A multitude of biological processes requires the participation of specific cations, such as H+, Na+, K+, Ca2+, and Mg2+. Many of these processes can take place only when proteins have the ability to discriminate between different ions with a very high fidelity. How this is possible is a fundamental question that has fascinated scientists for a long time. At the most fundamental level, it is anticipated that ion selectivity must result from a delicate balance of strong interactions. Yet, identifying and quantifying the key microscopic factors is difficult, as many of these cannot be directly measured by experiments. Theory and computations can contribute by providing a virtual route to complement the missing information. Because ion selectivity is often dominated by thermodynamic factors, detailed molecular dynamics (MD) free energy simulations become important tools. This was vividly illustrated early on with studies of ion solvation (Straatsma and Berendsen, 1988; Åqvist, 1990) and ion-selective systems (Lybrand et al., 1986; Grootenhuis and Kollman, 1989; Åqvist, 1992). These pioneering studies inspired our own efforts.
In this Perspective, we aim to present our understanding of ion selectivity as it has evolved over approximately 15 years from studies based on various specific structures: gramicidin A channels (e.g., Roux, 1996; Allen et al., 2006), the KcsA channel (e.g., Bernèche and Roux, 2000, 2001, 2003, 2005; Noskov et al., 2004a; Egwolf and Roux, 2010), the NaK channel (Noskov and Roux, 2007), the LeuT transporter (Noskov and Roux, 2008), and the Na/K pump (Ratheal et al., 2010). We also discuss the insights that can be gained from simple models, which can be used to illustrate and clarify fundamental physical principles governing ion selectivity in channels and transporters (Noskov et al., 2004a; Noskov and Roux, 2007, 2008; Yu and Roux, 2009; Yu et al., 2009, 2010b; Roux, 2010a).
In Section 1, we briefly present some of the historical background that motivated and shaped our studies of ion selectivity in the last decade, which were prompted by the determination of the x-ray structure of the KcsA K+ channel. We then proceed with a discussion of the phenotype of selectivity (Section 2), free energy landscapes (Section 3), the theoretical formulation of reduced models (Section 4), and issues of accuracy in force fields (Section 5). The review ends with a brief conclusion in Section 6.
1. Historical context
The x-ray structure of the KcsA K+ channel offered a first chance to visualize the architecture of a highly selective biological channel (Doyle et al., 1998). The structure (Fig. 1 A) showed that K+ ions entering the narrowest region of the pore, the selectivity filter, are coordinated by backbone carbonyl oxygens (Fig. 1 B). The x-ray structure appeared to offer direct support for a simple and appealing structural explanation of ion selectivity in close correspondence with the snug-fit mechanism (Bezanilla and Armstrong, 1972), which thereafter became implicitly taken for granted (Hille et al., 1999). The general idea, borrowed from host–guest chemistry, is that the narrow pore is perfectly suited (at the sub-angstrom level) to provide a cavity of the appropriate size to fit K+, but is unable—for structural reasons—to adapt to the slightly smaller Na+ (Dietrich, 1985). Very early on, however, MD simulations based on all-atom models suggested that the selectivity filter of the KcsA channel might be too flexible to satisfy the requirement of the traditional snug-fit mechanism (Guidoni et al., 1999; Bernèche and Roux, 2000; Biggin et al., 2001). In MD simulations performed at room temperature, the fluctuating filter appeared to distort easily to cradle Na+ with little energetic cost, an observation that did not jibe with the proposed structural explanation of selectivity (Hille et al., 1999).
Figure 1.
Reduced model of the S2 site in the selectivity filter of the KcsA channel used by Yu et al. (2010b). (A) The full-length KcsA channel is shown with its four subunits. (B) The selectivity filter is represented with three K+ ions occupying the sites S0, S2, and S4. (C) The reduced model of the site S2 based on four diglycines is shown. This is adapted from Yu et al. (2010b).
MD simulations and multi-ion potential of mean force (PMF) computations were used to predict the position of all the K+ binding sites along the pore, showing that the free energy barriers opposing ion conduction were not larger than a few kBT and that the pore was selective for K+ over Na+ (Bernèche and Roux, 2001). All-atom alchemical free energy perturbation (FEP) MD (FEP/MD) simulations were performed to characterize ion selectivity in quantitative terms. The relative affinity of K+ and Na+, ΔΔGNa,K, was calculated for the five cation binding sites located along the narrow pore (S0–S4; Fig. 1 B),
| (1) |
where (By this definition, a binding site is K+selective when ΔΔGNa,K is positive and Na+selective when it is negative.) Such FEP/MD computations assume that ion selectivity can be understood on the basis of thermodynamic binding equilibrium, leaving out any kinetic considerations (see Section 3). The computations showed that the cation binding sites in the dynamical selectivity filter observed in the MD simulations were indeed selective for K+over Na+, particularly the site S2 located at the center of the narrow pore. At the time, the suggestion of a K+-selective flexible filter undergoing angstrom-scale fluctuations seemed to be conceptually incompatible with the available high-resolution x-ray structures.
To resolve this dilemma, a series of computer experiments were then performed on a wide range of atomic models of a KcsA binding site (Noskov et al., 2004a). The goal was, first, to present a convincing demonstration that ion selectivity was possible in a loosely restricted flexible binding site in which the conditions for the classical snug-fit mechanism were not realized, and second, to delineate the microscopic elements giving rise to selectivity under such conditions. The analysis led to a surprising and provocative conclusion. Robust free energy differences controlling selectivity were observed to arise principally from the number and type of ion-coordinating ligands, without the need to enforce the position of those ligands to sub-angstrom precision (Noskov et al., 2004a). Selectivity for K+ over Na+ was significant (i.e., >4–5 kcal/mol) only with eight carbonyl-like ligands. The relative free energy of K+ and Na+ in the pore resulted from a local competition between the favorable ion–ligand interactions and the unfavorable ligand–ligand interactions. This pointed to the importance of local energetics in flexible binding sites, a rather novel concept.
In this framework, the ion coordination by the carbonyl ligands was described as “dynamic” and “liquid-like,” as a contrast to the snug-fit picture with its geometry enforced to sub-angstrom positional precision. This has apparently led to some unfortunate confusion, as some have taken the semantic too literally. By liquid-like, it was meant that the carbonyls coordinating the ion are fluctuating like the ligands in the first coordination shell of an ion solvated in a liquid (i.e., the first peak in the radial distribution function of the ligands around the ion have similar widths). Furthermore, the relative free energy of K+ and Na+ in the pore arises from a balance of dynamical local interactions, which bears some similarities with ion solvation in a liquid (see Section 4). Describing the dynamics of proteins as being fluid-like (McCammon et al., 1977; Sheinerman and Brooks, 1998) or liquid-like (Caspar et al., 1988; Kneller and Smith, 1994; Lindorff-Larsen et al., 2005) is actually quite common. Nevertheless, semantic shortcuts can generate confusion because drawing a clear distinction between a material that is stiff and rigid versus one that is dynamic and flexible is partly subjective and context dependent. Notions of rigidity and flexibility in the case of ion channels have quantifiable meaning only once scales of lengths and energies have been specified (Allen et al., 2004). In physical chemistry, there is a long tradition that relies on the Lindemann melting criterion (Lindemann, 1910), according to which a material is declared to be more liquid-like than solid-like when the root-mean-square fluctuations of the atoms exceed ∼10–15% of the nearest neighbor distance. This type of analysis has often been used to contrast the folded and the so-called molten globule states of a protein (Zhou and Karplus, 1997, 1999; Jang et al., 2002). With root-mean-square fluctuations being ∼1 Å and ion-carbonyl distances ∼3 Å, the carbonyl ligands are liquid-like on atomic length scales according to the Lindemann criterion. But the analogies with ion solvation in a liquid should not be overstated. One should never lose sight that the atomic fluctuations from MD are at most on the order of ∼1 Å (Noskov et al., 2004a), and that the protein unequivocally plays an essential structural role by keeping the ion and the carbonyls restricted within some small region.
