Abstract
Computational prediction of microRNA targets remains a challenging problem. The existing rule-based, data-driven and expression profiling approaches to target prediction are mostly approached from the gene-level. The increasing availability of RNA-seq data provides a new perspective for microRNA target prediction on the isoform-level. We hypothesize that the splicing isoform is the ultimate effector in microRNA targeting and that the proposed isoform-level approach is capable of predicting non-dominant isoform targets as well as their targeting regions that are otherwise invisible to many existing approaches. To test the hypothesis, we used an iterative expectation maximization (EM) algorithm to quantify transcriptomes at the isoform-level. The performance of the EM algorithm in transcriptome quantification was examined in simulation studies using FluxSimulator. We used joint evidence from isoform-level down-regulation and seed enrichment to predict microRNA-155 targets. We validated our computational approach using results from 149 in-house performed in vitro 3′-UTR assays. We also augmented the splicing database using exon–exon junction evidence, and applied the EM algorithm to predict and quantify 1572 cell line specific novel isoforms. Combined with seed enrichment analysis, we predicted 51 novel microRNA-155 isoform targets. Our work is among the first computational studies advocating the isoform-level microRNA target prediction.
INTRODUCTION
The regulation of gene expression by microRNAs is a fundamental mechanism for controlling many biological processes. Thus far, more than 1000 microRNA’s have been discovered in human cells using either computational or experimental approaches [miRBase (1), release 16 September 2010]. The gene encoding the microRNA, microRNA-155, was classified as an oncogene many years before it was identified as a microRNA and is now among the most highly implicated microRNAs in cancer. Despite its link to hematologic and other cancers, there is currently little information regarding direct isoform targets or pathways through which microRNA-155 signals to promote the tumor phenotype.
Over the years, an array of computational approaches have been developed to predict microRNA target sites and these methods have been useful for guiding investigations towards the function of microRNAs (2). These approaches are roughly divided into rule-based and data-driven approaches (3). Earlier methods are largely rule-based, predicting microRNA targets as a function of simple discriminative rules derived from features of experimentally validated targets. For example, miRanda (4), DIANA-microT (5), TargetScan (6) and PicTar (7) are mainly based on scanning for conserved 7-/8-mer seeds combined with free energy calculations of the RNA–RNA duplex. Latter methods were developed which are more data-driven, such as miTarget (8) and NBmiRTar (9), where machine learning-based approaches were applied to train a classifier that is able to discriminate true microRNA targets from false targets using sequence features.
An alternative data-driven approach is to use 3′-expression microarrays to quantify transcriptomes. In this approach, microRNA targets are predicted by calling significantly down-regulated genes between microRNA over-expressing cell lines and the respective isogenic wild-type cell lines (10–12). Gene expression-based target prediction approaches [e.g. GenMiR++ (13)], were found to outperform many rule-based approaches, such as TargetScan (3). More importantly, the gene expression-based approach allows for the discovery of context specific (cell type specific) microRNA target repertoires and this context specific targetome can be related back to the biological processes implicated by the global analysis of the respective microarray experiments. Despite this advantage over purely computational approaches, the intrinsic limitations of the 3′-expression microarrays (such as non-specific hybridization, signal saturation and excessive noise) significantly compromise the performance of microarray-based microRNA target prediction.
The advent of next-generation sequencing (NGS) technologies provides new opportunities to profile transcriptomes and microRNA targetomes at base-wise resolution. In our recent work (14), we sequenced the transcriptome of microRNA-155 expressing cells using an Illumina Genome Analyzer II. Our RNA-seq data contains more than 100-million single-ended 50-mer short reads generated from both wild-type Mutu I cells (control) and Mutu I cells expressing microRNA-155 (case). We then developed a computational pipeline to analyze microRNA-155 transcriptome and targetome regulation by performing gene-level down-regulation analysis combined with 7-/8-mer seed evidence in 3′-UTR regions. Our analysis yielded a much larger targetome than was previously described using microarray experiments; many predicted microRNA-155 targets were verified by in vitro 3′-UTR reporter assays. Although this analysis was among the first to use RNA-seq data for microRNA target prediction, this approach did not sufficiently exploit the full value that RNA-seq data has to offer—that is, using gene-structure information derived from the RNA-seq data to assess isoform specific microRNA regulation. Based on the isoform-level analysis described here, we propose that microRNA targets are more appropriately predicted and characterized at the isoform-level.
On a more general level, we believe that the term, ‘isoform’ may be a more appropriate concept than ‘gene’ in transcriptome studies since the isoform is the ultimate effecter of microRNA responses (as well as many other biological processes). Further, recent studies have shown that microRNA targeting is not limited to the 3′-UTR (15), further emphasizing the need for microRNA target prediction based on the isoform-level.
Genome-wide analysis of transcriptomes and targetomes at the isoform-level is needed, not only for microRNA target prediction but also for many other genomics research areas, such as biomarker discovery, cancer classification, biological pathway analysis and network reconstruction. The problem itself can be quite challenging since the base-wise gene expression signal from RNA-seq data is often accumulated from a mixture of coexisting isoforms in the living cell. The development of computational algorithms to deconvolve the gene expression signal emitted from each splicing isoform is not a trivial task.
