Abstract
EmrD is a multidrug transporter from the Major Facilitator Superfamily that expels amphipathic compounds across the inner membrane of Escherichia coli. Here, we report the x-ray structure of EmrD determined to a resolution of 3.5 angstrom. The structure reveals an interior that is composed mostly of hydrophobic residues, which is consistent with its role transporting amphipathic molecules. Two long loops extend into the inner leaflet side of the cell membrane. This region can serve to recognize and bind substrate directly from the lipid bilayer. We propose that multi-substrate specificity, binding, and transport are facilitated by these loop regions and the internal cavity.
While the advent of medicinal antibiotics heralded an unprecedented breakthrough in the treatment of infectious disease, the emergence of drug resistant bacteria is threatening to undermine this achievement. Multidrug resistance (MDR) develops partially through direct drug efflux by integral membrane transporters. There are two classes of MDR transporters: ATP-binding cassette (ABC) proteins that directly couple drug efflux to adenosine 5′triphosphate (ATP) hydrolysis, and secondary transporters that use energy derived from electrochemical gradients across the cell membrane. The secondary transporters comprise four families: the Resistance/Nodulation/Division superfamily (RND), Multiple Antimicrobial Toxin Extrusion family (MATE), Small Multidrug Resistance family (SMR), and the Major Facilitator Superfamily (MFS). The MDR transporters from the MFS family (MDR MFS) are among the most prevalent in microbial genomes and diverse in their substrate specificities (1).
One MDR MFS transporter, EmrD, is a proton-dependent secondary transporter from Escherichia coli. EmrD was first identified as an efflux pump for uncouplers of oxidative phosphorylation (2), which can rapidly arrest growth in bacteria by depleting the H+ gradient (3). Some of these uncouplers are structurally unrelated such as meta-chloro carbonylcyanide phenylhydrazone (CCCP) and tetrachlorosalicylanilide (TSA). It was later discovered that EmrD could also transport detergents such as benzalkonium and sodium dodecylsulfate (4). Sequence alignment suggests that EmrD is a close homolog to other MDR MFS transporters (5), including NorA from S. aureas (with 19% identity and 41% similarity), LmrP (22% and 40%) from L. lactis, FlorR from S. enterica (24% and 45%), Bmr from B. subtilis (20% and 40%) and the E. coli transporters MdfA (26% and 39%), and Bcr (24% and 44%) (Fig. S1). EmrD E. coli has 394 amino acids, and a molecular weight of ∼42.2 kD. Hydropathy analysis indicates that EmrD has 12 transmembrane α-helices, and phylogenetic studies have suggested that it is a H+:drug antiporter from the DHA12 drug efflux subfamily within the MFS (6).
The general model for substrate efflux by secondary transporters involves an alternating access mechanism, and most non-MDR MFS transport systems such as the lactose (LacY) and glycerol-3-phosphate (GlpT) permeases typically transport a relatively narrow range of structurally related substrates (7, 8). MDR MFS transporters, such as EmrD, differ significantly in that they are able to export a broad spectrum of hydrophobic compounds (9). How do they recognize this wide range of structurally distinct substrates and what are the conformational rearrangements within the MFS necessary for hydrophobic drug efflux? To elucidate the molecular basis of MDR MFS transport, we have determined the x-ray structure of EmrD to 3.5 Å resolution by anomalous dispersion methods.
Crystals of EmrD were grown in the presence of β-dodecyl-maltoside. X-ray diffraction data was collected from a native crystal and a gold thiomalate derivative (Table S1). After density modification and phase extension, the electron density map clearly showed two identical molecules in the asymmetric unit with densities corresponding to side chains (Fig. 1A). The crystal lattice contacts between the two copies of EmrD are small (< 250 Å2) and we believe that the packing arrangement of the dimer is non- physiological. We designate the transmembrane helices in each monomer as H1 to H12 and the connecting loops L1-2 to L11-12.
Fig. 1.
