Abstract
The mammalian hairless (hr) gene plays a critical role in the maintenance of hair growth. Although the hr gene has been identified, the biochemical function of its encoded protein (Hr) has remained obscure. Here, we show that Hr functions as a transcriptional corepressor for thyroid hormone receptors (TRs). We find that two independent regions of Hr mediate TR binding and that interaction requires a cluster of hydrophobic residues similar to the binding motifs proposed for nuclear receptor corepressors (N-CoR and SMRT). Similarly, we show that Hr binds to the same region of TR as known corepressors. We show that Hr interacts with histone deacetylases (HDACs) and is localized to matrix-associated deacetylase (MAD) bodies, indicating that the mechanism of Hr-mediated repression is likely through associated HDAC activity. Thus, Hr is a component of the corepressor machinery, and despite its lack of sequence identity with previously described corepressors, its mode of action is remarkably conserved. On the basis of its thyroid hormone-inducible and tissue- and developmental-specific expression, Hr likely defines a new class of nuclear receptor corepressors that serve a more specialized role than ubiquitous corepressors. The discovery that Hr is a corepressor provides a molecular basis for specific hair loss syndromes in both humans and mice.
Keywords: Nuclear receptor, thyroid hormone, transcriptional repression, histone deacetylase
Genetic analysis is a powerful way to discern the biological role of a given gene and its encoded protein. In mammals, the advent of technology that allows manipulation of the mouse genome has resulted in the generation of loss-of-function mutations for many genes that often encode proteins of known biochemical function. Conversely, for many spontaneous mutations, the biological role of a given gene is known whereas the biochemical function of its encoded protein remains unclear. A number of spontaneous mutations in mice fall into the latter category, including the mammalian hairless (hr) gene.
Mutation of the hr gene was first recognized in the mouse 75 years ago (Brooke 1926). The original mutation was caused by insertion of an endogenous retrovirus, and the murine hr gene was cloned by mapping the retroviral insertion site (Stoye et al. 1988; Cachon-Gonzalez et al. 1994). The human ortholog was subsequently found to be associated with congenital hair disorders (alopecia universalis and papular atrichia; Ahmad et al. 1998, 1999; Cichon et al. 1998; Kruse et al. 1999; Sprecher et al. 1999). Multiple murine and human alleles have been characterized, and all share a distinctive phenotype in which initial hair growth is normal, but after shedding, the hair does not regrow (Lyon and Searle 1989; Panteleyev et al. 1998; Ahmad et al. 1999). The phenotype is variable and can include complete loss of hair follicles, papular rash, and excessive wrinkling of the skin (Panteleyev et al. 1998; Ahmad et al. 1999). The precise role of hr in hair follicle biology is unknown, although there is evidence that reduced hr function causes an increase in intrafollicular apoptosis (Panteleyev et al. 1999). The hr gene also has an important role in the brain, as hr expression is directly regulated by thyroid hormone in developing rat brain (Thompson 1996; Thompson and Bottcher 1997; Thompson and Potter 2000). Consistent with this idea, phenotypic analysis of hr mutant (the hrrh allele) mice has revealed alterations in neuronal morphology and inner ear defects (Lyon and Searle 1989; Garcia-Atares et al. 1998; Cachon-Gonzalez et al. 1999). In humans, neurological deficits are sometimes correlated with the hair loss phenotype of hr mutants, as there are multiple cases in which concurrent mental retardation is observed (del Castillo et al. 1974; Aita et al. 2000).
Despite the importance of hr gene function in the skin and brain, little is known about the biochemical role of the protein encoded by hr (Hr). The function of Hr cannot be predicted on the basis of homology, as the primary amino acid sequence is not related to proteins of known function. Hr does not have known structural motifs with the exception of a cluster of conserved cysteine residues that has been proposed to form a noncanonical zinc finger (Cachon-Gonzalez et al. 1994). We have shown previously that Hr interacts directly and specifically with thyroid hormone receptor (TR) (Thompson and Bottcher 1997). TR is a transcription factor that influences the transcription rate of target genes in response to hormone binding. TR has been shown to interact with a number of proteins that mediate or modify its transcriptional activity. In the absence of thyroid hormone (TH), TR represses transcription through interaction with corepressors such as nuclear receptor corepressor (N-CoR) and silencing mediator for retinoic acid and thyroid hormone receptors (SMRT), which form multiprotein complexes that often include histone deacetylases (HDACs) (Chen and Evans 1995; Horlein et al. 1995; Sande and Privalsky 1996; Alland et al. 1997; Heinzel et al. 1997; Nagy et al. 1997). When hormone is bound, TR interacts with distinct multiprotein complexes that include coactivators, resulting in transcriptional activation (Xu et al. 1999; Glass and Rosenfeld 2000).
Evidence that Hr can mediate transcriptional repression suggested that Hr might function as a corepressor (Thompson and Bottcher 1997). To determine whether Hr is a corepressor, we identified Hr domains that specify TR interaction. We show here that the regions of Hr that interact with TR are similar in hydrophobicity and predicted secondary structure to motifs shown previously to mediate corepressor–receptor interaction (Moras and Gronemeyer 1998; Hu and Lazar 1999; Nagy et al. 1999; Perissi et al. 1999; Webb et al. 2000). Furthermore, we demonstrate that Hr can mediate transcriptional repression by unliganded TR and identify multiple domains that confer repression. Finally, we show that, like other corepressors, Hr interacts with HDACs in vivo, indicating that the mechanism by which Hr mediates transcriptional repression is likely through associated HDAC activity. On the basis of its receptor interaction specificity and both tissue-restricted and TH-inducible expression, Hr is representative of a new class of nuclear receptor corepressors that likely perform a more specialized role than previously characterized, ubiquitous corepressors.
Results
Hr interacts with TR via two independent domains
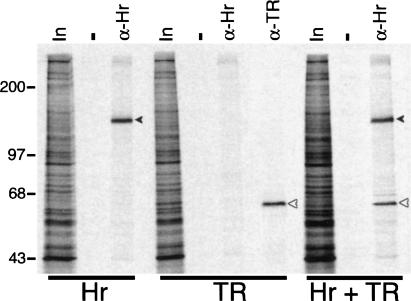
We have shown previously that Hr interacts directly and specifically with TR in vitro (Thompson and Bottcher 1997). To confirm that Hr interacts with TR in vivo, we tested interaction using a coimmunoprecipitation assay. Hr and TRα were expressed in cultured cells both individually and together and extracts were prepared and used for immunoprecipitation with Hr- and TR-specific antisera (Fig. 1). Both Hr and TR were detected by immunoprecipitation with their respective antiserum, as bands corresponding to the molecular mass of each protein were detected. When Hr was immunoprecipitated in the presence of TR, an additional band corresponding to the size of TR was observed. Thus, in the presence of coexpressed TR, Hr and TR coimmunoprecipitate, indicating that they associate in vivo.
Figure 1.
Hr interacts with TR in vivo. COS cells transfected with expression vectors for TRα and Hr both individually and together (Hr + TR) were metabolically labeled with [35S]methionine and protein extracts were prepared and used for immunoprecipitation analysis. Hr-specific antiserum (α-Hr) immunoprecipitates Hr (closed arrowhead) and, in the presence of TR, coprecipitates TR (open arrowhead). Immunoprecipitation with TR-specific antiserum (α-TR) shows position of TR (open arrowhead). (In) Two percent of extract used for immunoprecipitation. Sizes of molecular mass markers (kD) are indicated.
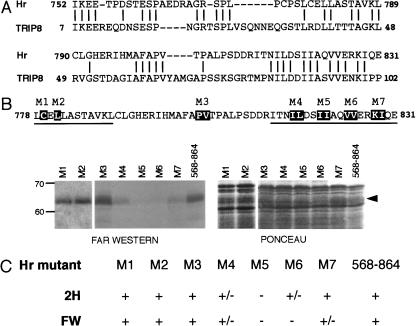
To analyze further the Hr–TR interaction, we mapped the regions of Hr required for interaction with TR. Deletion derivatives of Hr were constructed and tested for interaction by use of both Far Western and yeast two-hybrid assays (Fig. 2). The part of Hr originally shown to interact with TR included amino acids 568–1207. Deletion of the amino- (724–1207) and carboxy- (568–1048) terminal regions of 568–1207 did not abolish interaction. An internal domain (amino acids 750–1084) was found to support interaction in both Far Western and two-hybrid assays. Subdivision of this domain into smaller regions revealed two potential interaction domains. The minimal region found to interact in both assays was amino acids 750–864, indicating that this region is a primary site of interaction (TR-ID1). The derivative including amino acids 980–1084 was strongly positive in the Far Western assay, suggesting that this region includes a second site of interaction (TR-ID2). Previous results have shown that Hr interacts with TR in the absence of TH (Thompson and Bottcher, 1997). Therefore, we determined whether the separate interaction domains also interact with TR in a TH-dependent manner (Fig. 2C). Far Western analysis in the absence and presence of TH showed that both TR-ID1 and TR-ID2 interact preferentially with TR in the absence of TH.