A number of computational approaches have recently been developed to characterize and quantify transcriptomes at the isoform-level [e.g. (5,16–21)]. These approaches quantify isoform levels of transcripts either annotated in the alternative splicing databases such as those from the UCSC (University of California, Santa Cruz) and alternative splicing and transcript discovery (ASTD) resources or predicted by short-read assembly. However, to the best of our knowledge, no computational approach has been developed to predict microRNA targets at the isoform-level. With this type of approach, we will be able to answer the following interconnected and relevant research questions that could not be fully answered previously: (i) Which set of genes are microRNA-155 targets? (ii) For each target gene, which annotated or novel (not annotated) isoform is the true microRNA-155 target? (iii) What is the targeting region and the mechanism? E.g. does the microRNA bind to the 3′-UTR, to exons and/or to the 5′-UTR?
Here we present an expectation maximization (EM) type of algorithm that can be generally used for characterizing and quantifying transcriptomes at the isoform-level. Applying this algorithm, we also present a comprehensive isoform-level analysis of the microRNA-155 targetome and an isoform-level analysis of microRNA-155 mediated transcriptome changes. Combining this isoform specific expression analysis with 7-/8-mer seed enrichment analysis, we attempt to answer the above-mentioned questions. Using both a simulation system that in silico emulates the experimental pipeline for RNA-Seq technology (the flux simulator, http://flux.sammeth.net/index.html), and a real-world in vitro validation system of 149 3′-UTR reporters that we have assayed (14), we are able to show that the isoform-level approach (this work) significantly outperforms the recently published gene-level approach (14). Extending the analysis to the whole transcriptome, our target prediction results identify a larger targetome at a base-wise resolution and indicate novel microRNA-155 targeting mechanisms. Through an exon–exon junction analysis, we discovered many cell type specific isoform transcripts that were not previously annotated in the Ensembl database. Many of these are likely to be novel microRNA-155 targets.
MATERIALS AND METHODS
In Figure 1a, we summarize the workflow of our transcriptome and targetome quantification pipeline. It begins with short-read alignment, followed by transcript prediction and quantification. An isoform is predicted as a microRNA target by joint evidence from differential expression/splicing analysis and seed enrichment analysis.
Figure 1.
(a) The workflow of our transcriptome and targetome analysis pipeline. Solid arrows represent annotated transcript quantification and dotted arrows represent novel transcript quantification. (b) Novel transcript discovery illustrated using splicing graph. One source of exon–exon junctions (solid lines) is available from alternative splicing database, and another source (dotted lines) of junctions is available from computational prediction using TopHat.
Short-read alignment
The Burkitt’s lymphoma cell line, Mutu I, was retrovirally transduced in duplicate with either a control or a microRNA-155 expressing retrovirus. microRNA-155 real time RT–PCR analysis showed at least 100 000-fold higher expression in microRNA-155 transduced pools relative to control transduced pools (14). Despite these elevated levels, microRNA-155 expression in transduced Mutu I cells was slightly less than that observed in several activated B-cell lines that naturally express microRNA-155 (14); arguing against supra-physiological expression of microRNA-155 in transduced Mutu I cells. The transcriptomes of the wild-type Mutu I cell line and the microRNA-155 expressing Mutu I cell line were deep sequenced using Illumina Genome Analyzer II with a read length of 50 (NCBI Short Read Archive, Accession Number SRA011001) (14). For each biological or technical replicate, around 10-million single-ended short reads were generated. Short reads were initially aligned to the reference genome (hg19/GRCh37) using Novoalign (http://www.novocraft.com). We used standard parameter settings to build an index (novoindex) and to run Novoalign. The alignment results were saved in the SAM format and parsed using SAMMate (http://sammate.sourceforge.net/) (22) to calculate gene-level abundance.
Annotation table augmentation via splicing junction analysis
Splicing isoform annotation databases are often incomplete and not cell type/condition-specific. The 100 000 some annotated isoforms in the ASTD database is not adequate to characterize the vast diversity of the human transcriptome. We augmented the ASTD annotation table using the cell type-specific exon–exon junction evidence, i.e. specific to the microRNA-155 targeting event (Figure 1b). In particular, we performed junction analysis using Tophat (23), a widely used junction mapper, and augmented the ASTD annotation table by adding novel isoform transcripts that are supported by cell line specific junctions (18).
EM algorithm to quantify transcriptomes at the isoform-level
For each gene, our algorithm infers an unobserved cDNA fragment-originating matrix (Z and Z′) from the observed cDNA fragment-compatible matrix (Y and Y′) (Figure 2a). For each matrix, a row represents cDNA fragments where (single- or paired-end) short reads were generated using the novel ‘sequence by synthesis’ technology from Illumina Inc. Columns represent sets of possible splicing isoforms either annotated in ASTD Ensembl database (Homo Sapiens.GRCh37.57) or supported by exon–exon junction analysis (23). Based on short-read alignment to the reference genome and the annotated or inferred isoform structures, the matrix Y or Y′ is directly observed but not of direct interest to us since each cDNA fragment can be compatible with multiple isoforms. For example, a mapped cDNA fragment spanning an exon–exon junction is compatible or possibly derived from a number of isoforms containing the junction, whether the junction is annotated or not. In Figure 2a, the first inferred cDNA fragment is compatible with the first, third and fourth isoforms. What we need to infer is a cDNA fragment-originating matrix Z or Z′ where each cDNA fragment is unambiguously originated from one isoform. The relevant proportion of isoforms can be easily derived by summing up the counts of each column, therefore, make it possible to characterize and quantify a transcriptome at the isoform-level.
Figure 2.