To further illustrate how size selectivity could be supported in a flexible binding site in the absence of any precise structural geometry, simple models (“toys”) were introduced with carbonyl-like groups confined to a spherical region (Noskov et al., 2004a). By construction, no restraint forces prevent the collapse of the ligands onto the ion. Yet, FEP/MD calculations showed that robust selectivity for K+ could occur. That such a simple model would yield selectivity for K+ over Na+ was unanticipated and quite surprising to many. After this work, several theoretical studies have examined analogous reduced toy models to explore how they could display size selectivity (Asthagiri et al., 2006; Bostick and Brooks, 2007, 2009, 2010; Thomas et al., 2007, 2011; Varma and Rempe, 2007, 2008; Dudev and Lim, 2009). Although differing in emphasis, the emerging consensus from these and our own studies has broadly confirmed that, depending on the number and type of ligands, robust ion selectivity is possible in a loosely restricted flexible binding site for which the classical notion of snug-fit does not apply.
2. The functional concept of ion selectivity
It is useful to note that the notion of ion selectivity can mean different things operationally, depending on the molecular system that is being considered (e.g., an ion channel or a transporter) and on the experimental conditions that are used to probe the system (e.g., equilibrium binding or nonequilibrium flux and ionic current measurements). In the case of a transporter, it is reasonable to expect that ion binding selectivity will be primarily governed thermodynamically by equilibrium binding, because the lifetime of the various conformational states of the protein is extremely long. This situation is unambiguously amenable to FEP/MD computations. In the case of ion channels, selectivity can mean different things, although ultimately it is understood that the wrong kinds of ions should encounter more difficulty than the right kind of ions to permeate.
Classically, ion channel selectivity has been characterized by determining the permeability ratio from the reversal potential (zero net current) under bionic conditions. Computational studies are able to simulate this situation and explain the origin of the reversal potential in an asymmetric ionic concentration with near-quantitative accuracy for OmpF porin (Im and Roux, 2002) and α-hemolysin (Noskov et al., 2004b; Egwolf et al., 2010; Luo et al., 2010). However, measurements of reversal potentials become impractical for highly selective channels such as the K+ channels. In this case, alternate methods such as Ba2+ blockade relief (Neyton and Miller, 1988a,b) or Na+ punchthrough (Nimigean and Miller, 2002) provide more effective strategies to characterize selectivity with quantitative accuracy. Ba2+ block is more sensitive to the depth of the free energy minima of the binding sites (i.e., equilibrium binding), whereas Na+ punchthrough is more sensitive to the height of free energy barriers (i.e., nonequilibrium rates).
Implicitly, it is assumed that the phenomenon of ion permeation and selectivity through an open conductive pore can be discussed independently from gating and inactivation. In reality, the situation is more complicated. For instance, the filter of some K+ channels is unable to remain in a conductive conformation when exposed to an ionic solution with little or no K+ ions. Under such conditions, the filter rapidly converts to a nonconductive state because K+ ions are needed to maintain the conductive state of the channel, giving rise to some kind of conformationally driven super-selectivity. An example of this is provided by wild-type KcsA channel (Lockless et al., 2007). Of possible relevance to these observations, the selectivity filter of the bacterial KcsA channel recapitulates most of the hallmarks of C-type inactivation displayed by eukaryotic voltage-dependent K+ channels (Cordero-Morales et al., 2006, 2007; Cuello et al., 2010). The complex phenomenon of super-selectivity must be distinguished from the simpler and perhaps more traditional notion of conductive-state selectivity, which is the primary subject of this Perspective. Examples of channels that unambiguously display the phenomenon of conductive-state selectivity by remaining in the conductive state even in the complete absence of K+ are the MthK channel (Ye et al., 2010) and the synthetic KcsAD-Ala77 (Valiyaveetil et al., 2006). Computational studies of selectivity in K+ channels have mainly focused on the phenomenon of conductive-state selectivity (Bernèche and Roux, 2001; Luzhkov and Åqvist, 2001; Noskov et al., 2004a; Thompson et al., 2009; Egwolf and Roux, 2010). Less attention has been devoted to the characterization of the complex coupling between selective ion binding and the conformational stability of the filter involved in super-selectivity using computational methods. Previous studies of the relative stability of the conductive and so-called low-K+ structure of the wild-type KcsA channel and how it depends on ion occupancy provide examples of the challenges and difficulties that might lie ahead (Domene and Furini, 2009; Boiteux and Bernèche, 2011). Addressing these issues systematically will require advanced computational techniques able to describe complex conformational changes in macromolecules (Domene et al., 2008; Pan and Roux, 2008; Pan et al., 2008; Maragliano et al., 2009).
3. Selectivity from the perspective of the multi-ion free energy landscape
The simplest view of conductive-state selectivity starts with the assumption that there exist well-defined binding sites that can be occupied by either K+ or Na+. Within this context, the problem can be treated with FEP/MD simulations to compute the relative free energy of K+ and Na+ in the binding site of interest via Eq. 1. But conductive-state selectivity can also be a result of kinetic factors caused by differences in free energy barriers (Thompson et al., 2009; Nimigean and Allen, 2011). Furthermore, it is good to recall that ion channels are not really designed to keep ions in place, but to allow them to diffuse rapidly across the membrane. Therefore, it is likely that binding sites inside the pore, even when they exist, should be rather shallow and separated by small free energy barriers.
A more complete analysis of conductive-state selectivity requires the knowledge of the entire free energy surface governing the multi-ion permeation process, with all its free energy wells and barriers (Bernèche and Roux, 2001; Roux et al., 2004; Furini and Domene, 2009). This can be achieved by computing the PMF for the ions in the pore, W(Z1,Z2,Z3), using umbrella sampling MD simulations (Bernèche and Roux, 2001, 2003). For example, the multi-ion PMF of K+ ions translocating through the KcsA selectivity filter displays a series of cation binding sites separated only by small free energy barriers (Bernèche and Roux, 2001), yielding a maximum channel conductance of ∼300–500 pS, in good accord with experiments (Bernèche and Roux, 2003). The predominant knock-on ion conduction mechanism extracted from the PMF calculations is supported by the results from long unbiased MD trajectories (Jensen et al., 2010).
Under physiological conditions, the functional consequence of the high selectivity of K+ channels is primarily to prevent the entry of Na+ ions from the extracellular side. To learn more about this process, all-atom MD simulations with an explicit membrane were used to determine the multi-ion PMFs corresponding to the entry of an extracellular ion, Na+ or K+, into the selectivity filter of the KcsA channel while it is in its conductive state and occupied by two K+ ions (Egwolf and Roux, 2010). The computations, summarized in Fig. 2, were able to capture key differences between the entry of a K+ or Na+ ion into the pore. Most importantly, the PMFs in Fig. 2 A show that the region corresponding to the binding site S2 near the center of the narrow pore emerges as the most selective for K+ over Na+. In particular, although there is a stable minimum for K+ in the site S2, the Na+ ion faces a steeply rising free energy landscape with no local well in this region. Although S2 is a stable location for K+, it may be a stable binding site for Na+ only in the context of a multi-ion configuration, with other ions bound in the sites S0 and S4. This has important implications for alchemical FEP/MD studies of ion selectivity in the KcsA channel. It is also possible to monitor the average number of coordinating oxygens from backbone carbonyl groups or water molecules around the K+ and Na+ ion during the entry process to see how it correlates with the free energy landscape. The results are shown in Fig. 2 B. As expected, the number of coordinating water molecules decreases as the ion moves from the bulk phase into the narrow selectivity filter, for Z1 < 7 Å. Significantly, the increase in selectivity for K+ over Na+ that is observed near the site S2 appears to be predominantly correlated with changes in the type of coordinating ligands (carbonyls vs. water molecules).