Figure 2.
Mapping of the interaction domains of Hr with TR. (A) Deletion derivatives of Hr tested in the Far Western assay for interaction with TR. Bacterially expressed fusion proteins of various Hr deletion derivatives were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with 35S-labeled TRβ1. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show loading of bacterial protein extracts and sizes of fusion proteins. (B) Summary of yeast two-hybrid (2H) and Far Western (FW) assay results. Yeast two-hybrid results were scored by growth on medium lacking histidine; (+) growth equal to 568–1207, (+/−) positive but weaker growth, (−) no detectable growth. All experiments were repeated at least twice with the same results. (C) Hr interacts with TR in the absence of ligand. Derivatives of Hr encoding TR interaction domains tested in the Far Western assay of duplicate blots incubated with 35S-labeled TRβ1 in the absence (−T3) or presence (+T3) of 10−6 M L-T3. Derivative 864–981 is included as a negative control. (Left and middle) Autoradiographs of Far Western filters; (right) Ponceau red stain. (D) Coimmunoprecipitation of individually expressed TR-IDs. COS cells were cotransfected with expression vectors for TRα and epitope-tagged Hr deletion derivatives that included the indicated amino acids. Protein extracts were used for immunoprecipitation analysis with TR-specific antiserum (α-TR) or mouse IgG (negative control). Hr derivatives were detected by Western analysis with Myc-specific antiserum. (Right) Western analysis of extracts with TR-specific antiserum to show expression of TR. Hr fragments expressed in COS cells alone did not precipitate with TR-specific antiserum (data not shown). (In) One percent of extract used for immunoprecipitation except for 750–1085 (5%). Sizes of molecular mass markers (in kD) are indicated.
To determine whether TR-ID1 and TR-ID2 could interact independently with TR in vivo, we performed coimmunoprecipitation assays (Fig. 2D). Vectors expressing regions of Hr with an epitope (Myc) tag were constructed and cotransfected with an expression vector for TR into COS cells. Hr deletion derivatives encompassing both interaction domains (amino acids 750–1084), TR-ID1 alone (amino acids 750–864), TR-ID2 alone (amino acids 980–1084), and a noninteracting region (amino acids 864–981) were tested. Extracts prepared from transfected cells were used for immunoprecipitation with TR-specific antiserum, followed by Western analysis with Myc-specific antiserum to detect the various Hr derivatives. The Hr derivative that included both interaction domains (750–1085) was detected by Western analysis, indicating coimmunoprecipitation with TR. Hr derivatives that contained only TR-ID1 or TR-ID2 were found to coimmunoprecipitate with TR as well, indicating that both domains can interact independently with TR. As expected, a region shown previously to lack TR interaction (864–981) does not coprecipitate. Hr derivatives expressed without coexpression of TR did not immunoprecipitate with TR-specific antiserum (data not shown). Thus, Hr has two separable domains that mediate TR interaction.
Within TR-ID1 is a stretch of amino acids related to TRIP8, a protein of unknown function that was shown previously to interact with TR (Lee et al. 1995; Fig. 3). Because both Hr and TRIP8 interact with TR, the conserved amino acids are likely important for mediating interaction. Therefore, to determine which amino acids in this region are essential for interaction, we used site-directed mutagenesis to change pairs of conserved amino acids to alanine in a derivative containing TR-ID1 (amino acids 568–864). Mutants were tested for interaction in Far Western and yeast two-hybrid assays. Mutation of the isoleucine residues at positions 820 and 821 (M5) and valine residues at positions 824 and 825 (M6) abolished interaction in both assays. Mutation of adjacent residues 816I, 817L (M4) and 828K, 829I (M7) decreased but did not abolish interaction, whereas mutations outside this hydrophobic region (M3) did not affect interaction. These results indicate that a small hydrophobic subdomain of the conserved region (IIAQVV) is critical for interaction. TR-ID1 also includes a motif (LXXLL, Hr amino acids 778–782) shown previously to mediate binding of coactivators to nuclear receptors (Heery et al. 1997). Mutation of the leucine residue at position 781 (M2) had no effect on interaction in the two-hybrid or Far Western assays (Fig. 3). Similarly, mutation of a second LXXLL motif outside of this region (amino acids 586–590) did not affect interaction (data not shown), demonstrating that Hr does not interact with TR via these coactivator-type binding domains.
Figure 3.
Hydrophobic amino acids in TR-ID1 mediate Hr–TR interaction. (A) Region of homology between Hr and TRIP8. Identical residues are indicated by dashes. (B) Far Western analysis of Hr mutants. (Top) Residues in TR-ID1 changed to alanine by site-directed mutagenesis (within deletion derivative 568–864) indicated by black boxes. Regions predicted to form alpha-helical secondary structure are underlined. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Arrowhead) Position of mutated Hr fusion proteins. (C) Summary of yeast two-hybrid (2H) and Far Western (FW) assays for interaction of mutated Hr derivatives. Yeast two-hybrid results were scored by growth on medium lacking histidine. (+) Growth equal to 568–864; (+/−) positive but weaker growth; (−) no detectable growth. For Far Western analysis: (+) signal equal to 568–864; (+/−) significantly weaker signal than 568–864; (−) no detectable signal. All experiments were repeated at least twice with the same results.
The carboxy-terminal interaction domain (TR-ID2) was defined further by construction and testing of additional deletion derivatives in the Far Western assay (Fig. 4A). The minimal region contained within deletions positive for interaction was between amino acids 1024 and 1040. This region contains a hydrophobic-rich core similar to the domain that mediates interaction by TR-ID1 (Fig. 4B). Residues in this region were changed to alanine and tested by use of the Far Western assay. Only mutation of amino acids LI1031/1032 (M11) abolished interaction, whereas mutation of V1026 (M8) and V1036 (M14) resulted in weak interaction. LI1031/2 is part of a motif (LISIL) that resembles a motif predicted previously to mediate corepressor–TR interaction (Hu and Lazar 1999; Nagy et al. 1999). However, mutation of the second isoleucine residue in this motif (I1034) resulted in normal binding, suggesting that this is not the critical motif. On the basis of the requirement for LI1031/2 and reduced interaction when residues L1035 and V1036 are mutated, LISILV is likely the minimal interaction domain, although adjacent residues are involved, as other mutants showed intermediate effects on TR binding.
Figure 4.
Analysis of Hr carboxy-terminal interaction domain (TR-ID2). (A) Far Western analysis of deletion derivatives from Hr amino acids 980–1084. (Top left) autoradiograph of Far Western filter; (top right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Open arrowheads) Positions of Hr fusion proteins. (Bottom) Schematic representation of deletion derivatives from Hr amino acids 980–1084. The minimal region in both interacting dervatives is between amino acids 1024 and 1040. (B) Far Western analysis of point mutants in TR-ID2. Site-directed mutatgenesis was used to change the indicated residues to alanine (within deletion derivative 980–1084). Underlined residues are predicted to form alpha-helical secondary structure. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Arrowhead) Position of mutated Hr fusion proteins, mutant M11 migrates slightly slower than other mutants; (++) interaction equivalent to 980–1084; (+) interaction weaker than 980–1084; (+/−) weak but detectable interaction; (−) no detectable interaction. (C) Summary of site-directed mutagenesis results for Hr TR-IDs. Boxed residues showed no interaction when changed to alanine, underlined residues showed partial interaction. Below is shown a comparison of Hr consensus with consensus sequences defined for receptor binding by N-CoR and SMRT. Consensus 1 is from Nagy et al. (1999), Hu and Lazar (1999), and Webb et al. (2000); consensus 2 is from Perissi et al. (1999). Bold residues indicate Hr consensus extended to match consensus 2.