(a) Illustration of the EM algorithm. Deriving the observed short-read compatible matrix Y or Y′ from short-read alignment (left panel) Applying EM algorithm to infer the short-read originating matrix Z or Z′ (middle panel). Calculating relative isoform proportions in case and control (right panel). Note the referred gene is differentially spliced between case and control but not differentially expressed. It is also known as dichotomy of regulation. (b) Simulation studies to evaluate the accuracy of isoform quantification using the FluxSimulator. For single-end data (b) and paired-end data (Supplementary Figure S1), we plot predicted isoform abundance scores against true abundance scores. R2 calculated by robust linear regression analysis were shown in each figure. Fifteen-millions 50-mer single-end short reads and 30-millions 50-mer paired-end short reads were generated and used for this simulation studies.
More formally, assume  is the mapped cDNA fragment (row) index,
is the mapped cDNA fragment (row) index,  is the isoform (column) index,
is the isoform (column) index,  is the length of jth isoform, and
is the length of jth isoform, and  , where
, where  is the mixture proportion for the isoform
is the mixture proportion for the isoform  Initializing all the compatible
Initializing all the compatible  to be the same, and add up to 1, i.e.
to be the same, and add up to 1, i.e.  The EM type algorithm is as follows (24): E-step:
The EM type algorithm is as follows (24): E-step:
 |
where  is the normalized indicator having value of
is the normalized indicator having value of  if the
if the  isoform is compatible, 0 otherwise. M-step: Let
isoform is compatible, 0 otherwise. M-step: Let
 |
The EM algorithm iterates between E and M steps until convergence, i.e.  is an arbitrarily small positive number, i.e. 0.00001. We denote the converged isoform proportion as
is an arbitrarily small positive number, i.e. 0.00001. We denote the converged isoform proportion as  Assuming the single gene expression abundance is quantified using RPKM (for single-ended reads) or FPKM (paired-ended reads) is
Assuming the single gene expression abundance is quantified using RPKM (for single-ended reads) or FPKM (paired-ended reads) is  , then the expression abundance for each isoform is:
, then the expression abundance for each isoform is:
 |
where  is the sum of total exon length of a gene, and
is the sum of total exon length of a gene, and  is the sum of total exon length of a transcript in this gene.
is the sum of total exon length of a transcript in this gene.
We make note that the EM type algorithms have been used to solve multiple problems in bioinformatics. In particular, similar algorithms have been designed and applied to infer full-length isoforms using expressing sequence tags (ESTs) data (24) and RNA-seq data (17,21). We expect that the RNA-seq data works better with the EM type algorithm due to a much larger sample size and a much reduced number of compatible splicing isoforms. We include additional mathematical details of the EM algorithm to the Supplementary Data (Section 1).
Genome-wide seed enrichment analysis
Although there are exceptions, microRNA targeting is primarily guided by 7- or 8-mer seeds in a gene region (usually within the 3′-UTR but sometimes within the 5′-UTR, or an exon). For our analysis, we consider seed enrichment as a necessary condition for target prediction. A single 7- or 8-mer seed in a long genome region tends to be less likely to be a microRNA target than many seeds in a short genome region. We also provide additional justification of using seed enrichment as opposed to seed presence, as well as combining isoform down-regulation with seed enrichment criteria for microRNA-155 target prediction in the Supplementary Data (Supplementary Table S7, Figures S3 and S4; Sections 2 and 3).
We used Pearson’s Chi-square test to quantify the seed enrichment for each genome region. Basically, we calculate a Chi-square test statistic, which quantifies how much the observed seed counts deviate from the expected seed counts in a given genome region. Larger values of the chi-square test statistic (small P-values) will reject the null hypothesis of non-enrichment. The Excel function ‘CHISQUARE’ was used to perform the genome-wide seed enrichment analysis. The raw P-values of the chi-square test will then be adjusted using the stringent Bonferroni’s procedure.
3′-UTR-luciferase reporter analysis
The 3′-UTR assay results were initially reported in our recent publication (14), and we used it in this article as a reference to compare the gene-level (14) and the isoform-level (this work) approaches to microRNA target prediction. For the sake of completeness, we repeat experimental protocols here: 3.75 µg of either the control (pMSCV-puro-GFP-miR-CNTL) or microRNA-155 (pMSCV-puro-GFP-microRNA-155) expression vector was cotransfected with 0.25 µg of the appropriate pMIR-REPORT-dCMV or pGL4.11 3′-UTR reporter plasmid into 1 × 106 MutuI cells using Lipofectamine (Invitrogen). Cells were harvested 48 h post-transfection and analyzed using Promega firefly luciferase assay. Values reported are expression change of a given 3′-UTR relative to change in the control reporter.
Quantitative RT–PCR
Novel exon–exon junctions were validated by PCR using primers indicated in Table 1. RT-PCR was performed on an Eppendorf Mastercycler® ep. Total RNA was reverse-transcribed with the SuperScript III First-Strand Synthesis System (Invitrogen) according to manufacturer’s instructions by using random hexamer primers (50 ng/1.5 µg total RNA).
Table 1.