Figure 2.
Summary of the main computational results for the entry of K+ and Na+ in the selectivity filter of the KcsA channel from the extracellular side (Z > 12 Å) in the presence of two other K+ ions in the pore in a K+/K+/K+ or a Na+/K+/K+ configuration. (A) The PMFs for the entry of K+ and Na+ in the selectivity filter (the thin red line is the original PMF for Na+ before it was shifted by 0.7 Å along Z to match the wells and barriers of the PMF for K+). (B) Coordination numbers for the ion entering the selectivity filter. The average numbers of water, carbonyl, and total oxygen atoms closer than 3.2 Å from an entering K+ or 2.8 Å for the entering Na+ are shown. The number of coordinating water molecules decreases as the ion moves from the bulk phase into the narrow selectivity filter for Z < 7 Å. However, the total coordination number displays only small variations: over the entire entry process, the average coordination number of K+ increases slightly from 6 to 7, whereas that of Na+ remains constant around 5.5. This is adapted from Egwolf and Roux (2010).
The critical role of the central region near the binding site S2 of KcsA is also reflected in the pore structure of homologous channels. For example, one of the main differences between the nonselective cationic NaK channel and the K+-selective KcsA channel is the widening of the pore at the level of the central binding site S2 in NaK (Shi et al., 2006). This led to the suggestion that loss of selectivity at the level of the central site S2 could be the principal reason why the NaK channel is able to conduct Na+ unlike KcsA (Zagotta, 2006), which was correlated with MD simulations (Noskov and Roux, 2007). Results from MD about the NaK channel (Noskov and Roux, 2007) were subsequently confirmed by additional x-ray structures (Alam and Jiang, 2009, 2011).
4. Studies of ion selectivity based on reduced models
Although computations accounting for the multi-ion free energy surface are desirable (see Section 3), theoretical studies based on reduced models comprising only the ion and the nearest coordinating ligands can provide meaningful insight when selectivity is concerned with thermodynamic binding equilibrium in a well-defined site. Typically, long-range interactions from distant parts of the protein or the solvent are not expected to directly affect relative free energy differences if, for example, the charge of the ion remains unchanged, as is the case with Na+ and K+. However, long-range effects can have an indirect influence when, for example, the structural stability of a binding site is affected by a mutation in a distant residue, or it could trigger the onset of some conformation-driven super-selectivity (see Section 2). Nonetheless, describing aspects of ion selectivity from local energetics, even if there are conformational transitions, is formally possible by considering a step-by-step free energy decomposition (Deng and Roux, 2009).
(a) Formulation of reduced models.
By construction, reduced models only treat the ion and the surrounding coordinating ligands explicitly. The effect of the missing atoms from the protein or the solvent must be incorporated indirectly via spatial constraints that are acting on the ion and ligands. Obviously, the results can be strongly affected by the character of those constraints (e.g., see Yu et al., 2009), and reduced models with either very loose or very stiff constraints can lead to very different conclusions about ion selectivity. Such ambiguity and arbitrariness increase the difficulty in drawing meaningful conclusions from studies based on reduced models.
To address this issue, a statistical mechanical framework for the local structural determinant of ion selectivity in protein binding sites was formulated (Yu et al., 2010b). First, a local subsystem comprising the ion and the N coordinating ligands is defined. Then, the influence of the rest of the system onto the subsystem is rigorously expressed as the sum of two separate contributions. The first incorporates all the molecular forces keeping the ion and the coordinating ligands loosely restricted to a microscopic sub-volume, but it does not prevent the ligands from adapting to a smaller or larger ion, whereas the second regroups all the remaining forces that control the precise geometry of the coordinating ligands best adapted to a given ion. An effective spring constant, λg, that is determined from the thermal fluctuations of the ligands is introduced to report on the magnitude of the geometrical precision forces.
(b) Two mechanisms of ion selectivity.
The framework reveals the existence of two distinct and idealized limiting mechanisms that are able to give rise to ion selectivity in protein binding sites (Yu et al., 2010b). In one limit, the geometric forces are dominant (λg→∞) and the binding site is nearly rigid. Then, it can be shown that the free energy difference ΔΔGNa,K is set by the difference between the mean ion–ligand interactions,
| (2) |
and that ion selectivity is sterically controlled by the cavity size, according to the familiar snug-fit mechanism. In the other limit, the geometric forces are negligible (λg→0), and the ion and ligands are free to fluctuate and adapt to ions of different sizes. We called this situation, which is somewhat less familiar, the confined microdroplet. In this case, it can be shown rigorously from a statistical mechanical development that free energy difference ΔΔGNa,K is set by the interplay between the mean ion–ligand and ligand–ligand interactions,
| (3) |
Two key physical factors control the relative free energy in the confined microdroplet limit: the number of available ligands N and their physical properties. The latter includes electrostatic (dipole, charge, and polarizability) as well as non-electrostatic (shape, radius, and size) factors. Free energy differences are dominated by enthalpic contributions, as −TΔS is often negligible (Noskov and Roux, 2006; Roux, 2010a). However, it is important to consider thermally averaged energies because an analysis based on energy minima from optimized geometries can lead to significant errors (Yu and Roux, 2009; Dixit and Asthagiri, 2011).
According to Eq. 3, both the favorable (negative) ion–ligand and unfavorable (positive) ligand–ligand interactions contribute and compete, in a nontrivial manner, to control selectivity in the confined microdroplet. The ligand–ligand interaction acts like an energetic strain. Similar conclusions have been obtained by Asthagiri and co-workers (Asthagiri et al., 2006; Dixit et al., 2009) using an analysis of all-atom MD and reduced models. Differences in the mean ion–ligand interaction, , do not give rise to selectivity in the confined microdroplet limit, in contrast with the host–guest mechanism with fixed cavity radius as described by Eq. 2, further illustrating the importance of the ligand–ligand interactions in the analysis of flexible systems. A clear discussion of this issue has been provided in Asthagiri et al. (2006). One may note that with one ion and N ligands, there are N ion–ligand pairs and N(N-1)/2 ligand–ligand pairs. Thus, is not expected to typically display a simple dependence with respect to N.
It is worth noting that there are no explicit contributions from protein forces to the free energy difference controlling the ion selectivity in Eq. 3. Nonetheless, it is important to keep in mind that the crowded environment imposed by the confinement is a crucial feature of the context that underlies selectivity in such systems. For example, it is well understood that completely free and unrestricted ligands (as in a bulk liquid) do not typically display high selectivity. Nevertheless, the spatial confinement does not need to be precise to give rise to ion selectivity. Nontrivial free energy patterns can emerge, even in the context of a relatively generic and featureless confinement. This is in striking contrast with the snug-fit mechanism, which requires protein forces dictating a precise geometry. Therefore, in the confined microdroplet limit, the protein structure plays a critical role to support selectivity, albeit not by enforcing a specific cavity size with a ligand geometry to sub-angstrom precision, nor by contributing directly to the free energy differences. Although there are differences in emphasis, these observations are consistent with the analysis presented by Varma et al. (2011).
In light of this theoretical analysis, the snug-fit and the confined microdroplet now appear as two extreme and idealized physical mechanisms of ion selectivity. Real systems are expected to take on a mixed character between these two idealized views. Some binding sites cannot be selective without additional architectural forces stabilizing the ligand geometry (e.g., the site Na2 in LeuT), whereas others can be robustly selective in the confined microdroplet limit (e.g., the site S2 of KcsA or the site Na1 in LeuT). Such robust trends appearing in the confined microdroplet limit correspond to an inherent selectivity, which is an emergent property set by the number and type of ligands forming the binding site (see Section 4c below).