Comparison of the essential residues in the two Hr interaction domains yields a consensus motif for Hr binding to TR of I/LIXXL/VV (Fig. 4C). This consensus is similar to that identified for N-CoR and SMRT binding to TR and retinoic acid receptor (RAR), defined by different investigators as either I/LXXV/II or LXXH/IIXXXI/L (Hu and Lazar 1999; Nagy et al. 1999; Perissi et al. 1999; Webb et al. 2000). Our results are in general agreement with both motifs if conservative substitutions are allowed (valine at position 9 in Hr TR-ID1 and positions 1 and 9 in Hr TR-ID2). The Hr consensus defines a motif in which the spacing but not the identity of hydrophobic residues is well conserved. These results support models suggesting that corepressor–TR interaction domains form an amphipathic alpha helix, and it is this secondary structure that is critical for receptor interaction (Nagy et al. 1999; Perissi et al. 1999). Consistent with this idea, both Hr interaction domains are in regions of predicted alpha helical structure (PSIPRED, McGuffin et al. 2000), and a helical plot shows hydrophobic amino acids lie on one face of an alpha helix (positions 1, 4, 5, and 8; data not shown). The spacing of hydrophobic residues in the Hr interaction domains fits well with the extended helix predicted by Perissi et al. (1999), including the hydrophobic residues in positions 1 and 9, indicating that the Hr-binding motif may include these residues.
Hr interacts with the TR ligand-binding domain
To map the domain(s) of TR that interact with Hr, deletion derivatives of TR were tested in the Far Western and yeast two-hybrid assays (Fig. 5). As shown previously, Hr interacts with both TRα and TRβ and not with RARα or RXRβ in the Far Western assay (Thompson and Bottcher 1997). Interaction with Hr is not affected by deletion of 57 amino acids from the carboxyl terminus of TRα (TRα 1–353). This region includes the AF-2 domain/helix 12, a region shown to mediate interaction with transcriptional coactivators but that is not required for corepressor binding (Barettino et al. 1994; Durand et al. 1994; Chen and Evans 1995; Horlein et al. 1995; Heery et al. 1997; Moras and Gronemeyer 1998). The amino terminus and DNA-binding domain alone do not interact with Hr (TRα 1–126), whereas the Hr–TR interaction is unaffected by deletion of the amino terminus and DNA-binding domain (TRβ 199–456). Taken together, these results suggest that the site of interaction lies in the ligand-binding domain between amino acids 150 and 353 (numbering per TRα). Because Hr is a putative corepressor, we tested a TRα derivative that has a point mutation (amino acid 160 from arginine to proline) shown previously to abolish interaction with a known corepressor, SMRT (Chen and Evans 1995). Indeed, we found that TRα160 failed to interact with Hr in both assays. Although this mutation has been shown to be a structural determinant rather than an interaction surface, these results indicate that the same mutation that disrupts interaction of TR with other corepressors also disrupts interaction with Hr. Together, these results are consistent with the TR-binding properties of previously described corepressors (N-CoR, SMRT), suggesting that Hr binds the same region of TR as corepressors (Chen and Evans 1995; Horlein et al. 1995).
Figure 5.
Mapping of the TR–Hr interaction domain. Deletion derivatives of TR were tested in the yeast two-hybrid and Far Western assays for interaction with Hr. (A) TR derivatives tested in Far Western assay. Protein extract from bacteria expressing GST-568–1207 was separated by SDS-PAGE, transferred to nitrocellulose, and individual lanes incubated with 35S-labeled TR derivatives (see schematic in B) as indicated. Panels are autoradiographs of Far Western filters. (Far right) Ponceau red staining to show loading of extract. (Arrowhead) Position of GST-568–1207. (Bottom) SDS–polyacrylamide gel showing 35S-labeled in vitro translated proteins used as probes. (B) Summary of yeast two-hybrid (2H) and Far West-ern (FW) assays for interaction of TR derivatives. Yeast two-hybrid results were scored by growth on medium lacking histidine. (+) Interaction; (−) no interaction; (ND) not done. Shaded region of TRα2 indicates a divergent carboxyl terminus; TRα160 has a point mutation changing a proline residue at position 160 to arginine. (DNA) DNA-binding domain; (T3) hormone-binding domain; (AF-2) coactivator-interaction domain in helix 12. All experiments were repeated at least twice with the same results.
Hr mediates transcriptional repression by unliganded TR
We have shown previously that Hr (amino acids 568–1207) can repress basal transcription when expressed as a fusion protein with the GAL4 DNA-binding domain (DBD), indicating that Hr likely functions in transcriptional repression (Thompson and Bottcher 1997). Together with evidence that Hr interacts preferentially with TR in the absence of TH (Thompson and Bottcher 1997; Fig. 2C), this result suggests that Hr may mediate transcriptional repression by unliganded TR. Attempts to demonstrate directly the role of Hr and other corepressors in TR-mediated transcriptional repression have shown only modest effects (twofold; Li et al. 1997; Dressel et al. 1999; data not shown), likely because corepressors such as N-CoR and SMRT are ubiquitously expressed. Therefore, we tested the influence of Hr on TR transcriptional activity using pituitary-derived cell lines (GH1, GC) that endogenously express high levels of TR and low levels of N-CoR and SMRT (Yaffe and Samuels 1984; Misiti et al. 1998; data not shown; Fig. 6A). These cells also express endogenous Hr in a TH-dependent manner (Thompson 1996). TH-responsive (MLV tk-luc) and control (tk-luc) reporter genes were introduced into cells by transient transfection and activity was measured in the absence and presence of thyroid hormone. As expected, transcriptional activity is induced from the TH-responsive reporter gene in the presence of TH. The effect of Hr on TR transcriptional activity was measured by cotransfection of an expression vector for Hr. When Hr is expressed, transcriptional activation by TH is slightly reduced (<twofold). Strikingly, in the presence of Hr, transcriptional activity in the absence of TH is reduced by ∼fivefold only for the TH-responsive reporter gene. This result indicates that Hr can enhance transcriptional repression by unliganded TR. To demonstrate that the effect of Hr on TR-mediated repression requires TR binding, we used a derivative of Hr (ΔHr) that lacks both interaction domains. As expected, the mutant Hr was unable to enhance repression by TR. Expression of the mutant Hr was equivalent to wild type as assessed by Western analysis (data not shown).
Figure 6.
Hr mediates transcriptional repression by unliganded TR. (A) TH-responsive (MLV TRE-tk-luc) and control (tk-luc) reporter genes were cotransfected into GH1 cells together with an expression vector for Hr (+Hr) or expression vector alone (−Hr). Luciferase activity was measured in the absence (−T3) and presence (+T3) of 10–7 M L-T3. ΔHr is a Hr derivative that lacks both TR-IDs and consists of amino acids 1–568.Relative luciferase activity (RLU) is luciferase activity normalized to β-gal activity from CMX β-gal. Shown is the mean of two experiments done in duplicate. (Right) Fold repression is luciferase activity in the absence of Hr divided by activity in the presence of Hr. Shown are the mean values from three experiments done in duplicate. (B) Transcriptional repression by Hr is specific for TR. A TH-responsive reporter gene which can be activated by both TR and retinoic acid receptor (RAR) (TREpal-tk-luc) was cotransfected into GC cells together with an expression vector for TR (left) or RAR (right). The effect of Hr on transcription by these receptors was assayed by cotransfection of an expression vector for Hr (+Hr) or expression vector only (−Hr). Luciferase activity was measured in the absence and presence of 10–7 M L-T3 (T3) or 10–6 M all-trans retinoic acid (RA). Relative luciferase activity (RLU) is luciferase activity normalized to β-gal activity from CMX β-gal. Experiments were done three times with similar results; shown is a representative experiment. Fold repression is luciferase activity in the absence of Hr divided by activity in the presence of Hr and is the mean of three experiments done in duplicate.
To further demonstrate the specificity of Hr on TR-mediated repression, we used RAR in the cotransfection assay (Fig. 6B). TR and RAR can both bind to the same response element (TREpal), and both repress transcription in the absence of ligand. Because Hr does not bind to RAR, expression of Hr should have no effect on RAR-mediated repression. So that receptor levels would be comparable, TR and RAR were both expressed by transient transfection. As expected, both TR and RAR induce transcription in the presence of their respective ligands. When TR is expressed, addition of Hr results in a fourfold reduction of basal transcription in the absence of TH. In contrast, when RAR is expressed, basal level transcription is unaffected by coexpression of Hr. Thus, Hr does not mediate repression by a nuclear receptor to which it does not bind (RAR) and can mediate transcriptional repression by TR on two different TH-responsive genes. Together, these results show that Hr mediates TR-dependent transcriptional repression in the absence of ligand, the functional definition of a nuclear receptor corepressor.