Quantitative RT–PCR experiments to verify isoform target TAF5L
| TABLE I | Forward primer | Reverse primer | ||
|---|---|---|---|---|
| TAF5L LTE | ENST00000366676 | Last two exons | 5′-AGCCCCACCAAGTAGACGTGT-3′ | 5′-TCTCCGTGCCTGCATTATCAT-3′ |
| TAF5L EX | ENST00000366676 | Last two exons | 5′-AGTAGACGTGTCCCGCATCCATTT-3′ | 5′-AACAAGAGAGCAACCCTGAGCTGT-3′ |
| TAF5L Iso | ENST00000366675 | Last exon | 5′-CACAGGAAGTAGAGTTGCCAGCT-3′ | 5′-AACGGTTACAAGCCAAACAAGATT-3′ |
| Iso TAF5L | ENST00000366675 | Last exon | 5′-CCCACAGAAGGTTGTGCCATTTCA-3′ | 5′-ACATGGAGCCACAGGATATGCACT-3′ |
| TBP | Housekeeping | 5′-GATGGATGTTGAGTTGCAGGGTGT-3′ | 5′-AGCACGGTATGAGCAACTCACAGT-3′ |
The resulting cDNA was subjected to PCR using sequence specific forward and reverse primers (Inetgrated DNA Technologies) (Table 1). Platinum Taq polymerase (Invitrogen) was used following manufacturer’s recommendations. The reaction mixure consisted of 1× High Fidelity PCR buffer, 0.2 mM dNTP each, 2 mM MgSO4, 0.2 µM primer each, 2 µl cDNA and 1 U Platinum Taq High Fidelity per 50 µl reaction volume. Amplification was carried out using the following conditions: 2 min at 94°C, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 68°C for 60 s. PCR products were analyzed on a 1.2% agarose gel and fragment size was determined by comparison to the TrackIt 100bp DNA Ladder (Invitrogen). qRT-PCR (TAF5L) was performed using the isoform specific primers shown in Table 2. A total RNA of 250 ng was reverse-transcribed with the SuperScript III First-Strand Synthesis System (Invitrogen) according to manufacturer’s instructions, using Oligo(dT) primers. The resulting cDNA was subjected to quantitative (real-time) PCR using specific primers (Integrated DNA Technologies) (Table 2). A master mix was prepared for each PCR run which included Platinum SYBR green SuperMix plus UDG (Invitrogen), 50 nM fluorescein-NIST traceable dye, 250 nM forward and reverse primer and nuclease-free water. Amplification was carried out using the following conditions: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 95°C for 30 s and 60°C for 30 s.
Table 2.
Quantitative RT–PCR experiments to verify novel junctions (9 out of 10 were verified)
| TABLE II | Reference transcript ID | Junction assessed | Forward primer | Reverse primer | |
|---|---|---|---|---|---|
| PLDN | ENST00000220531 | Exons 1–4 | 5′-CACACGTTTGCTTCTTCCCTGTGT-3′ | 5′-GGCATGATAGTGTTTAGCCTCAGC-3′ | 379 bp |
| TMEM126A | ENST00000304511 | Exons 1–3 | 5′-CCCAGGTAATTTGAGCAAAGGCCA-3′ | 5′-CTATGAGGCCACAAAGAGCAGCAT-3′ | 244 bp |
| PGPEP1 | ENST00000269919 | Exons 3–5 | 5′-TCCGGTTGAGTACCAAACAGTCCA-3′ | 5′-CGTGACTCTGGTACAAAGAGGTGT-3′ | 344 bp |
| NOL9 | ENST00000377705 | Exons 3–7 | 5′-TAACCAGCTATCCGGGTTCATCCT-3′ | 5′-TGTGGAGTCCTCAGGTGAGTGAAA-3′ | 403 bp |
| C15orf17 | ENST00000357635 | Exons 1–3 | 5′-AGATCGGTAATAGAGCCCTCCGTCT-3′ | 5′-ATCTGGACTCTGGCTAAGAGCAGT-3′ | 242 bp |
| YEATS4 | ENST00000247843 | Exons 2–5 | 5′-GGGCACACTCATCAGTGGACAGTAT-3′ | 5′-CCCAGCATTGCATTGGTGTCTGAT-3′ | 266 bp |
| NARS2 | ENST00000281038 | Exons 12–14 | 5′-GCTGTTGATCTTCTGGTTCCTGGAGT-3′ | 5′-AAGATGCACTGCAGGTAGCGTTCA-3′ | 200 bp |
| SLC7A11 | ENST00000280612 | Exons 1–3 | 5′-GCACCATCATTGGAGCAGGAATCT-3′ | 5′-TGTAGCGTCCAAATGCCAGGGATA-3′ | 285 bp |
| ARL1 | ENST00000261636 | Exons 3–5 | 5′-TAGGAGGACAGACAAGTATCAGGCCA-3′ | 5′-TCCTTCAAGGCAGGTAACCCAAGT-3′ | 247 bp |
The novel splice junction tested for each gene spanned the exons indicated in the ‘Junction assessed’ column of the indicated ‘reference transcript ID’.
Melt curve analysis was performed at the end of every qRT–PCR run. Samples were tested in triplicate. Real-time PCR was performed on Bio-Rad MyiQ iCycler and data analysis was performed using Bio-Rad IQ5 v2.0 software. No-template controls and no-reverse transcription controls were performed with each PCR run. Relative quantification was calculated by using the ΔΔCt method.