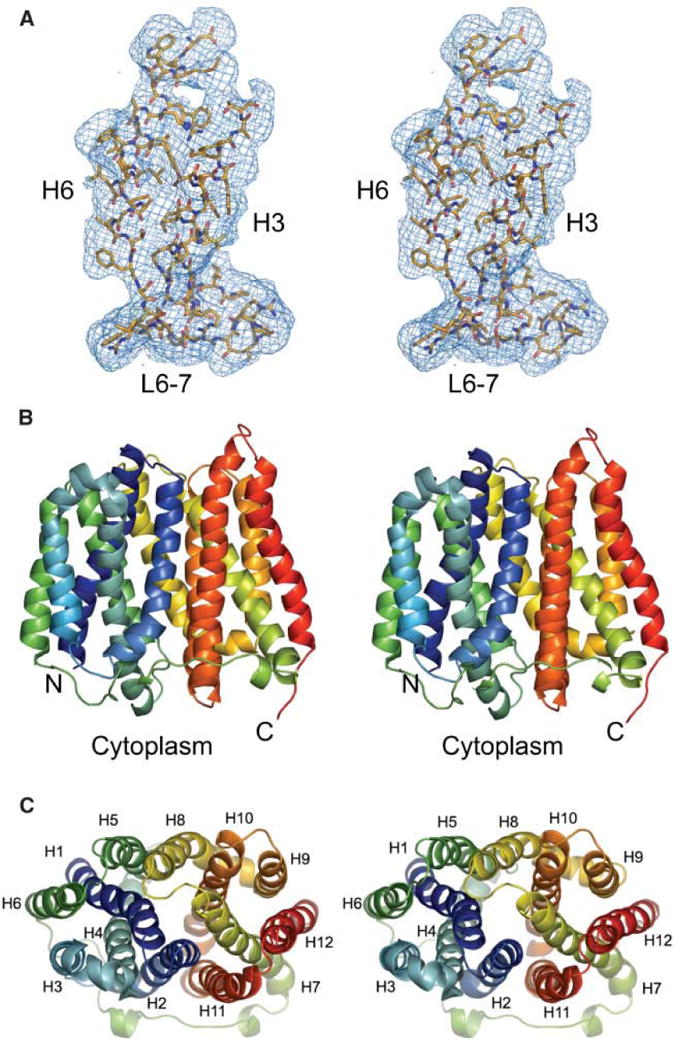
Crystallography and structure of EmrD. (A) A portion of the experimental electron density map is shown for H3, H6, and L6-7. The map is contoured to 1 σ. (B) Side view of EmrD. The N- and C-terminus are indicated. (C) View of EmrD looking towards the cytoplasm showing the pseudo two-fold axis relating the N- and C-terminal halves. Transmembrane helices are indicated. The images were created by PyMol.
The overall structural topology of EmrD is reminiscent of LacY and GlpT. Twelve transmembrane helices form a compact structure that spans ∼50 Å in the plane of the lipid bilayer, and ∼45 Å along the membrane normal (Fig. 1B and 1C). The transmembrane helices facing away from the interior (H3, H6, H9, and H12) demonstrate a similar organization to LacY and GlpT. The remaining transmembrane helices form the internal cavity and their relative orientations, however, show significant deviation from those observed in LacY and GlpT. This structural arrangement may reflect a general architecture of MDR MFS transporters.
Unlike the LacY and GlpT structures, which are both in the cytoplasmic facing configuration, this EmrD structure is not in a V-shape and probably represents an intermediate state. The periplasmic loops in EmrD are more imbedded in the cell membrane and the central loop L6-7 is also considerably shorter. A pseudo two-fold relates the N- and C-terminal halves of EmrD (H1-H6 and H7-H12; average RMS deviation of 0.78 Å for 116 Cα positions) and supports the notion that the MFS arose from recurrent gene duplication of an ancestral six-helix domain (9, 10). The two halves of EmrD, however, are less symmetric than those of LacY or GlpT and the most significant internal asymmetry is localized to the loop regions on the periplasmic side. For example, L3-4 (residues 92 to 99) is actually a bent helix while L9-10 (residues 285 to 289) is a short loop. Compared to LacY, H6 in EmrD is shorter while H11 is significantly longer.