Mapping of the Hr repression domains
To identify specific regions of Hr that function in transcriptional repression, deletion derivatives of Hr were expressed as fusion proteins with the GAL4 DBD and tested in a cotransfection assay with a GAL4-responsive reporter gene (Fig. 7). This assay allows the assessment of repressor activity without dependence on receptor binding by the various deletion derivatives. Expression of full-length Hr repressed transcription by 11-fold, whereas derivatives encompassing the amino- and carboxy-terminal halves of the protein (amino acids 31–568, and 568–1207, respectively) repressed transcription as well, suggesting that there are at least two distinct repression domains. Further deletion derivatives were constructed and tested. Within the amino-terminal region, the minimal deletion derivative capable of repressing transcription included amino acids 236–450 (RD1). Deletion derivatives of the carboxy-terminal region (568–1207) were also tested, and multiple derivatives could mediate repression. Amino acids 750–864 (RD2) were as effective as the parental construct (14-fold) and overlap with one of the regions that interacts with TR. Amino acids 864–980 (RD3) repressed transcription to a smaller but significant degree (7.5-fold). Thus, like previously described corepressors, Hr has multiple, separable repression domains (Horlein et al. 1995; Li et al. 1997; Ordentlich et al. 1999).
Figure 7.
Hr has multiple independent repression domains. (A) Fusion proteins of Hr with the GAL4 DNA-binding domain (DBD) were used in a cotransfection assay with a GAL4-responsive promoter (GALp3-tk luc). Values were calculated by normalization of luciferase activity to β-gal activity from CMX β-gal; fold repression is activity of GAL4 DBD divided by the activity of the indicated deletion derivative. Results are the mean of four independent experiments done in duplicate. (B) Schematic representation of Hr functional domains defined for TR interaction (TR-ID) and transcriptional repression (RD). Repression domains are shaded, ZF indicates region proposed to form a zinc finger. (TR-ID1) Amino acids 816–830; (TR-ID2) amino acids 827–838; (RD1) amino acids 236–450; (RD2) amino acids 568–864; (RD3) amino acids 864–981.
Interaction of Hr with histone deacetylases
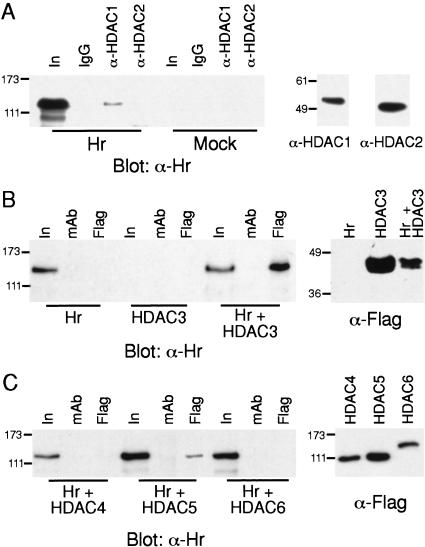
Having shown that Hr can repress transcription via multiple domains, we next addressed the mechanism by which Hr functions as a repressor. In many cases, transcriptional repression results from the association of corepressors with histone deacetylases (Pazin and Kadonaga 1997; Burke and Baniahmad 2000; Glass and Rosenfeld 2000). There are eight known HDACs, which are divided into two classes (I and II) on the basis of structural homology (Grozinger et al. 1999; Gray and Ekstrom 2001). Corepressors have been shown to be components of multiprotein complexes that include histone deacetylases and other proteins such as Sin3A (Alland et al. 1997; Heinzel et al. 1997; Nagy et al. 1997; Laherty et al. 1998; Guenther et al. 2000; Wen et al. 2000). Because it is likely that Hr functions through a similar mechanism, we examined whether Hr interacts with HDACs. Using the two-hybrid and Far Western assays, we did not detect interaction of Hr with several HDACs or Sin3A (data not shown). However, because corepressors form large, multiprotein complexes, it is feasible that Hr could be found in a similar complex without directly contacting HDACs. Therefore, we used coimmunoprecipitation assays to determine whether Hr is found in an in vivo complex with HDACs (Fig. 8). To test for interaction of Hr with endogenous HDAC1 and HDAC2, immunoprecipitation with antisera specific for HDAC1 and HDAC2 was performed on protein extracts prepared from Hr-transfected cells. Western blot analysis with Hr-specific antiserum showed that when HDAC1 is immunoprecipitated, a proportion of Hr is precipitated as well (Fig. 8A). In contrast, immunoprecipitation of HDAC2 does not result in detectable precipitation of Hr. The presence of both HDAC1 and HDAC2 in the extract was verified by Western analysis. To determine whether Hr interacts with other HDACs, epitope (FLAG)-tagged derivatives of HDAC3–HDAC6 were transfected into cultured cells along with Hr (Fig. 8B,C). FLAG-specific antiserum was used to immunoprecipitate the various HDACs, followed by Western analysis with Hr-specific antiserum. When extracts from cells expressing only Hr or only HDAC3 were immunoprecipitated with FLAG-specific antiserum, Hr protein was not detected by Western analysis. However, when HDAC3 was present together with Hr, immunoprecipitation with FLAG-specific antiserum resulted in the coprecipitation of Hr, indicating that HDAC3 associates with Hr. Under the same conditions, we find that HDAC5 coprecipitated with Hr but HDAC4 and HDAC6 did not (Fig. 8C). The presence of the various HDACs in the extracts was verified by Western analysis. Therefore, Hr appears to associate with HDACs, coimmunoprecipitating preferentially with HDAC1, HDAC3, and HDAC5.
Figure 8.
Hr associates with HDACs. (A) Hr associates with endogenous HDAC1. (Left) Protein extracts prepared from COS cells transfected with an expression vector for Hr (Hr) or vector only (Mock) were used for immunoprecipitation with HDAC1- and HDAC2-specific antisera. Hr was detected by Western analysis with Hr-specific antisera. (In) 10% of extract used for immunoprecipitation; (IgG) nonspecific rabbit IgG (negative control). (Right) Western analysis detecting HDAC1 and HDAC2 in extract used for immunoprecipitation (10% of input). (B) Hr coimmunoprecipitates with HDAC3. (Left) Protein extracts prepared from COS cells transfected with expression vectors for Hr, FLAG-tagged HDAC3, or both (Hr + HDAC3). FLAG-specific antiserum or a nonspecific monoclonal antiserum (mAb) were used for immunoprecipitation. Hr was detected by Western analysis with Hr-specific antisera. (In) Ten percent of extract used for immunoprecipitation. (Right) Western analysis of extract used for immunoprecipitation with FLAG-specific antisera to detect HDAC3. (C) Hr associates with HDAC5. (Left) COS cells were doubly transfected with expression vectors for Hr and FLAG-tagged HDAC4–HDAC6. FLAG-specific antiserum or a nonspecific monoclonal antibody (mAb) were used for immunoprecipitation. Hr was detected by Western analysis with Hr-specific antiserum. (Right) Western analysis of extracts used for immunoprecipation with FLAG-specific antiserum to detect HDACs. Protein loaded is 10% of amount used for immunoprecipitation. Sizes of molecular mass markers (kD) are indicated.
Hr is localized to MAD bodies
Hr is a nuclear protein, and many of the known HDACs are also localized to the nucleus (Thompson and Bottcher 1997; Emiliani et al. 1998; Kao et al. 2000). To determine whether Hr colocalizes with HDACs in vivo, we first examined the distribution of Hr in the nucleus. Hr was expressed in cultured cells by transient transfection and detected by indirect immunofluorescence with Hr-specific antiserum (Fig. 9). As shown previously, we find that, in vivo, Hr expression is nuclear. In 5%–20% of cells, Hr nuclear expression is not uniform but rather is concentrated in discrete nuclear regions resulting in a punctate appearance. This expression pattern resembles that observed for HDAC5 (Downes et al. 2000). Within the nucleus, HDAC5 is found in discrete subnuclear domains termed matrix-associated deacetylase (MAD) bodies. MAD bodies are comprised of a complex of proteins that includes corepressors (SMRT) and HDACs (HDAC5 and HDAC7; Downes et al. 2000).
Figure 9.