RESULTS AND DISCUSSIONS
Simulation studies
To assess the accuracy of our EM algorithm in isoform quantification, we simulated RNA-seq experiments using FluxSimulator, a freely available software package that simulates whole transcriptome sequencing experiments with the Illumina Genome Analyzer. The software works by first randomly generating integer copies of each splicing isoform according to the annotation file provided by the user, followed by constructing an amplified, size-selected library and sequencing it in silico. The resulting cDNA fragments are then sampled uniformly at random for simulated sequencing, where the initial and terminal 25, 50 and 75 bp of each selected fragment are reported as reads. In our simulation studies, the human Ensembl ASTD database (version 57) was supplied to the software, along with the hg19 version of the human reference genome. In the ASTD annotation file, there are 100 297 protein coding isoforms, corresponding to 21 271 protein-coding genes. FluxSimulator then randomly assigned expression to 19 992 isoforms, corresponding to 10 343 genes. About 15-million single-end and 30-million paired-end RNA-seq 50-mer short reads were generated by size selection of fragments between 175 and 225 bases.
The estimated isoform abundance was plotted against the true isoform abundance using single-ended short reads (Figure 2b) and paired-end short reads (Supplementary Figure S1). A very good linear correlation was observed (over 0.97) for both single- and paired-end data. We make note that the EM algorithm fails to estimate the abundance level of a very small portion (∼10%) of the isoforms, which fall into such situations, as many isoforms within one gene, the length of the unique exon is shorter than the read length, and so on (detailed information can be found in Supplementary Data, Section 4) Simulation results obtained using other experimental factors, such as read length and counts, showed a similarly robust correlation. These simulation studies provide compelling evidence for the excellent accuracy of our algorithm in quantifying isoform transcripts.
Validation studies
Our validation studies were carried out using in-house results from 149 different 3′-UTR reporter plasmids containing a spectrum of microRNA-155 seed types, configurations and potency (11). The rationale of selecting these 3′-UTR’s for in vitro assays is based on current microRNA target database. The 149 genes analyzed in the current study at the 3′-UTR reporter level were selected from a wider panel of 170 such 3′-UTR reporters based on adherence to the following three criteria: (i) the expression estimated from RNA-seq experiments is above 0.5 RPKM at the gene-level; (ii) the expression estimated from our isoform-level approach is above 0.2 RPKM; (iii) the genes exhibit a 7-/8-mer seed enrichment (adjusted P < 0.05) in their 3′-UTR region at the isoform-level. Using the corresponding 3′-UTR reporter data from this set of genes, we tested our isoform-level approach and compared the results to those obtained using the gene-level approach (14). We used stringent and loose relative expression cut-offs (0.6, 0.7 and 0.8; relative expression meaning expression in Mutu-microRNA-155 versus Mutu-control cells) to discriminate true targets from false targets. We also used a statistical criterion, i.e. q-value (30), as an auxiliary evaluation parameter. Results using different cut-offs are consistent and are included in the Supplementary Table S1.
We compared the microRNA-155 targets predicted by gene-level, isoform-level approaches and 3′-UTR assay. In Figure 3a, the set of 149 targets were divided into eight distinct categories. Because a full list of true microRNA-155 targets is not available as a gold standard, the eight categories essentially represent all possible outcomes of comparing three approaches to microRNA target prediction, i.e. gene-level, isoform-level and 3′-UTR assay. We provide biological interpretation and/or insight for each category. In particular, categories (1) and (2) are to support the notion that the isoform-level approach dominates the gene-level approach. The detailed gene information for each category is available as Supplemental Data (Supplementary Table S1).
Figure 3.
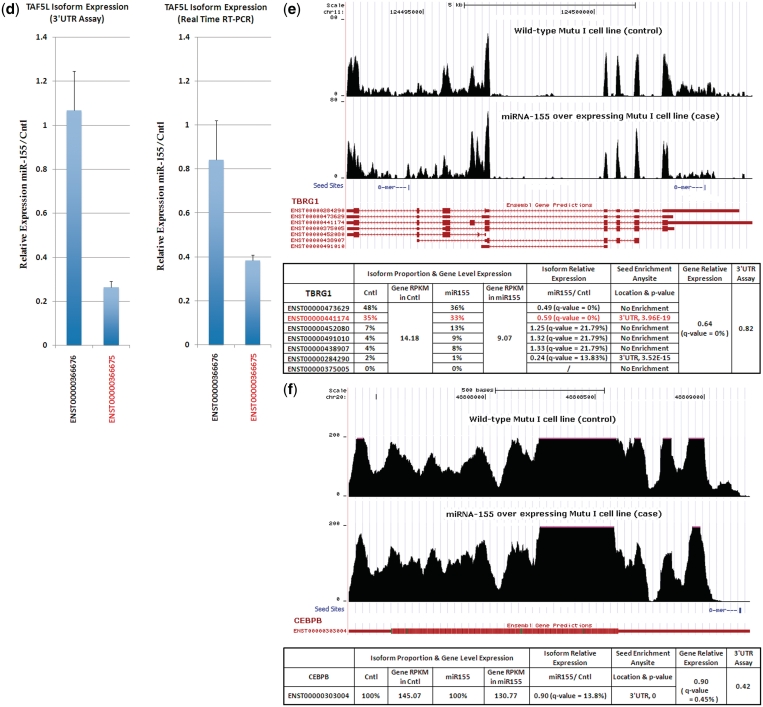
(a) Venn Diagram of the microRNA targets predicted by the three approaches at 0.8 cut-off level of relative expression. (b) An example of isoform target exclusively predicted by the isoform-level approach (Gene PHF17). It represents a group of genes with dichotomy-regulated isoforms and the down-regulated isoform (potential target) was not tested in 3′-UTR assay. (c). An example of isoform target predicted jointly by the isoform-level approach and the 3′-UTR assay (Gene TAF5L). It represents a group of genes with dichotomy-regulated isoforms where the down-regulated isoform was also tested in 3′-UTR assay. (d) Quantitative RT–PCR and 3′-UTR reporter assay of the TAF5L isoform relative expression. (e) An example of target predicted by both the isoform- and gene-level approaches, but not by the 3′-UTR assay (Gene TBRG1). (f) An example of drop out in the 8-mer seed region of the 3′-UTR (Gene CEBPB).