The results of Fig. 3 show that it is exceedingly difficult to counteract the inherent selectivity in the case of the KcsA site S2 or the site Na1 of the LeuT with small alterations to the structure or the architectural forces. It is possible to adapt the local geometry of the ligands to construct a putative Na+-selective KcsA binding site or a K+-selective LeuT-Na1 site, but the sites become selective only when the protein is made unrealistically rigid (λg>>10 kcal/mol/Å2). Similar results were shown previously in an initial study (see Fig. 3 from Noskov et al., 2004a). This analysis suggests that stabilizing the local geometry to build upon the inherent selectivity set by the number and type of ion-coordinating ligands is probably an important design principle of ion-selective binding sites in proteins and other biological macromolecules. For example, some inherent selectivity could be displayed by a loosely restricted binding site existing within an imperfectly folded protein at an early stage of its evolution. Then, the amino acid sequence could subsequently undergo mutations to increase the architectural stiffness of the binding site toward a snug-fit mechanism if this confers some functional advantage; e.g., see the interesting evolutionary analysis of cation-regulated proteases by Page and Di Cera (2006). From this perspective, the stiffening of the local architecture may serve to enhance (rather than oppose) the inherent selectivity that might already exist within a loosely restricted site.
Figure 3.
Selectivity in reduced models in which the optimal geometry is adapted to best-fit K+ or Na+ (a binding site is K+ selective when ΔΔGNa,K is positive and Na+ selective when it is negative). The curves with solid lines correspond to realistic models of KcsA and LeuT, and the curves with dashed lines correspond to putative KcsA and LeuT sites stabilizing an optimal geometry adapted by energy minimization to best-fit Na+ or K+, respectively. Except for the site Na2 of LeuT, the onset of selectivity is established only for very large values of the architectural rigidity λg, indicating that a fairly stiff geometry is required to counterbalance the inherent trend that is already observed in the confined microdroplet limit (λg = 0) with loosely restricted ligands. This is adapted from Yu et al. (2010b).
(c) Relation with classical field strength theory.
The thermodynamic behavior in the fluctuating confined microdropet bears some similarities with the classical field strength theory of Eisenman and Krasne (Eisenman, G., and S. Krasne. 1972. Further considerations on the ion selectivity of carrier molecules and membranes. Proceedings of the fourth International Biosphysics Congress Symposium on Membrane Structure and Function); e.g., low-field ligands tend to favor larger cations, whereas high-field ligands tend to favor small ones. For example, selectivity can shift markedly toward Na+ if carboxylate oxygens (high-field ligands) are incorporated in a binding site. But traditional ion–ligand field strength arguments do not explain why a flexible binding site of eight carbonyls remains robustly selective for K+, because the dipole of the backbone amide carbonyl is about twice as large as the dipole of a water molecule; the interaction between a cation and a single backbone carbonyl is significantly larger than that with a water molecule, and this difference is even more prominent in the case of Na+ than for K+ (Noskov and Roux, 2006). In other words, carbonyls are high-field ligands according to the traditional field strength arguments, and they would favor Na+ over K+ if it were not for the counter-effect (the electrostatic strain energy) from ligand–ligand interactions (Noskov and Roux, 2006; Dixit et al., 2009).
(d) A wide range of ionic selectivities.
Even reduced models with simple confinement can give rise to a wide range of ionic selectivities. To illustrate this, an extensive collection of 1,077 reduced models was characterized using FEP/MD simulations (Roux, 2010a). The models comprised four to nine oxygen ligands of different types, with zero to eight N-methylacetamide, methanol (CH3OH), or water, zero to four diglycines (each with two backbone carbonyls), and zero to two neutral or ionized acetic acids (CHCOOH or CHCOO−, each with two oxygen ligands). The results confirmed that nontrivial selectivity can arise in those reduced models, without the need for precise geometry for the ligands. The notion that selectivity is an emergent property of self-organized confined microdroplets is similar to the charge/space competition mechanism that has been used to model calcium channels, where selectivity arises from a balance of electrostatics and the excluded volume of ions in the crowded selectivity filter (Gillespie et al., 2005; Gillespie, 2008).
(e) The issue of coordination numbers.
The number of ligands as well as their properties is expected to play important roles in the selectivity (Noskov et al., 2004a). However, some investigators propose that selectivity is controlled primarily by the number of coordinating ligands, their physical properties being less important (Bostick and Brooks, 2007, 2009, 2010; Thomas et al., 2007; Varma and Rempe, 2007, 2008; Dudev and Lim, 2009). A detailed point-by-point discussion of the diverging views requires a critical examination of their statistical mechanical underpinnings and goes beyond this Perspective. Among the specific issues that need to be considered are the utilization of energy minimization to estimate free energies in reduced models (Yu and Roux, 2009), the underlying assumptions used to infer the selectivity of protein binding sites based on the coordination numbers of ions in bulk water (Yu et al., 2009), and the accuracy (and applicability) of an approximate statistical mechanical framework based on the primitive quasi-chemical theory for ion solvation in bulk liquids for modeling protein binding sites (Roux and Yu, 2010). Although there appears to be no consensus on the best approach to dissect the various contributions to ion selectivity at the present time, an important take-home message is that the various studies concur that nontrivial trends can emerge in reduced models with loosely restricted ligands, depending both on the number and the type of ligands. The importance of chemistry for ion selectivity—hardly a radical notion—is clearly on display whether one considers an analysis of ion binding sites in proteins (Noskov and Roux, 2008), a characterization of 1,077 reduced models from FEP/MD simulations (Roux, 2010a), or a comparison of experimental ion solvation free energies in various organic liquids (Fig. 2 in Varma et al., 2011).secc
5. Force field, induced polarization, and quantum mechanics (QM)
Conclusions derived from computational studies of ion selectivity depend on the accuracy of underlying computational methods. Simulations of biomolecular systems are essentially limited by the accuracy of the potential energy surface and statistical sampling (Roux, 2010b). To obtain meaningful results, one must seek a balance between more accurate but more expensive representations of the energy surface and the obligation to adequately sample the relevant configurations. Most simulations of biomolecular systems are based on potential functions with fixed effective atomic charges (Åqvist, 1990; Cornell et al., 1995; MacKerell et al., 1998), which account for many-body polarization effects in an average way. Such computationally inexpensive potential functions make it possible to extensively explore conformational space with long MD trajectories (Jensen et al., 2010). But with increasingly powerful computers, simulations based on more sophisticated representations are becoming affordable.
A promising strategy to improve the accuracy of MD studies is the development of potential functions that explicitly account for induced polarization (Grossfield et al., 2003; Patel et al., 2004; Lamoureux and Roux, 2006; Whitfield et al., 2007; Yu et al., 2010a,c). Although simulations of full proteins with polarizable force fields are still not routinely performed, there are already some interesting results concerning ion solvation in water (Lamoureux et al., 2006; Yu et al., 2010c) and in liquid amides (Grossfield et al., 2003; Yu et al., 2010a). For example, according to FEP/MD computations, there is an extremely sharp variation in the relative free energy of Na+ and K+ as a function of the number of N-methylacetamide molecules in reduced models, whereas the corresponding free energy variations with water molecules are much smaller (Yu et al., 2010a). This observation highlights the quantitative differences presented by water and carbonyls as ion-coordinating ligands.
