Abstract
The classical nuclear export signal (NES), also known as the leucine-rich NES, is a protein localization signal often involved in important processes such as signal transduction and cell cycle regulation. Although 15 years has passed since its discovery, limited structural information and high sequence diversity have hampered understanding of the NES. Several consensus sequences have been proposed to describe it, but they suffer from poor predictive power. On the other hand, the NetNES server provides the only computational method currently available. Although these two methods have been widely used to attempt to find the correct NES position within potential NES-containing proteins, their performance has not yet been evaluated on the basic task of identifying NES-containing proteins. We propose a new predictor, NESsential, which uses sequence derived meta-features, such as predicted disorder and solvent accessibility, in addition to primary sequence. We demonstrate that it can identify promising NES-containing candidate proteins (albeit at low coverage), but other methods cannot. We also quantitatively demonstrate that predicted disorder is a useful feature for prediction and investigate the different features of (predicted) ordered versus disordered NES’s. Finally, we list 70 recently discovered NES-containing proteins, doubling the number available to the community.
INTRODUCTION
Among the complicated ‘route map’ of protein sub-cellular localization, the nucleocytoplasmic traffic of proteins occurs through the nuclear pore complexes, which allow passive diffusion of small proteins (<60 kDa) but require active transport for larger proteins. This transport is mostly mediated by karyopherin proteins and the specific sequence signals of cargo molecules; nuclear localization signals (NLSs) and nuclear export signals (NESs), for each direction, respectively. Compared to classical NLSs, the classical ‘leucine-rich’ NESs are more difficult to identify correctly because the NES consensus sequence often spuriously matches regions forming the hydrophobic core of proteins (1). The karyopherin Exportin 1/CRM1 (chromosomal region maintenance 1) mediates the export of many cellular and viral proteins containing leucine-rich NESs. To date, more than 75 proteins containing this leucine-rich NES have been experimentally verified. Many of them are related to signal transduction, cell-cycle regulation or the export of unspliced or partially spliced viral messenger RNA (mRNA) such as the HIV-1 Rev protein. Recently, this export pathway has also been suggested to be involved in the mechanism inducing the abnormal localization of many tumor suppressors containing leucine-rich NES’s, p53 for instance, in various cancer cells (2).
Despite its importance, we know little about this CRM1-meditated leucine-rich nuclear export signal (NES, hereafter), other than the abundance of hydrophobic residues, mostly leucine, and the specific spacing between them. Limited structural information is one factor which hampers further characterization of the NES. Based on secondary structure prediction and eight structures (six determined by X-ray crystallography) of NES-containing proteins, previous research has suggested a strong preference of α-helical structure and a bias against β-strands in the N-terminal end of NESs. However, in 2007, the first NES located on a β-strand was reported in fibroblast growth factor-1 (FGF-1) (3).
Unfortunately, no complete structures are available for CRM1 bound to classical NES containing proteins. However, in 2009, the crystal structure of CRM1 in complex with snurpotin 1 (SNUPN), an export substrate previously considered to be exported through an NES-independent interaction with CRM1, was reported (4,5). This complex structure revealed some details of the binding interface including a minor binding patch near the N-terminus of SNUPN resembling the classical NES. However, this NES mimic may not be sufficient to understand the classical NES, because of its much lower binding affinity. Furthermore, the multipartite recognition and the number of critical hydrophobic residues within this NES mimic are different than what is known about the classical NESs and so far observed only in this CRM1–SNUPN complex.
The first proposed consensus sequence of the classical NES is L-x-(2,3)-[LIVFM]-x(2,3)-L-x-[LI] where x is any amino acid, defined from analysis of mutant variants of HTLV-1 Rex and HIV-1 Rev (6) following the discovery of NESs in the human immunodeficiency virus type 1 (HIV-1) Rev protein (7) and cyclic adenosine monophosphate (cAMP)-dependent protein kinase inhibitor (PKI) (8). This consensus sequence was widely used until la Cour et al. (9) showed that the majority of NESs (63%) in NESbase, a database of experimentally verified NESs, deviated from this consensus sequence. By allowing a more general consensus sequence, [LIVFM]-x-(2,3)-[LIVFM]-x(2,3)-[LIVFM]-x-[LIVFM], sensitivity can be improved (from 37% to 72%) at a cost of greatly increasing false positives (precision drops from 52% to 16%) (10). In practice, this ‘tolerant’ consensus sequence has then been commonly accepted, though such a trade-off seems to be unsatisfactory.
Based on NESbase, la Cour et al. (10) provided the NetNES web server aiming to solve this condition. The prediction of the NetNES server is performed from primary sequence and implemented by the combination of a hidden Markov model (HMM) and a neural network. Tested on a small set of independent NES-containing proteins, three out of five NESs were correctly located by NetNES. Unfortunately, despite the growing number of experimentally verified NES-containing proteins in recent years, NESbase has not been updated since 2003. Thus, there is an urgent need to collect recently discovered NES-containing proteins, both to re-evaluate NetNES and to provide a more complete data set for public use.
Kosugi et al. (11) developed an assay to detect NESs and proposed an alternative set of consensus sequences. However, they did not evaluate the trade-off between sensitivity and precision, and, in fact, their precision is even lower than the more tolerant consensus sequence mentioned above (see ‘Materials and Methods’ section, Supplementary Figure S1).