(1) Targets exclusively predicted by the isoform-level approach (four predicted targets). The seeming paradox of this category is why there is discordance between the isoform-level analysis and the 3′-UTR reporter analysis. The discrepancy for at least two of these genes, PHF17 and MALT1, can probably be explained by differences in the 3′-UTR’s tested in these two approaches (with the different 3′-UTR’s containing different microRNA-155 seed site repertoires). For example, PHF17 has two expressed isoforms (ENST00000413543 and ENST00000226319). The 3′-UTR of the isoform ENST00000413543 that was tested in our 3′-UTR assay contains two 7-mer microRNA-155 seeds and was found to be a false target based on the 3′-UTR reporter analysis (fc = 0.81) (in Figure 3b). On the other hand, the 3′-UTR of the isoform, ENST00000226319 contains one 8-mer microRNA-155 seed and was predicted by our isoform-level approach as the true target (fc = 0.53) (in Figure 3b). While we have not tested the 3′-UTR for this isoform in a 3′-UTR reporter assay, we anticipate that the 8-mer microRNA-155 seed contained within the 3′-UTR of this isoform is, in fact, responsive to microRNA-155.
Looking more carefully into the gene- and the isoform-level expression, the differential expression ratio of ENST00000226319 is 0.53 and that of ENST00000413543 is 1.67 (the two isoforms are roughly equally abundant). The differential expression ratio observed at the gene-level (0.85) is a composite of the down-regulated ENST00000226319 isoform (predicted target) and the up-regulated ENST00000413543 isoform.
(2) Targets predicted by both the isoform-level approach and the 3′-UTR reporter assays but not by the gene-level approach (19 predicted targets). This category best highlights the importance of performing isoform-based assessment of microRNA targeting. Here, the differential expression ratio of the target isoform calculated from the isoform-level analysis is more consistent with the 3′-UTR reporter assay results than it is with the results from the gene-level analysis. A good illustration of where this could have important biological significance is the case of TAF5L. TAF5L has three expressed isoforms (ENST00000366676, ENST00000366675 and ENST00000258281). The 3′-UTR of the isoform ENST00000366675 (abundance proportion 11–20%, non-dominant isoform) was tested in our 3′-UTR reporter assay, and was predicted by both the isoform-level approach and the 3′-UTR reporter assay as a microRNA-155 target (Figure 3c). It was not detected by the gene-level approach because this isoform accounts for only 20% of the total gene-level expression in control cells. To validate that the predominant, unregulated isoform (ENST00000366676) is not responsive to microRNA-155 (as negative control of no repression), we cloned the 3′-UTR of this isoform into a reporter vector and tested it for responsiveness to microRNA-155. As shown in Figure 3d, while ENST00000366675 again showed inhibition by microRNA-155, the ENST00000366676 isoform was not responsive. To further validate the isoform specific differences in expression at the endogenous RNA level, real time RT–PCR analysis was carried out on microRNA-155 expressing versus control cells using isoform specific PCR primers (Table 1). As shown in Figure 3d, RT–PCR demonstrated concordance with the isoform-level analysis of the RNA-seq data. Since the amino acid composition of the proteins expressed from these two isoforms is different at the carboxyl terminus, the isoform specific regulation of one of these isoforms can have a significant regulatory impact on the TAF5L interactome and consequently, TAF5L function.
(3) Targets predicted by both the isoform- and gene-level approaches, but not the 3′-UTR assays (five predicted targets). Genes within this group are likely to have non-functional microRNA-155 seed sequences within their 3′-UTR’s as evidenced by a lack of a response in the 3′-UTR reporter assay. In these cases, the regulation observed at the gene- and isoform-level likely occurs through indirect mechanisms. There is now good evidence that microRNA-155 targets an abundance of transcription factors (14,26–30). It is reasonable that the expression of at least some of the genes within this group is suppressed through the microRNA-155 mediated inhibition of one or more of these factors. For example, gene TBRG1 has two highly expressed isoforms (ENST00000473629 and ENST00000441174). The 3′-UTR of the isoform ENST00000441174 (abundance proportion 33–35%, non-dominant isoform) was tested in 3′-UTR assay, but not predicted by 3′-UTR assay as the microRNA-155 target (Figure 3e). One possible reason includes that these are genes where regulation probably occurs through indirect mechanisms but not through targeting of 3′-UTR.
(4) Targets predicted by all three approaches (90 predicted targets). This group largely represents genes whose dominant isoform contains functional microRNA-155 seeds within the 3′-UTR region. The finding that this category contains a largest number of targets is expected and represents genes that may be discovered by conventional gene-level approaches.
(5) Targets predicted exclusively by the gene-level approach (one predicted target). Only one case exists in the validation studies, supporting the notion that the isoform-level approach dominates the gene-level approach.
(6) Targets predicted by the 3′-UTR assay and the gene-level approach but not the isoform-level approach (five predicted targets). These cases represent a relative minority of genes from our validation studies and may arise from a variety of different experimental error types including those that arise from low-expression values or by incorrect/inaccurate isoform assessment.