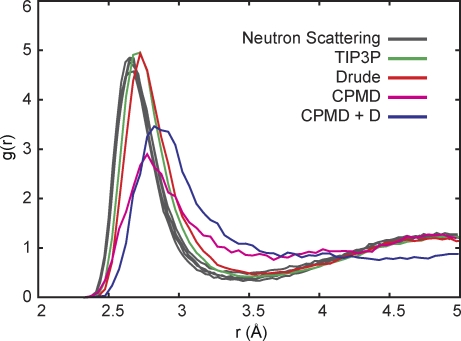
Another area of considerable progress concerns simulations based on QM electronic structure theories (Bucher and Rothlisberger, 2010). Those can be performed in the context of Car–Parrinello MD (CPMD; http://www.cpmd.org/), which is an algorithm for generating a MD trajectory of a periodic system that approximates the Born–Oppenheimer surface of a density functional theory (DFT) method. Typically, CPMD simulations are limited to systems of only a few hundred atoms and a few tens of picoseconds and are several thousand times slower than simulations using polarizable force fields. CPMD has been used extensively to calculate the solution structure of hydrated ions (Whitfield et al., 2007) and has recently been used to model the coordination modes of ions in the KcsA selectivity filter (Bucher et al., 2007, 2010). As a word of caution, conventional DFT methods are not exact and often underestimate intermolecular attractions resulting from van der Waals dispersive forces. This can lead to limitations in representing the solvation of ions such as K+ in liquid water; the simulations lead to a K+–O radial distribution function that is broader and shifted to larger distances than the experimental and molecular mechanical force field distribution functions (Whitfield et al., 2007). As illustrated in Fig. 4, there is some improvement with CPMD simulations with an empirical correction to account for van der Waals dispersion, but the agreement with the experimental distribution functions estimated from neutron scattering and anomalous diffraction x-ray absorption-fine structure (XAFS) is poorer than that with simulations based on a polarizable force field (Whitfield et al., 2007). Notably, the coordination number of K+ (at r = 3.5 Å) is considerably affected by the dispersion correction. The coordination number increases from 6.15 in the uncorrected CPMD simulations to 7.45 in the dispersion-corrected CPMD simulations—a value larger than the one obtained from the non-polarizable force field (6.77) and the polarizable force field (6.80). This illustrates that the approximations currently used in CPMD simulations can substantially affect ion solvation and must be considered with caution. It cannot be assumed that a strategy based on QM methods is inherently superior to one based on a well-calibrated force field for modeling ion channel selectivity, but they remain valuable tools to parameterize and validate these simulations.
Figure 4.
Average solvation structure of K+ in liquid water (the radial distribution function g(r) of the ion–oxygen pair) at 298 K. Results are shown for the non-polarizable TIP3P/CHARMM force field and Drude polarizable force field (SWM4-NDP water model; Lamoureux et al., 2006) as well as two ab initio CPMD models. CPMD (version 3.13.3) simulations were performed using a cubic periodic cell with a length of 12.41 Å containing 64 water molecules and one K+ ion. The electronic structure was calculated with the BLYP functional (Becke, 1988; Lee et al., 1988), Troullier and Martins pseudopotentials (Troullier and Martins, 1991), and a plane-wave cutoff of 80 Ry. Also included are CPMD results with the BLYP function with an empirical correction (+D) for van der Waals dispersion (Grimme, 2006). CPMD results were generated by a 20-ps equilibration followed by a 30-ps trajectory for analysis, using a time step of 0.12 fs. Each CPMD simulation was initiated from an equilibrated Drude force field simulation, as described by Whitfield et al. (2007). The multiple gray lines for the neutron scattering correspond to experiments on solutions of different potassium-halide salts (Soper and Weckström, 2006).
Ultimately, what matters is determining the degree of confidence regarding the conclusions that are drawn from any given computation. A good strategy is to assess the validity of a dominant trend using high-level methods. For example, the importance of the carbonyl–carbonyl repulsion in the selectivity filter of KcsA, first noted in FEP/MD simulations based on a non-polarizable force field, was subsequently assessed using high-level ab initio computations to show that the repulsion between water molecules is smaller than that between carbonyl groups (see supplemental material in Noskov et al., 2004a). More recently, the importance of the carbonyl–carbonyl repulsion was demonstrated again in the CPMD simulations of the KcsA channel performed by Bucher et al. (2010). It is also of interest to revisit and validate some previous conclusions about the high K+ selectivity of the binding site S2 in KcsA using FEP/MD computations with a polarizable model or a fully QM calculation based on DFT (Fig. 1 C). The results given in Table I indicate that in all cases, the relative free energy of Na+ and K+ is fairly similar, indicating that meaningful conclusions were drawn from the FEP/MD computations based on the non-polarizable force field.
Table I.
Free energy in reduced model of the binding site S2 in KcsA (kcal/mol)
| Systems | |||
| Non-polarizable force field; Yu et al., 2010ba | −18.3 | −12.1 | 6.2 |
| Polarizable force fielda | −17.3 | −8.8 | 8.5 |
| DFTb | −25.6 | −21.5 | 4.1 |
Details about the features of the reduced model and the computations for the non-polarizable force field are given in Yu et al. (2010b). The model system is comprised of one ion (K+ or Na+) surrounded by four diglycine residues representing the carbonyl groups of Val76 and Gly77. The protein atoms are confined to small spherical volumes deduced from an analysis of all-atom MD trajectories of the full-length KcsA in a lipid bilayer. The FEP/MD simulations were carried out via the PERT module of the program CHARMM (Brooks et al., 2009) using Langevin dynamics. The computations with the Drude polarizable force field were carried out using the same procedure with the ion parameters of Yu et al. (2010c).
DFT FEP calculations were performed at the PBE/DZVP level of theory (Perdew et al., 1996) using the CHARMM/DeMon2k interface (Lev et al., 2010; Zhang et al., 2010). The DFT relative free energy difference for the ions in bulk was calculated using 16 water molecules restrained within a spherical volume around the ion, mimicking the bulk density. The FEP/MD simulations were carried out using 11 evenly spaced windows for a thermodynamic coupling parameter varying from 0 to 1, with 10 ps of sampling per window.
6. Conclusion
Early MD simulations of the KcsA channel brought to the forefront the provocative idea that ion selectivity could robustly be achieved despite considerable structural flexibility, prompting a reexamination of the microscopic factors giving rise to ion selectivity using a wide range of computational experiments. Some of those computational experiments represented fairly artificial situations (e.g., switching off the carbonyl–carbonyl repulsion in KcsA), whereas others involved reduced models (e.g., a few confined carbonyl-like ligands). Ideas about the need for precise rigid binding sites were reassessed, and issues previously believed to be settled were reopened.
Classical notions about conductive-state selectivity such as the snug-fit mechanism, field strength, and free energy profiles are still important, but they are now sharing the stage with additional and less familiar concepts relevant to flexible and loosely restricted systems. The provocative notion that ion selectivity in a flexible binding site can emerge from local energetics has now been firmly grounded. This analysis has helped delineate the existence of two different idealized mechanisms able to support ion selectivity (Yu et al., 2010b).
Much of the progress has relied on a combination of all-atom MD simulations on known structures with explicit solvent and membrane and the analysis of reduced models. It is our contention that the reduced models, although useful for identifying and illustrating fundamental physical principles, will always be subservient to studies on real protein structures. Although features of reduced models can be a matter of debate, it is important to recall that the results from all-atom MD computations on KcsA, NaK, and LeuT have, thus far, been in good accord with experiments. This relative success of computer simulations suggests that they provide a realistic representation of these molecular systems, and that they can contribute to insight about ion selectivity.
Until now, most efforts have implicitly been focused on the factors governing conductive-state selectivity, with a fair amount of attention devoted to notions of thermodynamic equilibrium. More efforts will be required to better understand the complex phenomenon of conformation-driven super-selectivity, which remains poorly understood. Research into ion channel selectivity will continue to advance, with additional channel structures, along with improvements in computational models.
Appendix
Eq. 3, taken from Yu et al. (2010b), is closely related to Eq. 2 of Varma et al. (2011). The difference between the average ion–ligand interactions for Na+ and K+, , in Eq. 3 is equivalent to , and the difference between the average ligand–ligand interactions for Na+ and K+, , in Eq. 3 corresponds to . Differences in entropy between ions, , are often small in practice and can be neglected for the sake of simplicity (Roux, 2010a). The remaining term is the average internal energy contribution from the external field imposed by the rest of the protein . Assuming that the external field from the protein corresponds to some harsh confining potential acting on the ligands, , its average is proportional to , which is expected to be small because the Boltzmann factor is negligible at all the points x where is nonzero (x represents all the relevant degrees of freedom).