Hr localizes to MAD bodies. (A) Hr localizes to subnuclear structures. COS cells were transfected with an expression vector for Hr, plated on coverslips and used for indirect immunofluorescence with Hr-specific antiserum. (Left) DAPI staining to show nucleus; (center) staining with Hr-specific antiserum detected with Cy3-conjugated secondary antiserum; (right) differential interference contrast (DIC) image of cell. Bar, 20 μm. (B) Hr and HDAC5 colocalize to the same subnuclear bodies. COS cells were transfected with expression vectors for Hr (Hr), FLAG-tagged HDAC5 (HDAC5), or both (Hr + HDAC5) and detected by indirect immunofluorescence with Hr- or FLAG-specific antiserum. α-Hr and α-FLAG were detected with Cy3- and FITC-conjugated secondary antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow color indicates regions of overlap. (C) Hr colocalizes with SMRT in MAD bodies. COS cells were transfected with the indicated expression vectors and used for indirect immunofluorescence. (Top) Hr and SMRT detected by use of myc- and SMRT-specific antisera, respectively. α-myc and α-SMRT were detected with Cy3- and FITC-conjugated antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow indicates regions of overlap. (Middle) SMRT and HDAC5 detected with SMRT- or FLAG-specific antiserum. α-SMRT and α-FLAG were detected with Cy3- and FITC-conjugated secondary antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow indicates regions of overlap. (Bottom) Hr and RNA splicing factor SC-35 detected with Hr- and SC-35-specific antisera. α-Hr and α-SC-35 were detected with Cy3- and FITC-conjugated secondary antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow indicates regions of overlap.
To determine whether Hr localization is the same as HDAC5 and that, therefore, Hr is a component of MAD bodies, cells were transfected with expression vectors for Hr and FLAG–HDAC5 both individually and together. Indirect immunofluorescence with Hr-specific and FLAG-specific antisera was used to detect Hr and HDAC5, respectively (Fig. 9B). Both Hr and HDAC5 showed punctate, nuclear staining in a subset of cells. When both Hr and HDAC5 were present, the staining pattern of the two proteins largely overlapped. Because MAD bodies also contain SMRT, indirect immunofluorescence was used to detect Hr and SMRT. As for HDAC5, we find an overlapping staining pattern for Hr and SMRT, indicating that MAD bodies may contain multiple corepressors. To verify that the punctate staining pattern was similar to that previously observed in MAD bodies, antibody staining for HDAC5 and SMRT was used (Fig. 9C). As expected, the staining pattern of SMRT and HDAC5 was coincident. To assess the specificity of the subnuclear structures detected by the Hr antiserum, cells were stained for Hr and SC-35, a protein found in RNA splicing bodies that also has a punctate staining pattern (Fig. 9C). Both Hr and SC-35 detect punctate, nuclear expression; however, staining for Hr and SC-35 does not overlap. Together, these results indicate that Hr is a component of MAD bodies, providing further evidence that Hr and HDACs associate in vivo.
Discussion
Hr defines a new class of specialized nuclear receptor corepressors
Hr shares little sequence identity with previously described nuclear receptor corepressors such as N-CoR and SMRT. Yet, dissection of Hr domains involved in TR interaction and transcriptional repression has revealed striking functional similarities between Hr and corepressors. Common features include (1) binding to TR via two independent domains, (2) binding to TR in the absence of hormone, (3) the presence of multiple, separable repression domains, (4) interaction with HDACs, and (5) localization to MAD bodies. Notably, we demonstrate that Hr can mediate transcriptional repression by unliganded TR. Thus, this analysis has revealed that Hr is a nuclear receptor corepressor.
Because Hr shares many functional similarities with N-CoR and SMRT, what is the precise role of each of these factors? N-CoR and SMRT are ubiquitously expressed and likely have a general role in mediating repression by TR and RAR (Horlein et al. 1995; Chen et al. 1996). In contrast, expression of hr is largely restricted to skin and brain (Cachon-Gonzalez et al. 1994; Thompson 1996). In addition, hr expression in the brain is developmentally regulated, reaching maximal levels during the first three postnatal weeks, the period within which TH is known to be important for brain development. On the basis of its spatially and temporally restricted expression, Hr may impart a specialized function to the corepressor complex not provided by ubiquitous corepressors. Receptor binding specificity is consistent with this notion, as results demonstrating that Hr influences transcriptional repression by unliganded TR but not RAR indicate that Hr preferentially influences TR signaling.
Perhaps the most unique property of hr is that its expression is TH-dependent in developing brain (Thompson 1996; Thompson and Potter 2000). Hr may be part of a novel autoregulatory pathway in which the Hr protein subsequently influences its own expression by mediating transcriptional repression by TR. Additionally, because TH inhibits Hr–TR binding, Hr may modify TH-regulated gene expression preferentially in tissues in which Hr expression is not TH-dependent, such as skin or adult brain (Thompson 1996; C.C. Thompson, unpubl.). In either scenario, Hr likely affects the expression of other, downstream TH-responsive genes, playing an integral role in mediating the effects of TH on gene expression.
Detailed analysis of Hr interaction with TR and HDACs has revealed both similarities and differences in Hr function compared with previously characterized corepressors. The consensus generated for Hr–TR binding is similar in hydrophobicity but differs in sequence identity from that identified for N-CoR and SMRT binding to TR and RAR (Hu and Lazar 1999; Nagy et al. 1999; Perissi et al. 1999; Webb et al. 2000). In fact, the Hr-interaction domains share little sequence identity with each other or with those defined for N-CoR and SMRT (3/19 identities). This lack of sequence similarity is in contrast to the interaction domains of N-CoR and SMRT, which are highly related (11/19 identities in the carboxy-terminal interaction domain and 14/17 identities in the amino-terminal interaction domain). Thus, our results for Hr define a minimal motif in which the spacing of hydrophobic residues is well conserved, supporting the proposal that the secondary structure of this region (amphipathic alpha helix) is important for specifying receptor interaction (Nagy et al. 1999; Perissi et al. 1999). In addition, the spacing of interaction motifs is conserved between Hr and N-CoR/SMRT, as, in all cases, the interaction motifs are separated by ∼200 amino acids. This spacing may be critical as it has been proposed that the N-CoR interaction domains act cooperatively (Webb et al. 2000).
Although the predicted secondary structure of the Hr interaction domains resembles that of other corepressors, the identity of the hydrophobic residues in the core consensus is likely significant and may reflect binding specificity. Hr binds to TR but unlike N-CoR and SMRT, Hr does not bind to RAR (Thompson and Bottcher 1997; Fig. 4). Together with the analysis of other putative TR-selective corepressors such as Alien (Dressel et al. 1999), the Hr-binding motif will help define the mechanism of corepressor–receptor binding specificity.
Transcriptional repression has been correlated with associated HDAC activity for nuclear receptors as well as for other transcription factors such as Mad (Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997; Laherty et al. 1997; Nagy et al. 1997; Glass and Rosenfeld 2000). We find that Hr interacts with both class I and class II HDACs, indicating that the mechanism by which Hr mediates transcriptional repression is likely by recruiting HDAC activity. Unlike N-CoR and SMRT, which have been shown to associate either directly or indirectly with all known HDACs except HDAC6, Hr interacts preferentially with HDAC1, HDAC3, and HDAC5 (Alland et al. 1997; Heinzel et al. 1997; Nagy et al. 1997; Guenther et al. 2000; Huang et al. 2000; Kao et al. 2000; Wen et al. 2000). This interaction was not detected in the yeast two-hybrid or Far Western assays, suggesting the interaction is indirect and is likely part of a larger protein complex. This result is not surprising, as HDACs and corepressors are components of large, multiprotein complexes (Downes et al. 2000; Guenther et al. 2000; Li et al. 2000; Wen et al. 2000).
Within the nucleus, we find that Hr is localized to subnuclear structures termed MAD bodies. MAD bodies were recently identified as subnuclear structures associated with the nuclear matrix that contain SMRT and HDAC5 and HDAC7 (Downes et al. 2000). The dependence of MAD body integrity on HDAC activity suggests that it is a functionally significant unit. Coimmunoprecipitation of SMRT and HDAC5 with a number of other proteins suggested that these other proteins are components of MAD bodies as well (Downes et al. 2000). The presence of Hr in MAD bodies indicates that Hr is likely part of a large, multiprotein complex involved in regulating transcriptional repression and predicts that Hr interacts with other proteins in addition to TR and HDACs. Evidence that N-CoR and SMRT reside in large protein complexes (apparent molecular mass of 1–2 MD; Downes et al. 2000; Li et al. 2000; Jones et al. 2001) raises the question of whether corepressors and other proteins form a single multiprotein complex that includes multiple corepressors and HDACs, or whether N-CoR, SMRT, and Hr subserve the same function in a collection of independent complexes.