(7) Targets exclusively predicted by 3′-UTR assay (20 predicted targets). There are a good number of predicted targets falling into this category and many of these likely represent tissue specific differences in 3′-UTR structure. For example, shortened transcript extension through the 3′-UTR in vivo is a hallmark of lymphocyte activation and cancer cells (31). Such truncated 3′-UTR’s obviously cannot be targeted by microRNAs whose seeds are located in the truncated regions of the 3′-UTR’s. To illustrate this point, as shown in Figure 3f, we observed low read numbers across the microRNA-155 seed sequences at the 3′-end of CEBPB. A few more genes might be interpreted in a similar way, such as DCUN1D2 and KIAA1274. Another possible reason for a lack of response at the gene- or isoform-level includes the possibility that these genes are inhibited principally at the translational level but not at the transcriptional level. Whereas the output of the 3′-UTR reporter assay reflects changes at both of these levels, RNA-seq reflects only changes at the transcriptional level.
(8) There are five genes where all three approaches predict them to be non-targets. These genes represent accordant results on genes that have non-functional microRNA-155 seed sequences.
Genome-wide microRNA-155 target prediction at the isoform-level
Our validation studies have demonstrated the potential application of the proposed computational approach for quantifying transcriptomes and microRNA targetomes. We next proceeded to carry out genome-wide studies to predict microRNA-155 targets at the isoform-level. For this analysis, we used isoform-based differential expression information in combination with seed enrichment information to predict targeting isoforms and the respective targeting region. We attempted to answer the following important biological questions that were raised initially: ‘Which isoform is the target? What’s the targeting region? What might be the targeting mechanisms?’
There are a total of 100 297 annotated protein coding isoforms in Ensembl ASTD database (version 57), corresponding to 21 271 protein coding genes. Since our RNA-seq data is single-ended, we used RPKM to quantify the abundance level of each isoform. We first applied an RPKM cut off of 0.5 to the control wild-type Mutu I cell line to determine which genes need to be filtered out, and then remove these very low-abundance genes in both control and case conditions (we note that low-abundance gene expression under the case condition might represent the genuine repression effect). After selection using this criterion, 10 513 genes were left for the genome-wide isoform-level analysis. For both case and control conditions, we then filtered out very low-abundance isoforms, which have an average RPKM value less than 0.2 in wild-type Mutu I cells. Finally, the results of 42 093 isoforms, corresponding to 10 193 genes, were used to compare the isoform- and gene-level approaches.
Due to the up- and down-regulation that takes place within the same gene (differential splicing) for some cases, the variance of the isoform-level analysis is higher than that observed at the gene-level analysis since the up- and down-regulation of the isoforms can cancel each other out at the gene-level. In addition, the total number of statistical tests needed for the isoform-level analysis is much greater than what is required for the gene-level analysis, making the comparison using adjusted P-values less sensible. We therefore primarily used a biological criterion, differential expression ratio (or fold change), and used q-value (25) as an auxiliary statistical criterion in detecting significant down-regulation. Similar to the validation studies, we used a number of fold change cut-offs ranging from a strict 0.6 to a loose 0.8. Detailed results are included in Supplementary Tables S2–S4.
Since the isoform proportion and abundance results we obtained are in the format of continuous numbers, we used a shrinkage t-test (32) to call significantly down-regulated genes, and predicted them as microRNA targets based on the presence of seed enrichment. Among these 10 513 genes, 1176 genes were predicted as targets using a cutoff = 0.8, q-value = 0.0045 and microRNA-155 seed enrichment level of 0.05. We also applied the same test on the corresponding 42 093 isoforms and found 2828 of these (corresponding to 1722 genes) that were predicted as microRNA targets using the same down-regulation fold change and seed enrichment criteria. The overlapping information of the microRNA target prediction using the gene- and isoform-level approaches is presented in Figure 4a. In Figure 4a, 1056 microRNA gene targets were predicted by both the isoform- and gene-level approach, and the isoform-level approach further determines the specific isoform that is targeted. There are also 666 and 120 microRNA gene targets exclusively predicted by the isoform- and gene-level approaches, respectively.
Figure 4.
(a) Venn diagram of the microRNA gene targets prediction using the gene- and isoform-level approaches. (b) The percentage of gene targets represented by dominant and non-dominant isoforms predicted by both the gene- and the isoform-level approaches. (c) The percentage of isoform targeting regions predicted by both the gene- and the isoform-level approaches. (d) The percentage of gene targets represented by dominant and non-dominant isoforms predicted by the isoform-level approach exclusively. (e) The percentage of isoform targeting regions predicted by the isoform-level approach exclusively.
Figure 4b shows that for 77% of the 1056 genes predicted as targets by both the gene- and isoform-level approaches, the dominant isoform being specifically regulated. We further used seed enrichment region to predict the targeting region of each targeting isoform. Figure 4c shows the percentage of seed targeting regions predicted by both the gene- and isoform-level approaches, in which 53% of the predicted isoform targets fall in the 3′-UTR region, supporting the conventional notion that 3′-UTR region is the most common microRNA targeting region. Interestingly, we also predicted almost half of the isoform targets with targeting regions in exons and 5′-UTR’s. For the 666 gene targets, corresponding to 1065 isoforms, which were exclusively predicted as targets by the isoform-level approach, the trend was reversed in that 62% of targeting genes are represented by non-dominant isoforms (Figure 4d). Figure 4e shows the percentage of seed targeting regions predicted by the isoform-level approach exclusively. Similar to Figure 4c, we observed that almost half of the predicted targeting regions are not in the 3′-UTR. In summary, it is clearly seen from Figure 4 that the isoform-level approach is capable of predicting non-dominant isoform targets that otherwise is invisible to the gene-level down-regulation approach.