The most striking structural difference between EmrD and LacY/GlpT is in the internal cavity. Whereas LacY and GlpT have hydrophilic interiors, the internal cavity of EmrD comprises mostly hydrophobic residues, consistent with its function of transporting lipophilic compounds. Several of these residues are bulky and aromatic (Ile28, Ile217, Ile253, Tyr52, Tyr56, Trp300, and Phe249), and some are conserved in other MDR MFS transporters (Fig. S1 and 2 A, B). This type of hydrophobic core has been previously proposed and also observed in the recent x-ray structure of EmrE with substrate (11, 12). These residues likely contribute to a general mechanism of substrate translocation and may play a significant role in dictating a level of drug specificity either through steric or aromatic interactions. The internal cavity also has several uncharged polar residues such as glutamines (Gln21, Gln24, Gln46, and Gln60), and a basic arginine residue (Arg118) that is located closer to the cytoplasmic side. On the periplasmic side lie a theronine (Thr25), an aspartate (Asp33), and a glutamate residue (Glu227) that could easily reorient into the cavity during the transport cycle and may participate in H+ translocation.
The hydrophobic interior of EmrD provides a generalized pathway and mechanism for transporting a variety of different substrates in drug efflux systems (11). EmrD possesses two pairs of stacked aromatic groups (Tyr52/Tyr56 and Trp300/Phe 249) that could play a key role in multi-substrate binding, given their ability to stack with aromatic drugs (Fig. 2B). In Bmr, two phenylalanines have been implicated in substrate recognition (13), while the multidrug binding site of the transcriptional repressor QacR uses several aromatic and polar residues (14, 15). The energetic cost of transporting charged amphipathic compounds may be compensated by these types of hydrophobic interactions (11).
Fig. 2.

(A) Stereo cut-away view of the hydrophobic internal cavity of EmrD. For clarity, residues 43 to 67 were omitted. Hydrophobicity is shown as a gradation from low (wheat color) to high (brown). Regions that are positive and negative are shown in blue and red, respectively. (B) Inside view of the internal cavity of EmrD characterized by the lining of hydrophobic residues. The N- and C- terminal halves of EmrD and the corresponding residues are colored blue (H1-H6) and orange (H7-H12), respectively. (C) Close view of the selectivity filter region of EmrD. The position of residues that are involved in substrate recognition based on protein sequence homology to other MDR MFS transporters are highlighted (Fig. S1). Residues colored in blue correspond to those in MdfA that when mutated into cysteines either reduce or abolish resistance. Residues in yellow correspond to positions in LmrP that are important for substrate recognition. Val17, shown in red, corresponds to Glu26 in MdfA and Asp23 in FlorR, which are both important for drug recognition. Residues shown in green correspond to cysteine mutations in MdfA that are protected from NEM labeling by substrate. The relative position of the cytoplasm and the internal cavity are indicated.
The structure of EmrD reveals another region that could provide additional substrate specificity. There are two long helical regions (H4, L4-5, H5 and H10, L10-11, and H11) located on the cytoplasmic side that are arranged much closer to the central cavity and extend further into the cytoplasm compared to LacY and GlpT (Fig. 2C). The cytoplasmic end of H4 also has a number of charged residues (Arg118, Arg122, Asp123, Glu126, Arg127, and Arg131), which may play a role both in defining the topology of the transporter as well as substrate recognition (9). Here we will refer to this region as the selectivity filter. Functional studies of the EmrD homolog MdfA have implicated that several residues in this region are important for substrate recognition. For example, residues in the cytoplasmic halves of H4, H5, and H6 in MdfA are protected by substrate against alkylation by N-ethylmaleimide (NEM) ((16); Fig. 2C, S1). In addition, certain single site mutations in the cytoplasmic halves of H10 and H11 as well as L10-11 of MdfA either abolish or significantly reduce MDR ((17, 18); Fig. 2C, S1). Biochemical studies on LmrP, another close homolog of EmrD, also suggest that significant conformational changes occur in this region. In LmrP, single site mutations at positions corresponding to 133 and 313 in EmrD show that negative charges in this region are not critical for transport function but are important for drug recognition (Fig. 2C). Further studies on LmrP using fluorescein maleimide labeling upon substrate binding suggest movement of this region from a nonpolar to a polar environment (18).
Helix 1 in EmrD might also be important for drug recognition. Studies in MdfA and FlorR have implicated a key residue (Glu26 in MdfA and Asp23 in FlorR) corresponding to Val17 in EmrD that is important for recognizing substrate (19, 20). If this residue is changed to a valine in MdfA, the transporter looses its ability to recognize cationic compounds but still retains wild-type resistance to the neutral antibiotic chloramphenicol (16). Interestingly, some other MDR MFS transporters having small hydrophobic residues at this position are also known to transport neutral hydrophobic compounds. In this EmrD structure, Val17 points towards the internal cavity but is also partially accessible from the inner membrane leaflet side (Fig. 2B, C).