Hr function in vivo
Hr function as a corepressor provides evidence for the biochemical role of Hr. What then, is the biological role of Hr? The biological function of Hr can be inferred from the phenotype of humans and mice with mutations in the hr gene. Mutation of the human hr gene results in congenital hair loss disorders (papular atrichia and alopecia universalis) and, in some cases, associated neurological deficits (del Castillo et al. 1974; Ahmad et al. 1998, 1999; Cichon et al. 1998; Kruse et al. 1999; Sprecher et al. 1999; Aita et al. 2000). Thus, these disorders are an example of human disease resulting from aberrant corepressor function. The phenotypes of hr mutants likely result from disruption of Hr corepressor function, suggesting that the molecular basis of these disorders is perturbation of gene expression by TR and possibly other transcription factors.
Given its role in TH-regulated gene expression, we expect that Hr also has a role in regulating TH-dependent aspects of growth and development. If this is true, then the phenotype of hr mutants should resemble some of the phenotypes associated with TH deficiency. Consistent with this idea, many symptoms of TH deficiency in the skin are similar to those of hr mutants. For example, up to 90% of patients with TH deficiency show thickening of the skin and 30%–40% suffer hair loss (Bernhard et al. 1996). In the brain, morphological changes in hr mutant mice resemble those seen in TH-deficient animals. Changes in Purkinje cell morphology have been reported, which correlate with the effects of TH deficiency on Purkinje cells (Legrand 1979; Garcia-Atares et al. 1998). The hr (hrrh allele) mutant mice are also deaf and show cochlear defects similar to congenital hypothyroid mutant (Tshrhyt) mice (O'Malley et al. 1995; Cachon-Gonzalez et al. 1999). Inner ear defects also correlate with expression of TRβ in the inner ear and deafness in TRβ null mice (Bradley et al. 1994; Forrest et al. 1996). The phenotypic parallels between hr mutation and TH deficiency establish a link between the biochemical and biological roles of Hr.
Materials and methods
Two-hybrid assay
Yeast two-hybrid assays were performed as described previously (Hollenberg et al. 1995; Thompson and Bottcher 1997). Deletion derivatives of hr were subcloned into pLexA for use as bait in the two-hybrid assay. The resulting plasmids were transformed into yeast strain L40 and used to test interaction with TR (pVP16-TRα2).
Construction of Hr deletion derivatives
pLexA-Hr568 (568–1207) has been described (Thompson and Bottcher 1997). pLexA-724–1207 was made by digestion of the rat hr cDNA with SmaI, ligation of BamHI linkers, and digestion with BamHI. The resulting fragment was subcloned into the BamHI site of pLexA. pLexA-568–1048 was made by digestion of pLexA-568–1207 with Asp718 and SalI, formation of blunt ends with Klenow, and re-ligation. pLexA-N568-C784 was made by digestion of pLexA-Hr568 with NheI and SalI, formation of blunt ends with Klenow, and re-ligation. pLexA-568–864 was made by digestion of pLexA-N568 with SacI and SalI, formation of blunt ends with T4 DNA polymerase, and religation. pLexA-864–1207 was made by digestion of the rat hr cDNA with SacI and formation of blunt ends with T4 DNA polymerase followed by ligation of BamHI linkers. The resulting fragment was cloned into the BamHI site of pLexA. To construct pLexA 750–1084, 864–981, 981–1084, and 1084–1207, PCR was used to generate the indicated fragment from the rat hr cDNA with specific oligonucleotides that introduced an in-frame BamHI site. PCR fragments were subcloned into pCR2.1 (Invitrogen). Fragments were excised from pCR2.1 with BamHI and the isolated fragments subcloned into the BamHI site of pLexA. pLexA 750–864 was made by digestion of pLexA 750–1085 with SacI and SalI, formation of blunt ends with T4 DNA polymerase and religation.
Construction of TR deletion derivatives
pVP16-TRα2 and TRα1–353 were isolated in the original two-hybrid screen with pLexA-Hr (Thompson and Bottcher 1997). pVP16-TRα1 was made by deletion of an NcoI–BamHI fragment from pVP16-TRα2 and replacement with the corresponding NcoI–BamHI fragment of pGEM4-rTRα1. pVP16-TRβ199–456 was made by subcloning of the EcoRI–BamHI fragment from pCMX-ΔΔTRβ1 (kindly provided by K. Umesono) into the EcoRI–BamHI sites of pLexA. pVP16-TRα14–126 was made by digestion of pVP16-TRα2 with BamHI and XbaI, formation of blunt ends with Klenow, and religation. pVP16-TRα160 was made by replacement of a BamHI–XbaI fragment from pVP16-TRα2 with the corresponding BamHI–XbaI fragment from pCMX TR160 (Chen and Evans 1995). Presence of the point mutation at amino acid position 160 was confirmed by sequencing.
Far Western assay
The procedure for Far Western blotting was as described (Cavailles et al. 1994; Thompson and Bottcher 1997). The same fragments subcloned into pLexA for the two-hybrid assay were subcloned into pGEX3X (Pharmacia) or pATH21 (derivatives 568–1048, 568–784) to express these sequences as GST or trpE fusion proteins, respectively. pGEX 750–864 was made by digestion of pLexA 750–1085 with SacI, formation of blunt ends with T4 DNA polymerase followed by digestion with BamHI. The resulting fragment was ligated with pGEX3X digested with BamHI and SmaI. To construct pGEX 980–1027, 980–1040, 1024–1084, and 1041–1084, PCR was used to generate the indicated fragment from the rat hr cDNA with specific oligonucleotides that introduced an in-frame BamHI site. PCR fragments were subcloned into pCR2.1. Fragments were excised from pCR2.1 with BamHI and the isolated fragments subcloned into the BamHI site of pGEX3X. pATH568–1048 and 568–784 were made by insertion of restriction fragments from the rat hr DNA (HindIII–PstI and HindIII–NheI respectively) into pATH21.
All proteins were produced in bacterial strain DH10 and whole-cell lysates from IPTG- or indoleacrylic acid-induced samples were used for Far Western analysis. Extracts from bacteria expressing fusion proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose. 35S-labeled proteins generated by in vitro transcription/translation (Promega) were used as probes. Equivalent expression of proteins used as probes was verified by SDS–polyacrylamide gel electrophoresis followed by fluorography. pTZ18 (rTRβ1) was kindly provided by H. Towle (University of Minnesota).
Site-directed mutagenesis
Point mutations were introduced by use of specific oligonucleotides for site-directed mutagenesis (Quick-Change Kit, Stratagene). For the amino-terminal interaction domain (TR-ID1), the rat hr cDNA (2.2-kb HindIII fragment) in pBluescript was used as the template for mutagenesis. Mutagenized fragments were subcloned into pLexA and pGEX3X for use in two-hybrid and Far Western assays, respectively. For point mutations in the context of Hr amino acids 568–864, pLexA derivatives were digested with SacI and SalI, blunt ends were generated with Klenow, and the products were ligated. pGEX derivatives were constructed by digestion of the pLexA derivatives with SacI and creation of blunt ends with T4 DNA polymerase followed by digestion with BamHI. The resulting BamHI blunt fragments were subcloned into the SmaI–BamHI sites of pGEX3X. Mutations in the carboxy-terminal interaction domain (TR-ID2) were made by use of specific oligonucleotides with pGEX 980–1085 as template. For all constructs, mutagenesis was confirmed by sequence analysis.