This Perspectives series includes articles by Andersen, Alam and Jiang, Nimigean and Allen, Dixit and Asthagiri, and Varma et al. (scheduled for the June 2011 issue).
Acknowledgments
This work was supported by the National Institutes of Health (GM-062342). It was also supported by the Canadian Institutes of Health Research (200804MOP-186232 to S.Y. Noskov and H. Yu). S.Y. Noskov is an Alberta Heritage Foundation for Medical Research Scholar. B. Lev is supported by the Alberta Innovates: Technology Futures. The computational support for this work was provided by the West-Grid Canada through resource allocation award to S.Y. Noskov.
Footnotes
Abbreviations used in this paper:
- CPMD
- Car–Parrinello molecular dynamics
- DFT
- density functional theory
- FEP
- free energy perturbation
- MD
- molecular dynamics
- PMF
- potential of mean force
- QM
- quantum mechanics
References
- Alam A., Jiang Y. 2009. High-resolution structure of the open NaK channel. Nat. Struct. Mol. Biol. 16:30–34 10.1038/nsmb.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam A., Jiang Y. 2011. Structural study of ion selectivity in tetrameric cation channels. J. Gen. Physiol. 137:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.W., Andersen O.S., Roux B. 2004. On the importance of atomic fluctuations, protein flexibility, and solvent in ion permeation. J. Gen. Physiol. 124:679–690 10.1085/jgp.200409111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.W., Andersen O.S., Roux B. 2006. Molecular dynamics—potential of mean force calculations as a tool for understanding ion permeation and selectivity in narrow channels. Biophys. Chem. 124:251–267 10.1016/j.bpc.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Åqvist J. 1990. Ion-water interaction potentials derived from free energy perturbation simulations. J. Phys. Chem. 94:8021–8024 10.1021/j100384a009 [DOI] [Google Scholar]
- Åqvist J., Alvarez O., Eisenman G. 1992. Ion-selective properties of a small ionophore in methanol studied by free energy perturbation simulations. J. Phys. Chem. 96:10019–10025 10.1021/j100203a079 [DOI] [Google Scholar]
- Asthagiri D., Pratt L.R., Paulaitis M.E. 2006. Role of fluctuations in a snug-fit mechanism of KcsA channel selectivity. J. Chem. Phys. 125:024701 10.1063/1.2205853 [DOI] [PubMed] [Google Scholar]
- Becke A.D. 1988. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 38:3098–3100 10.1103/PhysRevA.38.3098 [DOI] [PubMed] [Google Scholar]
- Bernèche S., Roux B. 2000. Molecular dynamics of the KcsA K(+) channel in a bilayer membrane. Biophys. J. 78:2900–2917 10.1016/S0006-3495(00)76831-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S., Roux B. 2001. Energetics of ion conduction through the K+ channel. Nature. 414:73–77 10.1038/35102067 [DOI] [PubMed] [Google Scholar]
- Bernèche S., Roux B. 2003. A microscopic view of ion conduction through the K+ channel. Proc. Natl. Acad. Sci. USA. 100:8644–8648 10.1073/pnas.1431750100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S., Roux B. 2005. A gate in the selectivity filter of potassium channels. Structure. 13:591–600 10.1016/j.str.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Bezanilla F., Armstrong C.M. 1972. Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axon. J. Gen. Physiol. 60:588–608 10.1085/jgp.60.5.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin P.C., Smith G.R., Shrivastava I., Choe S., Sansom M.S. 2001. Potassium and sodium ions in a potassium channel studied by molecular dynamics simulations. Biochim. Biophys. Acta. 1510:1–9 10.1016/S0005-2736(00)00345-X [DOI] [PubMed] [Google Scholar]
- Boiteux C., Bernèche S. 2011. Absence of ion-binding affinity in the putatively inactivated low-[K+] structure of the KcsA potassium channel. Structure. 19:70.– [DOI] [PubMed] [Google Scholar]
- Bostick D.L., Brooks C.L., III 2007. Selectivity in K+ channels is due to topological control of the permeant ion’s coordinated state. Proc. Natl. Acad. Sci. USA. 104:9260–9265 10.1073/pnas.0700554104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D.L., Brooks C.L., III 2009. Statistical determinants of selective ionic complexation: ions in solvent, transport proteins, and other “hosts.” Biophys. J. 96:4470–4492 10.1016/j.bpj.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D.L., Brooks C.L., III 2010. Selective complexation of K+ and Na+ in simple polarizable ion-ligating systems. J. Am. Chem. Soc. 132:13185–13187 10.1021/ja106197e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Brooks C.L., III, Mackerell A.D., Jr, Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. 2009. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30:1545–1614 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D., Rothlisberger U. 2010. Molecular simulations of ion channels: a quantum chemist’s perspective. J. Gen. Physiol. 135:549–554 10.1085/jgp.201010404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D., Guidoni L., Rothlisberger U. 2007. The protonation state of the Glu-71/Asp-80 residues in the KcsA potassium channel: a first-principles QM/MM molecular dynamics study. Biophys. J. 93:2315–2324 10.1529/biophysj.106.102509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D., Guidoni L., Carloni P., Rothlisberger U. 2010. Coordination numbers of K(+) and Na(+) ions inside the selectivity filter of the KcsA potassium channel: insights from first principles molecular dynamics. Biophys. J. 98:L47–L49 10.1016/j.bpj.2010.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar D.L., Clarage J., Salunke D.M., Clarage M. 1988. Liquid-like movements in crystalline insulin. Nature. 332:659–662 10.1038/332659a0 [DOI] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Cuello L.G., Zhao Y.X., Jogini V., Cortes D.M., Roux B., Perozo E. 2006. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13:311–318 10.1038/nsmb1069 [DOI] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Jogini V., Lewis A., Vásquez V., Cortes D.M., Roux B., Perozo E. 2007. Molecular driving forces determining potassium channel slow inactivation. Nat. Struct. Mol. Biol. 14:1062–1069 10.1038/nsmb1309 [DOI] [PubMed] [Google Scholar]
- Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Jr, Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179–5197 10.1021/ja00124a002 [DOI] [Google Scholar]
- Cuello L.G., Jogini V., Cortes D.M., Pan A.C., Gagnon D.G., Dalmas O., Cordero-Morales J.F., Chakrapani S., Roux B., Perozo E. 2010. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 466:272–275 10.1038/nature09136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Roux B. 2009. Computations of standard binding free energies with molecular dynamics simulations. J. Phys. Chem. B. 113:2234–2246 10.1021/jp807701h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich B. 1985. Coordination chemistry of alkali and alkaline-earth cations with macrocyclic ligands. J.Chem. Edu. 62:954–964 10.1021/ed062p954 [DOI] [Google Scholar]
- Dixit P.D., Asthagiri D. 2011. The role of bulk protein in local models of ion-binding to proteins: comparative study of KcsA, its semisynthetic analog with a locked-in binding site, and valinomycin. Biophys. J. 100:1542–1549 10.1016/j.bpj.2011.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit P.D., Merchant S., Asthagiri D. 2009. Ion selectivity in the KcsA potassium channel from the perspective of the ion binding site. Biophys. J. 96:2138–2145 10.1016/j.bpj.2008.12.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domene C., Furini S. 2009. Dynamics, energetics, and selectivity of the low-K+ KcsA channel structure. J. Mol. Biol. 389:637–645 10.1016/j.jmb.2009.04.038 [DOI] [PubMed] [Google Scholar]
