Hoffman RM, Ruoslahti E. Proc. Natl. Acad. Sci. USA. 2006;103:2800–2804. doi: 10.1073/pnas.0511219103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Nat. Med. 2006;12:354–360. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmacci S, Josephson L. 5262176. Cambridge, MA: Advanced Magnetics, Inc.; US Patent. 1993
- 36.Grassi-Schultheiss PP, Heller F, Dobson J. Biometals. 1997;10:351–355. doi: 10.1023/a:1018340920329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.