Cell culture/transfection
Cell lines were obtained from ATCC. COS-1 and CV-1 cells were maintained in DMEM supplemented with 10% fetal calf serum. GH1 cells were maintained in MEM supplemented with 10% fetal calf serum. GC cells were maintained in DMEM supplemented with 10% horse serum and 5% fetal calf serum. CV-1 cells were transfected (per well of a 6-well plate) with 167 ng of reporter plasmid, 50 ng of expression plasmid, and 80 ng of CMX-βgal by use of Lipofectamine (Life Technologies). GH1 and GC cells were transfected (per well of a 12-well plate) with 150 ng of reporter plasmid, 90 ng of expression plasmid, and 75 ng of CMX-βgal. Cells were harvested in reporter or passive lysis buffer (Promega) and assayed for β-galactosidase and luciferase activity. Luciferase activity was divided by β-galactosidase activity to normalize for transfection efficiency. All experiments were done in duplicate and repeated at least three times with similar results. For hormone-induction experiments, serum was depleted of thyroid and steroid hormones by treatment with AG-1-X8 resin (Bio-Rad) and charcoal (Sigma) as described (Samuels et al. 1979). Cells were grown for 1 d in hormone depleted media before transfection. After transfection, thyroid hormone (L-T3) or all-trans retinoic acid was added to 10–7 or 10–6 M, respectively. CMX-rTRα, CMXhRARα, TREpal tk-luc, and MLV tk-luc have been described (Umesono et al. 1988, 1991). Construction of pRK5myc-rhr is described below; pRK5myc-rhr 1–568 (ΔHr) was made by isolation of a 1.6-kb HindIII fragment from pRK5myc-rhr and insertion of the fragment in the HindIII site of pRK5myc.
Construction of GAL4 DBD expression plasmids
pCMX-GAL4, GALpx3 tkluc, and pCMX-GAL-Hr568 have been described (Thompson and Bottcher 1997). pCMX GAL-Hr was made by isolation of the AvrII fragment spanning nucleotide positions 593–4239 of the rat hr cDNA, formation of blunt ends with Klenow, and insertion into the EcoRV site of pCMX-GAL4. pCMX GAL568–1207, 724–1207, 750–1084, 864–981, 981–1084 were made by insertion of the BamHI fragments from the corresponding pLexA derivatives into the BamHI site of pCMX GAL4. pCMX GAL 31–568 was made by digestion of pLexA31–568 with EcoRI and PstI and subcloning of the resulting fragment into the corresponding sites of pCMX GAL4. To construct pCMX GAL 1–236, 150–450, and 450–730, PCR was used to generate the indicated fragment from the rat hr cDNA with specific oligonucleotides that introduced an in-frame BamHI site. PCR fragments were subcloned into pCR2.1. Fragments were excised from pCR2.1 with BamHI and the isolated fragments subcloned into the BamHI site of pCMX GAL4. pCMX GAL 236–450 was made by digestion of pLexA 150–450 with BamHI followed by incubation with Klenow to generate blunt ends. The 600-bp blunt-ended fragment was subcloned into the EcoRV site of pCMX GAL4. pCMX GAL450–568 was made by digestion of pLexA 450–730 with HindIII and generation of blunt ends with Klenow followed by digestion with EcoRI. The 350-bp EcoRI blunt fragment was then subcloned into pCMX GAL digested with EcoRI and EcoRV. pCMX GAL568–724 was made by isolation of the 470-bp SmaI fragment from pLexA568–1207 and insertion into the SmaI site of pCMX GAL4. All constructs were sequenced to verify in-frame fusion.
Generation of Hr-specific antiserum
To generate Hr-specific antiserum, amino acids 450–730 of rat Hr were expressed as a GST fusion protein (described above). Protein was purified by SDS-PAGE, and acrylamide containing the fusion protein was used to immunize rabbits (Covance). Antiserum was affinity purified by use of a bacterially expressed fusion protein of Hr amino acids 450–730 with trpE.
Immunoprecipitation
Extracts for immunoprecipitation were prepared from transiently transfected COS cells. For immunoprecipitation experiments with TR and full-length Hr, COS cells were transfected by electroporation with 2.5 μg of each DNA per 10-cm plate. Cells were metabolically labeled by growth in methionine-free medium supplemented with 150 μCi/mL [35S]methionine for 4 h. Extracts were prepared by solubilizing cells in RIPA buffer (50 mM Tris at pH 7.5, 0.15 M NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS). Extracts were incubated with primary antisera overnight at 4°C, then incubated with protein A–Sepharose for 3 h. After being washed three times with RIPA buffer, pellets were boiled in 1× sample buffer and run on SDS–polyacrylamide gels. Gels were fixed, treated with AMPLIFY (Amersham), dried, and exposed to X-ray film. For immunoprecipitation experiments with Hr derivatives and HDACs, after transfection (48 h), cells were harvested in PBS, then homogenized in ice-cold IP buffer (50mM Tris at pH 7.5, 150 mM NaCl, 1% NP-40, 10% glycerol) containing protease inhibitors (Complete Mini, Roche). Extracts were incubated for 2 h at 4°C and centrifuged at 15,000g for 10 min. The soluble protein supernatant was incubated overnight at 4°C with primary antibody and then incubated with Protein A–Sepharose beads (Sigma) for 3 h at 4°C. Beads were washed 3 times with ice-cold IP buffer. Immunoprecipitates were fractionated by SDS-PAGE and analyzed by Western blotting. Western analysis was performed with the indicated antisera and detected by use of enhanced chemiluminescence (ECL) under conditions specified by the manufacturer (Amersham). FLAG-, HDAC1- and HDAC2-specific and control antisera were obtained from Sigma. TR-specific antiserum was obtained from Santa Cruz Biotechnology, Inc and Affinity Bioreagents. Anti-myc antiserum (9E10) was kindly provided by N. Marsh-Armstrong (Kennedy Krieger Research Institute).
pRK5myc-rhr was made by insertion of the XbaI–SpeI fragment of the rat hr cDNA into the XbaI site of pRK5-myc (kindly provided by B. Prasad and R. Reed, Johns Hopkins University School of Medicine). Then, the EcoRV–XbaI vector fragment of this plasmid was ligated to the SmaI–XbaI fragment of pLexA-Hr (1–224; C.C. Thompson, unpubl.). pRK5myc-rhr 750–1085 was made by insertion of a PstI–SmaI fragment from pLexA 750–1085 into the PstI and EcoRV sites of pRK5myc. pRK5myc-rhr 750–864 was made by digestion of pRK5myc-rhr 750–1085 with SacI, treatment with T4 DNA polymerase to create blunt ends, and digestion with HindIII. The resulting fragment was inserted into the HindIII and SmaI sites of pRK5myc. pRK5myc-rhr 864–981 and 980–1084 were made by digestion of the corresponding pCMX GAL derivatives with SmaI and NheI and ligation of the isolated fragments into the SmaI and XbaI sites of pRK5myc. Other expression plasmids were kindly provided as follows: FLAG-tagged HDAC3 by W.-M. Yang and E. Seto (Moffitt Cancer Center and Research Institute); FLAG-tagged HDAC4–HDAC6 by C. Grozinger and S. Schreiber (Harvard University); SMRT by R. Evans (The Salk Institute).
Immunofluorescence
COS-1 cells transfected with the indicated expression vectors were plated onto glass coverslips. After 2 d, cells were fixed with 4% paraformaldehyde for 20 min at room temperature, then incubated in PBS containing 10% goat serum and 0.1% Triton X-100 for 1 h. Cells were incubated with primary antisera for 1 h at room temperature. After washing with PBS, incubation with secondary antisera was for 1 h at room temperature. Cells were washed twice with PBS and once with PBS containing 0.1 μg/mL 4‘,6-diaminido-2-phenylindole (DAPI). Coverslips were dried and mounted with Vectashield (Vector Laboratories). Digital images were captured by use of a Zeiss Axiocam with a Zeiss Axioplan microscope and analyzed by use of Adobe Photoshop software. Primary antisera and concentrations used were as follows: affinity-purified Hr-specific antibody, 1:200; mouse monoclonal anti-myc (9E10), 1:2; mouse monoclonal anti-FLAG (Sigma), 1:400; mouse anti-SC-35 (Sigma), 1:200; rabbit anti-SMRT (Affinity Bioreagents), 1:300. Secondary antisera and concentrations used were as follows: anti-rabbit or anti-mouse indocarbocyanine (Cy3; Jackson ImmunoResearch Laboratory), 1:4000; anti-rabbit or anti-mouse FITC (Jackson ImmunoResearch Laboratory), 1:400.
Acknowledgments
We thank J. Sisk for technical assistance, A. Lanahan, G. Seydoux, K. Smith, and D. Ginty for comments on the manuscript, and our colleagues who generously provided the indicated plasmids. This work was supported by the Klingenstein Fund, National Alliance for Research on Schizophrenia and Depression and the National Institutes of Health (DK46074). This paper is dedicated to the memory of friend and colleague, Dr. Kazuhiko Umesono.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL thompsonc@kennedykrieger.org; FAX (410) 502-8093.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.916701.