Based on the structure and homology to other MDR MFS transporters, we propose that EmrD intercepts CCCP on the inner membrane leaflet as it crosses toward the cytoplasm (Fig. 3). In the absence of drug efflux, CCCP diffuses across the inner membrane from the periplasmic space in the protonated form, disrupting the pH differential as it moves into the cytoplasm (3). The molecule then quickly releases its proton to become a lipophilic soluble anion that rapidly diffuses back to the periplasm (21). Binding of CCCP on the inner leaflet side is likely facilitated by the selectivity filter and hydrophobic interactions within the internal cavity of EmrD. Structural rearrangement favoring the outward facing conformation would be coupled to H+ antiport by a rocker-switch mechanism similar to those previously proposed, but this remains to be proven (7, 8). Based on the structure of EmrD, we speculate that proton translocation and drug transport may occur at different locations, which has also been proposed for MdfA (11).
Fig. 3.
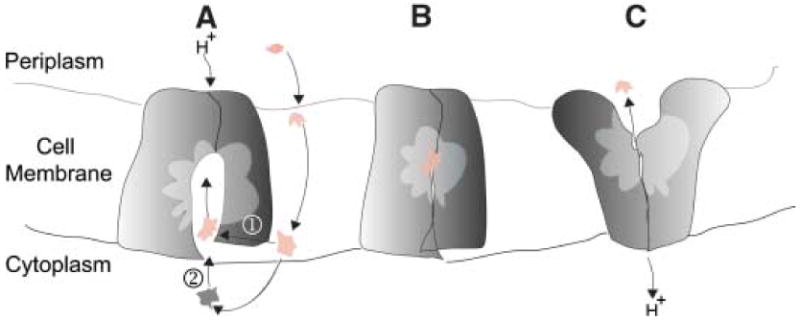
A potential mechanism for hydrophobic substrate transport by EmrD. (A) Drug can enter the internal cavity of the transporter either through the inner membrane leaflet (path ➀) or through the cytoplasm (path ➁). Substrate recognition and binding may be facilitated through the selectivity filter and the internal cavity containing hydrophobic residues. (B) Drug is transported through a rocker-switch alternating access model coupled with H+ antiport. (C) Drug is transported across the lipid bilayer.
What happens to CCCP when it enters the periplasmic space? There are at least two possibilities. Several MDR MFS systems have an adaptor protein that facilitates the transport of substrate through the periplasmic space; possibly using an apparatus similar to the TolC-adaptor-RND efflux systems (22-24). Perhaps the best known example is the EmrAB efflux system where EmrB operates as the MDR MFS transporter, and EmrA is an accessory protein (1, 25). In this case, the CCCP would be expelled out of the bacterial cell. However, no such adaptor protein or TolC-like apparatus has been identified associated with EmrD or any other 12-TMS MDR MFS transporter. If EmrD acts alone, like LmrP and Bmr in Gram positive bacteria, then CCCP would be expelled into the periplasmic space in E. coli.
The intracellular loop region of EmrD is reminiscent of the intracellular domain of MsbA, which is a bacterial homolog of MDR ABC transporters (26). In MsbA, these helices are thought to recognize head groups of the substrates as well as to transmit structural changes caused by ATP hydrolysis and substrate binding (27-29). Functional studies on the MDR ABC transporter LmrA suggest a model in which drug recognition by MDR transporters occurs in the inner leaflet of cell membrane bilayer (30). The lateral diffusion of hydrophobic substrate has also been proposed for the RND transporter AcrAB/TolC efflux system (22, 31) and access from the inner membrane leaflet is evident in the recent x-ray structures of both EmrE and MsbA with substrate (12, 28). Both of these structures also have hydrophobic pockets. In addition, mutational and biochemical studies on the cytoplasmic side of MdfA and LmrP suggest a model where drugs could diffuse laterally from the inner membrane leaflet (16-18, 32). This type of diffusion can be a common theme not only for the MDR MFS but also among all the MDR transporter families.