- Domene C., Klein M.L., Branduardi D., Gervasio F.L., Parrinello M. 2008. Conformational changes and gating at the selectivity filter of potassium channels. J. Am. Chem. Soc. 130:9474–9480 10.1021/ja801792g [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Dudev T., Lim C. 2009. Determinants of K+ vs Na+ selectivity in potassium channels. J. Am. Chem. Soc. 131:8092–8101 10.1021/ja900168k [DOI] [PubMed] [Google Scholar]
- Egwolf B., Roux B. 2010. Ion selectivity of the KcsA channel: a perspective from multi-ion free energy landscapes. J. Mol. Biol. 401:831–842 10.1016/j.jmb.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egwolf B., Luo Y., Walters D.E., Roux B. 2010. Ion selectivity of alpha-hemolysin with beta-cyclodextrin adapter. II. Multi-ion effects studied with grand canonical Monte Carlo/Brownian dynamics simulations. J. Phys. Chem. B. 114:2901–2909 10.1021/jp906791b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furini S., Domene C. 2009. Atypical mechanism of conduction in potassium channels. Proc. Natl. Acad. Sci. USA. 106:16074–16077 10.1073/pnas.0903226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D. 2008. Energetics of divalent selectivity in a calcium channel: the ryanodine receptor case study. Biophys. J. 94:1169–1184 10.1529/biophysj.107.116798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D., Xu L., Wang Y., Meissner G. 2005. (De)constructing the ryanodine receptor: modeling ion permeation and selectivity of the calcium release channel. J. Phys. Chem. B. 109:15598–15610 10.1021/jp052471j [DOI] [PubMed] [Google Scholar]
- Grimme S. 2006. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 27:1787–1799 10.1002/jcc.20495 [DOI] [PubMed] [Google Scholar]
- Grootenhuis P.D.J., Kollman P.A. 1989. Molecular mechanics and dynamics studies of crown ether-cation interactions: free energy calculations on the cation selectivity of dibenzo-18-crown-6 and dibenzo-30-crown-10. J. Am. Chem. Soc. 111:2152–2158 10.1021/ja00188a032 [DOI] [Google Scholar]
- Grossfield A., Ren P., Ponder J.W. 2003. Ion solvation thermodynamics from simulation with a polarizable force field. J. Am. Chem. Soc. 125:15671–15682 10.1021/ja037005r [DOI] [PubMed] [Google Scholar]
- Guidoni L., Torre V., Carloni P. 1999. Potassium and sodium binding to the outer mouth of the K+ channel. Biochemistry. 38:8599–8604 10.1021/bi990540c [DOI] [PubMed] [Google Scholar]
- Hille B., Armstrong C.M., MacKinnon R. 1999. Ion channels: from idea to reality. Nat. Med. 5:1105–1109 10.1038/13415 [DOI] [PubMed] [Google Scholar]
- Im W., Roux B. 2002. Ion permeation and selectivity of OmpF porin: a theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theory. J. Mol. Biol. 322:851–869 10.1016/S0022-2836(02)00778-7 [DOI] [PubMed] [Google Scholar]
- Jang H., Hall C.K., Zhou Y. 2002. Folding thermodynamics of model four-strand antiparallel beta-sheet proteins. Biophys. J. 82:646–659 10.1016/S0006-3495(02)75428-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.O., Borhani D.W., Lindorff-Larsen K., Maragakis P., Jogini V., Eastwood M.P., Dror R.O., Shaw D.E. 2010. Principles of conduction and hydrophobic gating in K+ channels. Proc. Natl. Acad. Sci. USA. 107:5833–5838 10.1073/pnas.0911691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller G.R., Smith J.C. 1994. Liquid-like side-chain dynamics in myoglobin. J. Mol. Biol. 242:181–185 10.1006/jmbi.1994.1570 [DOI] [PubMed] [Google Scholar]
- Lamoureux G., Roux B. 2006. Absolute hydration free energy scale for alkali and halide ions established from simulations with a polarizable force field. J. Phys. Chem. B. 110:3308–3322 10.1021/jp056043p [DOI] [PubMed] [Google Scholar]
- Lamoureux G., Harder E., Vorobyov I.V., Roux B., MacKerell A.D. 2006. A polarizable model of water for molecular dynamics simulations of biomolecules. Chem. Phys. Lett. 418:245–249 10.1016/j.cplett.2005.10.135 [DOI] [Google Scholar]
- Lee C., Yang W., Parr R.G. 1988. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter. 37:785–789 10.1103/PhysRevB.37.785 [DOI] [PubMed] [Google Scholar]
- Lev B., Zhang R., de la Lande A., Salahub D., Noskov S.Y. 2010. The QM-MM interface for CHARMM-deMon. J. Comput. Chem. 31:1015–1023 [DOI] [PubMed] [Google Scholar]
- Lindemann F.A. 1910. The calculation of molecular vibration frequencies. Physik. Z. 11:609–612 [Google Scholar]
- Lindorff-Larsen K., Best R.B., Depristo M.A., Dobson C.M., Vendruscolo M. 2005. Simultaneous determination of protein structure and dynamics. Nature. 433:128–132 10.1038/nature03199 [DOI] [PubMed] [Google Scholar]
- Lockless S.W., Zhou M., MacKinnon R. 2007. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5:e121 10.1371/journal.pbio.0050121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Egwolf B., Walters D.E., Roux B. 2010. Ion selectivity of alpha-hemolysin with a beta-cyclodextrin adapter. I. Single ion potential of mean force and diffusion coefficient. J. Phys. Chem. B. 114:952–958 10.1021/jp906790f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhkov V.B., Åqvist J. 2001. K(+)/Na(+) selectivity of the KcsA potassium channel from microscopic free energy perturbation calculations. Biochim. Biophys. Acta. 1548:194–202 10.1016/S0167-4838(01)00213-8 [DOI] [PubMed] [Google Scholar]
- Lybrand T.P., McCammon J.A., Wipff G. 1986. Theoretical calculation of relative binding affinity in host-guest systems. Proc. Natl. Acad. Sci. USA. 83:833–835 10.1073/pnas.83.4.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Jr, Brooks B., Brooks C.L., III, Nilsson L., Roux B., Won Y., Karplus M. 1998. CHARMM: the energy function and its parameterization with an overview of the program. The Encyclopedia of Computational Chemistry. Vol 1 Schleter P.V.R., Allinger N.L., Clark T., Gasteiger J., Kollman P.A., Schaefer H.F., III, Schreiner P.R., John Wiley & Sons, Chichester, England: 271–277 [Google Scholar]
- Maragliano L., Vanden-Eijnden E., Roux B. 2009. Free energy and kinetics of conformational transitions from Voronoi tessellated milestoning with restraining potentials. J. Chem. Theory Comput. 5:2589–2594 10.1021/ct900279z [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon J.A., Gelin B.R., Karplus M. 1977. Dynamics of folded proteins. Nature. 267:585–590 10.1038/267585a0 [DOI] [PubMed] [Google Scholar]
- Neyton J., Miller C. 1988a. Discrete Ba2+ block as a probe of ion occupancy and pore structure in the high-conductance Ca2+ -activated K+ channel. J. Gen. Physiol. 92:569–586 10.1085/jgp.92.5.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyton J., Miller C. 1988b. Potassium blocks barium permeation through a calcium-activated potassium channel. J. Gen. Physiol. 92:549–567 10.1085/jgp.92.5.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C.M., Allen T.W. 2011. Origins of ion selectivity in potassium channels from the perspective of channel block. J. Gen. Physiol. 137:405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C.M., Miller C. 2002. Na+ block and permeation in a K+ channel of known structure. J. Gen. Physiol. 120:323–335 10.1085/jgp.20028614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., Roux B. 2006. Ion selectivity in potassium channels. Biophys. Chem. 124:279–291 10.1016/j.bpc.2006.05.033 [DOI] [PubMed] [Google Scholar]
- Noskov S.Y., Roux B. 2007. Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels. J. Gen. Physiol. 129:135–143 10.1085/jgp.200609633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., Roux B. 2008. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J. Mol. Biol. 377:804–818 10.1016/j.jmb.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov S.Y., Bernèche S., Roux B. 2004a. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 431:830–834 10.1038/nature02943 [DOI] [PubMed] [Google Scholar]