References
- Ahmad W, Faiyaz ul Haque M, Brancolini V, Tsou HC, ul Haque S, Lam H, Aita VM, Owen J, deBlaquiere M, Frank J, et al. Alopecia universalis associated with a mutation in the human hairless gene. Science. 1998;279:720–724. doi: 10.1126/science.279.5351.720. [DOI] [PubMed] [Google Scholar]
- Ahmad W, Panteleyev AA, Christiano AM. The molecular basis of congenital atrichia in humans and mice: Mutations in the hairless gene. J Investig Dermatol Symp Proc. 1999;4:240–243. doi: 10.1038/sj.jidsp.5640220. [DOI] [PubMed] [Google Scholar]
- Aita VM, Ahmad W, Panteleyev AA, Kozlowska U, Kozlowska A, Gilliam TC, Jablonska S, Christiano AM. A novel missense mutation (C622G) in the zinc-finger domain of the human hairless gene associated with congenital atrichia with papular lesions. Exp Dermatol. 2000;9:157–162. doi: 10.1034/j.1600-0625.2000.009002157.x. [DOI] [PubMed] [Google Scholar]
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Barettino D, Vivanco Ruiz MM, Stunnenberg HG. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard JD, Freedberg IM, Vogel LN. The skin in hypothyroidism. In: Braverman LE, Utiger RD, editors. Werner and Ingbar’s the thyroid: A fundamental and clinical text. Philadelphia, PA: Lippincott-Raven; 1996. pp. 792–795. [Google Scholar]
- Bradley DJ, Towle HC, Young WS. Alpha and beta thyroid hormone receptor (TR) gene expression during auditory neurogenesis: Evidence for TR isoform-specific transcriptional regulation in vivo. Proc Natl Acad Sci. 1994;91:439–443. doi: 10.1073/pnas.91.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke HC. Hairless mice. J Heredit. 1926;17:173–174. [Google Scholar]
- Burke LJ, Baniahmad A. Co-repressors 2000. FASEB J. 2000;14:1876–1888. doi: 10.1096/fj.99-0943rev. [DOI] [PubMed] [Google Scholar]
- Cachon-Gonzalez MB, Fenner S, Coffin JM, Moran C, Best S, Stoye JP. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci. 1994;91:7717–7721. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon-Gonzalez MB, San-Jose I, Cano A, Vega JA, Garcia N, Freeman T, Schimmang T, Stoye JP. The hairless gene of the mouse: Relationship of phenotypic effects with expression profile and genotype. Dev Dyn. 1999;216:113–126. doi: 10.1002/(SICI)1097-0177(199910)216:2<113::AID-DVDY3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, Danielian PS, Parker MG. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Chen JD, Umesono K, Evans RM. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Anker M, Vogt IR, Rohleder H, Putzstuck M, Hillmer A, Farooq SA, Al-Dhafri KS, Ahmad M, Haque S, et al. Cloning, genomic organization, alternative transcripts and mutational analysis of the gene responsible for autosomal recessive universal congenital alopecia. Hum Mol Genet. 1998;7:1671–1679. doi: 10.1093/hmg/7.11.1671. [DOI] [PubMed] [Google Scholar]
- del Castillo V, Ruiz-Maldonado R, Carnevale A. Atrichia with papular lesions and mental retardation in two sisters. Int J Dermatol. 1974;13:261–265. doi: 10.1111/j.1365-4362.1974.tb05078.x. [DOI] [PubMed] [Google Scholar]
- Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. Identification of a nuclear domain with deacetylase activity. Proc Natl Acad Sci. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressel U, Thormeyer D, Altincicek B, Paululat A, Eggert M, Schneider S, Tenbaum SP, Renkawitz R, Baniahmad A. Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:3383–3394. doi: 10.1128/mcb.19.5.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: Presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest D, Erway LC, Ng L, Altschuler R, Curran T. Thyroid hormone receptor beta is essential for development of auditory function. Nat Genet. 1996;13:354–357. doi: 10.1038/ng0796-354. [DOI] [PubMed] [Google Scholar]
- Garcia-Atares N, San Jose I, Cabo R, Vega JA, Represa J. Changes in the cerebellar cortex of hairless Rhino-J mice (hr-rh-j) Neurosci Lett. 1998;256:13–16. doi: 10.1016/s0304-3940(98)00757-5. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes & Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes & Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Sachs LM, Rouse N, Wade PA, Shi YB. Multiple N-CoR complexes contain distinct histone deacetylases. J Biol Chem. 2001;276:8807–8811. doi: 10.1074/jbc.C000879200. [DOI] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Kruse R, Cichon S, Anker M, Hillmer AM, Barros-Nunez P, Cantu JM, Leal E, Weinlich G, Schmuth M, Fritsch P, et al. Novel Hairless mutations in two kindreds with autosomal recessive papular atrichia. J Invest Dermatol. 1999;113:954–959. doi: 10.1046/j.1523-1747.1999.00790.x. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR- mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- Legrand J. Morphogenetic actions of thyroid hormones. Trends Neurosci. 1979;2:234–236. [Google Scholar]
- Li H, Leo C, Schroen DJ, Chen JD. Characterization of receptor interaction and transcriptional repression by the corepressor SMRT. Mol Endocrinol. 1997;11:2025–2037. doi: 10.1210/mend.11.13.0028. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF, Searle AG. Genetic variants and strains of the laboratory mouse. Oxford, UK: Oxford University Press; 1989. [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Misiti S, Schomburg L, Yen PM, Chin WW. Expression and hormonal regulation of coactivator and corepressor genes. Endocrinology. 1998;139:2493–2500. doi: 10.1210/endo.139.5.5971. [DOI] [PubMed] [Google Scholar]
- Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: Structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Love JD, Li C, Banayo E, Gooch JT, Krishna V, Chatterjee K, Evans RM, Schwabe JW. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes & Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley BW, Jr, Li D, Turner DS. Hearing loss and cochlear abnormalities in the congenital hypothyroid (hyt/hyt) mouse. Hear Res. 1995;88:181–189. doi: 10.1016/0378-5955(95)00111-g. [DOI] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteleyev AA, Paus R, Ahmad W, Sundberg JP, Christiano AM. Molecular and functional aspects of the hairless (hr) gene in laboratory rodents and humans. Exp Dermatol. 1998;7:249–267. doi: 10.1111/j.1600-0625.1998.tb00295.x-i1. [DOI] [PubMed] [Google Scholar]
- Panteleyev AA, Botchkareva NV, Sundberg JP, Christiano AM, Paus R. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am J Pathol. 1999;155:159–171. doi: 10.1016/S0002-9440(10)65110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor–corepressor interaction. Genes & Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels HH, Stanley F, Casanova J. Depletion of L-3,5,3′-triiodothyronine and L-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology. 1979;105:80–85. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- Sande S, Privalsky ML. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- Sprecher E, Bergman R, Szargel R, Friedman-Birnbaum R, Cohen N. Identification of a genetic defect in the hairless gene in atrichia with papular lesions: Evidence for phenotypic heterogeneity among inherited atrichias. Am J Hum Genet. 1999;64:1323–1329. doi: 10.1086/302368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP, Fenner S, Greenoak GE, Moran C, Coffin JM. Role of endogenous retroviruses as mutagens: The hairless mutation of mice. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Thompson CC. Thyroid hormone-responsive genes in developing cerebellum include a novel synaptotagmin and a hairless homolog. J Neurosci. 1996;16:7832–7840. doi: 10.1523/JNEUROSCI.16-24-07832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Bottcher MC. The product of a thyroid hormone-responsive gene interacts with thyroid hormone receptors. Proc Natl Acad Sci. 1997;94:8527–8532. doi: 10.1073/pnas.94.16.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Potter GB. Thyroid hormone action in neural development. Cereb Cortex. 2000;10:939–945. doi: 10.1093/cercor/10.10.939. [DOI] [PubMed] [Google Scholar]
- Umesono K, Giguere V, Glass CK, Rosenfeld MG, Evans RM. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs) Mol Endocrinol. 2000;14:1976–1985. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Yaffe BM, Samuels HH. Hormonal regulation of the growth hormone gene. Relationship of the rate of transcription to the level of nuclear thyroid hormone-receptor complexes. J Biol Chem. 1984;259:6284–6291. [PubMed] [Google Scholar]