Discovery of novel transcripts
Similar to other biological processes, novel transcripts that are not annotated in splicing databases are likely to play an important role in microRNA-155 targeting. In order to discover these novel transcripts, we performed the same genome-wide analysis using the augmented ASTD annotation table. We provide more details and results in Tables 1 and 2, Supplementary Tables S5 and S6, Figures S2 and S5 and the Supplementary Data (Section 5).
CONCLUSION
Due to its importance, computational prediction of microRNA targets has been well-studied. However, the existing rule-based, data-driven and expression profiling approaches to target prediction are mostly approached from the gene-level. Gene is a unit of heredity in a living cell that is used extensively in genetics but is becoming a less appropriate concept in transcriptome and targetome research. Here we propose the use of splicing isoform as a more appropriate concept for microRNA target prediction, and other genomics research since it is the isoform that is the ultimate effector of biological outcomes.
Before the emergence of the deep-sequencing technology, exon and tiling microarrays allowed for the analysis of transcriptomes at the isoform-level. The widespread use of these two microarray platforms were limited, however, by intrinsic technological limitations such as resolution, coverage, and signal saturation etc. The advent of deep sequencing technology provides, for the first time, an opportunity to profile transcriptomes at base-wise resolution, making it possible to develop computational approaches to predict microRNA targets at the isoform-level. The abundant exon–exon junction evidence revealed in RNA-seq data enables novel transcript discovery. We believe this work to be one-of-its-kind, as it allows for the prediction of isoform targets that have not been possible with the gene-level approach that we developed previously (14). Our computational work has provided deeper biological insights into the microRNA targeting mechanisms as evidenced by in vitro 3′-UTR assay validation and in vivo genome-wide microRNA target prediction.
Going beyond microRNA target prediction, the gene as a concept has been extensively used in many transcriptome-based studies. Familiar examples are gene regulatory networks and cancer classification using high throughput data. These studies have given insights into important biological mechanisms. As illustrated here for microRNA targetome research, splicing isoform is a more appropriate object for transcriptome-based studies. Consequently, the abovementioned analyses are expected to be more biologically insightful when performed at the isoform-level. Our isoform-level approach can be potentially integrated with many transcriptome-based studies to open an avenue for new isoform-based bioinformatics analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R21LM010137 to D.Z.; R01CA138268 and R01CA130752-02S1 to E.K.F.; Research Centers in Minority Institutions grant 1G12RR026260-02(GD) to K.Z.). Louisiana Board of Regent award (LEQSF(2008-11)-RD-A-32) to K.Z.; Tulane Cancer Center Post-doctoral Matching Funds to N.D. Funding for open access charge: National Institutes of Health (Research Centers in Minority Institutions grant 1G12RR026260-02(GD) to K.Z.). Louisiana Board of Regent award (LEQSF (2008-11)-RD-A-32) to K.Z.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Claire Fewell for the 3′-UTR assays. We thank the reviewers for their thorough review and helpful comments.
REFERENCES
- 1.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database Issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajewsky N. microRNA target predictions in animals. Nature Genet. 2006;38(Suppl. 1):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 3.Yue D, Liu H, Huang Y. Survey of computational algorithms for microRNA target prediction. Current Genom. 2009;10:478–492. doi: 10.2174/138920209789208219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37(Web Server issue):W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Kim SK, Nam KW, Rhee JK, Lee WJ, Zhang BT. mitarget: microrna target-gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yousef M, Jung S, Kossenkov AV, Showe LC, Showe MK. Naive Bayes for microRNA target predictions–machine learning for microRNA targets. Bioinformatics. 2007;23:2987–2992. doi: 10.1093/bioinformatics/btm484. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creighton CJ, Nagaraja AK, Hanash SM, Matzuk M, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nature Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 14.Xu G, Fewell C, Taylor C, Deng N, Hedges D, Wang X, Zhang K, Lacey M, Zhang H, Yin Q, et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA. 2010;16:1610–1622. doi: 10.1261/rna.2194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTRand 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohnert R, Ratsch G. rQuant.web: a tool for RNA-seq-based transcript quantitation. Nucleic Acids Res. 2010;38(Suppl. 2):W348–W351. doi: 10.1093/nar/gkq448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincrnas. Nature Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard H, Schulz MH, Sultan M, Nurnberger A, Schrinner S, Balzereit D, Dagand E, Rasche A, Lehrach H, Vingron M, et al. Prediction of alternative isoforms from exon expression levels in RNA-seq experiments. Nucleic Acids Res. 2010;38:e112. doi: 10.1093/nar/gkq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol. Med. 2011;6:2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing Y, Yu T, Wu NY, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstruction of full-length isoforms from splice graphs. Nucleic Acids Res. 2006;34:3150–3160. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. PNAS USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne RJ. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin Q, Wang X, Fewell C, Cameron J, Zhu H, Baddoo M, Lin Z, Flemington EK. MiR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J. Virol. 2010;84:6318–6327. doi: 10.1128/JVI.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Z, Xu G, Deng N, Taylor C, Zhu D, Flemington EK. Quantitative and qualitative RNA-seq-based evaluation of Epstein-Barr virus transcription in type I latency Burkitt's lymphoma cells. J. Virol. 2010;84:13053–13058. doi: 10.1128/JVI.01521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs- with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.