- Noskov S.Y., Im W., Roux B. 2004b. Ion permeation through the alpha-hemolysin channel: theoretical studies based on Brownian dynamics and Poisson-Nernst-Plank electrodiffusion theory. Biophys. J. 87:2299–2309 10.1529/biophysj.104.044008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.J., Di Cera E. 2006. Role of Na+ and K+ in enzyme function. Physiol. Rev. 86:1049–1092 10.1152/physrev.00008.2006 [DOI] [PubMed] [Google Scholar]
- Pan A.C., Roux B. 2008. Building Markov state models along pathways to determine free energies and rates of transitions. J. Chem. Phys. 129:064107 10.1063/1.2959573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan A.C., Sezer D., Roux B. 2008. Finding transition pathways using the string method with swarms of trajectories. J. Phys. Chem. B. 112:3432–3440 10.1021/jp0777059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Mackerell A.D., Jr, Brooks C.L., III 2004. CHARMM fluctuating charge force field for proteins: II protein/solvent properties from molecular dynamics simulations using a nonadditive electrostatic model. J. Comput. Chem. 25:1504–1514 10.1002/jcc.20077 [DOI] [PubMed] [Google Scholar]
- Perdew J.P., Burke K., Ernzerhof M. 1996. Generalized gradient approximation made simple. Phys. Rev. Lett. 77:3865–3868 10.1103/PhysRevLett.77.3865 [DOI] [PubMed] [Google Scholar]
- Ratheal I.M., Virgin G.K., Yu H., Roux B., Gatto C., Artigas P. 2010. Selectivity of externally facing ion-binding sites in the Na/K pump to alkali metals and organic cations. Proc. Natl. Acad. Sci. USA. 107:18718–18723 10.1073/pnas.1004214107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. 1996. Valence selectivity of the gramicidin channel: a molecular dynamics free energy perturbation study. Biophys. J. 71:3177–3185 10.1016/S0006-3495(96)79511-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. 2010a. Exploring the ion selectivity properties of a large number of simplified binding site models. Biophys. J. 98:2877–2885 10.1016/j.bpj.2010.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. 2010b. Perspectives on: molecular dynamics and computational methods. J. Gen. Physiol. 135:547–548 10.1085/jgp.201010456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Yu H. 2010. Assessing the accuracy of approximate treatments of ion hydration based on primitive quasichemical theory. J. Chem. Phys. 132:234101 10.1063/1.3436632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Allen T., Bernèche S., Im W. 2004. Theoretical and computational models of biological ion channels. Q. Rev. Biophys. 37:15–103 10.1017/S0033583504003968 [DOI] [PubMed] [Google Scholar]
- Sheinerman F.B., Brooks C.L., III 1998. Molecular picture of folding of a small alpha/beta protein. Proc. Natl. Acad. Sci. USA. 95:1562–1567 10.1073/pnas.95.4.1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N., Ye S., Alam A., Chen L., Jiang Y. 2006. Atomic structure of a Na+- and K+-conducting channel. Nature. 440:570–574 10.1038/nature04508 [DOI] [PubMed] [Google Scholar]
- Soper A.K., Weckström K. 2006. Ion solvation and water structure in potassium halide aqueous solutions. Biophys. Chem. 124:180–191 10.1016/j.bpc.2006.04.009 [DOI] [PubMed] [Google Scholar]
- Straatsma T.P., Berendsen H.J.C. 1988. Free energy of ionic hydration: analysis of a thermodynamic integration technique to evaluate free energy differences by molecular dynamics simulations. J. Chem. Phys. 89:5876–5886 10.1063/1.455539 [DOI] [Google Scholar]
- Thomas M., Jayatilaka D., Corry B. 2007. The predominant role of coordination number in potassium channel selectivity. Biophys. J. 93:2635–2643 10.1529/biophysj.107.108167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Jayatilaka D., Corry B. 2011. Mapping the importance of four factors in creating monovalent ion selectivity in biological molecules. Biophys. J. 100:60–69 10.1016/j.bpj.2010.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.N., Kim I., Panosian T.D., Iverson T.M., Allen T.W., Nimigean C.M. 2009. Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore. Nat. Struct. Mol. Biol. 16:1317–1324 10.1038/nsmb.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troullier N., Martins J.L. 1991. Efficient pseudopotentials for plane-wave calculations. II. Operators for fast iterative diagonalization. Phys. Rev. B Condens. Matter. 43:8861–8869 10.1103/PhysRevB.43.8861 [DOI] [PubMed] [Google Scholar]
- Valiyaveetil F.I., Leonetti M., Muir T.W., Mackinnon R. 2006. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 314:1004–1007 10.1126/science.1133415 [DOI] [PubMed] [Google Scholar]
- Varma S., Rempe S.B. 2007. Tuning ion coordination architectures to enable selective partitioning. Biophys. J. 93:1093–1099 10.1529/biophysj.107.107482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Rempe S.B. 2008. Structural transitions in ion coordination driven by changes in competition for ligand binding. J. Am. Chem. Soc. 130:15405–15419 10.1021/ja803575y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S., Rogers D.M., Pratt L.R., Rempe S.B. 2011. Design principles for K+ selectivity in membrane transport. J. Gen. Physiol. 137:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield T.W., Varma S., Harder E., Lamoureux G., Rempe S.B., Roux B. 2007. Theoretical study of aqueous solvation of K+ comparing ab initio, polarizable, and fixed-charge models. J. Chem. Theory Comput. 3:2068–2082 10.1021/ct700172b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Li Y., Jiang Y. 2010. Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat. Struct. Mol. Biol. 17:1019–1023 10.1038/nsmb.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Roux B. 2009. On the utilization of energy minimization to the study of ion selectivity. Biophys. J. 97:L15–L17 10.1016/j.bpj.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Noskov S.Y., Roux B. 2009. Hydration number, topological control, and ion selectivity. J. Phys. Chem. B. 113:8725–8730 10.1021/jp901233v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Mazzanti C.L., Whitfield T.W., Koeppe R.E., Andersen O.S., Roux B. 2010a. A combined experimental and theoretical study of ion solvation in liquid N-methylacetamide. J. Am. Chem. Soc. 132:10847–10856 10.1021/ja103270w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Noskov S.Y., Roux B. 2010b. Two mechanisms of ion selectivity in protein binding sites. Proc. Natl. Acad. Sci. USA. 107:20329–20334 10.1073/pnas.1007150107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Whitfield T.W., Harder E., Lamoureux G., Vorobyov I., Anisimov V.M., Mackerell A.D., Roux B. 2010c. Simulating monovalent and divalent ions in aqueous solution using a Drude polarizable force field. J. Chem. Theory Comput. 6:774–786 10.1021/ct900576a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta W.N. 2006. Membrane biology: permutations of permeability. Nature. 440:427–429 10.1038/440427a [DOI] [PubMed] [Google Scholar]
- Zhang R., Lev B., Cuervo J.E., Noskov S.Y., Salahub D.R. 2010. A guide to QM/MM methodology and applications. Adv. Quantum Chem. 59:353–400 10.1016/S0065-3276(10)59010-5 [DOI] [Google Scholar]
- Zhou Y., Karplus M. 1997. Folding thermodynamics of a model three-helix-bundle protein. Proc. Natl. Acad. Sci. USA. 94:14429–14432 10.1073/pnas.94.26.14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Karplus M. 1999. Folding of a model three-helix bundle protein: a thermodynamic and kinetic analysis. J. Mol. Biol. 293:917–951 10.1006/jmbi.1999.2936 [DOI] [PubMed] [Google Scholar]