Supplementary Material
Acknowledgments
We thank S. Lieu for general lab support and the staff at the Stanford Synchrotron Radiation Laboratory (SSRL) and the Advanced Light Source (ALS) for assistance with data collection and crystal screening. We thank Drs. P. Wright, R. Milligan, M. Saier, C. Reyes, S. Aller, and A. Ward for carefully reading and valuable suggestions to the manuscript. This study was supported by grants from the NIH (GM70480 and GM65798), NASA (NAG8-1834), a NIH Presidential Early Career Award, and the Skaggs Institute for Chemical Biology. Coordinates have been deposited in the Protein Data Bank (PDB code: 2GFP).
Reference List
- 1.Putman M, van Veen HW, Konings WN. Microbiol Mol Biol Rev. 2000;64:672. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naroditskaya V, Schlosser MJ, Fang NY, Lewis K. Biochemical and Biophysical Research Communications. 1993;196:803. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 3.Krulwich TA, Quirk PG, Guffanti AA. Microbiological Reviews. 1990;54:52. doi: 10.1128/mr.54.1.52-65.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishino K, Yamaguchi A. J Bacteriol. 2001;183:5803. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulsen IT, Brown MH, Skurray RA. Microbiol Rev. 1996;60:575. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saier MH, Jr, Tran CV, Barabote RD. Nucl Acids Res. 2006;34:D181. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abramson J, et al. Science. 2003;301:610. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Science. 2003;301:616. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 9.Pao SS, Paulsen IT, Saier MH. Microbiol Rev. 1998;62:1. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saier MH. Molecular Microbiology. 2003;48:1145. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 11.Paulsen IT, Lewis K. Microbial Multidrug Efflux. Horizon Press; Norfolk: 2002. [Google Scholar]
- 12.Pornillos O, Chen YJ, Chen AP, Chang G. Science. 2005;310:1950. doi: 10.1126/science.1119776. [DOI] [PubMed] [Google Scholar]
- 13.Klyachko KA, Schuldiner S, Neyfakh AA. J Bacteriol. 1997;179:2189. doi: 10.1128/jb.179.7.2189-2193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray DS, Schumacher MA, Brennan RG. J Biol Chem. 2004;279:14365. doi: 10.1074/jbc.M313870200. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher MA, Miller MC, Brennan RG. Embo Journal. 2004;23:2923. doi: 10.1038/sj.emboj.7600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adler J, Bibi E. J Biol Chem. 2004;279:8957. doi: 10.1074/jbc.M313422200. [DOI] [PubMed] [Google Scholar]
- 17.Adler J, Bibi E. J Biol Chem. 2005;280:2721. doi: 10.1074/jbc.M412332200. [DOI] [PubMed] [Google Scholar]
- 18.Mazurkiewicz P, Konings WN, Poelarends GJ. J Biol Chem. 2002;277:26081. doi: 10.1074/jbc.M203141200. [DOI] [PubMed] [Google Scholar]
- 19.Edgar R, Bibi E. EMBO J. 1999;18:822. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braibant M, Chevalier J, Chaslus-Dancla E, Pages JM, Cloeckaert A. Antimicrob Agents Chemother. 2005;49:2965. doi: 10.1128/AAC.49.7.2965-2971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasianowicz J, Benz R, McLaughlin S. Journal of Membrane Biology (Historical Archive) 1984;82:179. doi: 10.1007/BF01868942. [DOI] [PubMed] [Google Scholar]
- 22.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Nature. 2002;419:587. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 23.Fralick JA. J Bacteriol. 1996;178:5803. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Nature. 2000;405:914. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 25.Nishino K, Latifi T, Groisman EA. Molecular Microbiology. 2006;59:126. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 26.Chang AB, Lin R, Studley WK, Tran CV, Saier MH. Molecular Membrane Biology. 2004;21:171. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 27.Chang G, Roth CB. Science. 2001;293:1793. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 28.Reyes CL, Chang G. Science. 2005;308:1028. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- 29.Woebking B, et al. J Bacteriol. 2005;187:6363. doi: 10.1128/JB.187.18.6363-6369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolhuis H, et al. EMBO J. 1996;15:4239. [PMC free article] [PubMed] [Google Scholar]
- 31.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr Science. 2003;300:976. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 32.Bolhuis H, et al. J Biol Chem. 1996;271:24123. doi: 10.1074/jbc.271.39.24123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