In their effort to better characterize NESs, la Cour et al. noted some correlations not included in the consensus sequence representation. In particular, they hypothesized that some protein attributes, such as flexibility, and a minor preference for negatively charged or polar amino acids around the NES, are potentially relevant to NES function. However, instead of directly using predicted flexibility as a feature of NESs, la Cour et al. built NetNES using primary sequence information alone, perhaps due to the lack of suitable predictors available at that time.
Recent research indicates that intrinsically disordered region of proteins are often involved in molecular recognition with both high specificity and low affinity (12,13). Interestingly, the binding affinities between NESs and CRM1 were found to be generally low and, furthermore, high-affinity artificial NESs impair the efficient release of export complexes from the NPC (14).
In this study, we first hypothesized that protein intrinsic disorder may be relevant to NES recognition. We therefore investigated the correlation between protein intrinsic disorder and NES sites and applied our findings to develop a new predictor, NESsential, which aims to not only find the correct position of NESs at the site level, but also find potential NES-containing proteins at the protein level.
MATERIALS AND METHODS
Features
Integrating new potentially relevant properties of NES function with those previously suggested, we extracted 22 biophysically inspired features from not only the region matching the pre-filter, but also its upstream and downstream 10-mer flanks. These features mainly consist of (i) simple primary sequence attributes, such as the frequency of some specific amino acids: proline, negatively charged and polar residues; (ii) predicted protein attributes [solvent accessibility and secondary structure by SABLE (15); protein intrinsic disorder by Poodle-L (16)]; and (iii) other properties such as the average hydrophobicity within the pre-filter matches (by the Kyte–Doolittle scale) and the distance in between the previous and next matches of the pre-filter (or to the N- or C-terminal when no such match exists) normalized by the protein length. We calculate flank disorder and solvent accessibility features based on a window of length 10, which requires special handling near the ends of sequences. For those matches close to the termini, we regard the ‘missing part’ of such flanks as extremely disordered and accessible, assigning a disorder score of 1 and solvent accessibility of 100 to each missing ‘virtual residue’. Tables 1, 2 and Supplementary Table S1 provide more detailed descriptions of these 22 features.
Table 1.
The top-10 features ranked by F-score on our combined (training + test) data set (disordered model of split NESsential)
| Rank | Feature description | F-score |
|---|---|---|
| 1 | # of leucines among the three hydrophobic positions | 0.130 |
| 2 | Distance to previous match of Φx(2,3)ΦxΦ divided by the protein length | 0.042 |
| 3 | Whether a hydrophobic residue exists in the upstream -3 (for 7-mer match) or -4 position (for 6-mer match) | 0.035 |
| 4 | # of negatively charged residues in the upstream flank | 0.032 |
| 5* | # of prolines within the pre-filter match Φx(2,3)ΦxΦ | 0.026 |
| 6 | # of negatively charged residues within the pre-filter match Φx(2,3)ΦxΦ | 0.013 |
| 7* | # of polar residues in the downstream flank | 0.009 |
| 8* | Whether the first two residues are involved in a β-strand based on second structure prediction | 0.009 |
| 9* | Whether the first residue is involved in a β-strand based on second structure prediction | 0.008 |
| 10 | Avg. predicted solvent accessibility of downstream flank | 0.008 |
*Indicates features that the mean value of spurious matches is greater than that of real NES sites.
Table 2.
The top-10 features ranked by F-score on our combined (training + test) data set (ordered model of split NESsential)
| Rank | Feature description | F-score |
|---|---|---|
| 1 | # of leucines among the three hydrophobic positions | 0.036 |
| 2 | Whether a hydrophobic residue exists in the upstream -3 (for 7-mer match) or -4 position (for 6-mer match) | 0.033 |
| 3 | Avg. predicted disorder score of the downstream flank | 0.025 |
| 4 | Avg. predicted disorder score of the pre-filter match Φx(2,3)ΦxΦ | 0.014 |
| 5 | Distance to next match of Φx(2,3)ΦxΦ divided by the protein length | 0.013 |
| 6 | Distance to previous match of Φx(2,3)ΦxΦ divided by the protein length | 0.012 |
| 7 | Avg. predicted disorder score of the upstream flank | 0.007 |
| 8 | Avg. predicted solvent accessibility of the downstream flank | 0.007 |
| 9 | # of negatively charged residues within the pre-filter match Φx(2,3)ΦxΦ | 0.006 |
| 10 | Avg. predicted solvent accessibility of the pre-filter match Φx(2,3)ΦxΦ | 0.005 |
Protein intrinsic disorder prediction
To investigate the correlation between protein intrinsic disorder and NES function, we applied POODLE-L (16) and DISOPRED (17), two of the best-performing tools for disorder prediction in the critical assessment of techniques for protein structure prediction (CASP7), to all protein sequences in the training data. In particular, POODLE-L is designed to predict long (≥40 aa) disordered regions. We used both tools to analyze the correlation between intrinsic disorder and NES function, but for prediction we only report results using POODLE-L, as this choice yielded better NES prediction performance.
Training
Training data
We selected 60 NES-containing proteins from NESbase as our training data after the removal of redundant sequences (with sequence identity >25%), and those lacking verified sensitivity to leptomycin B (LMB, an effective CRM1 inhibitor) or other experimental data showing CRM1 dependency. Note that due to using a stricter identity threshold, our training data contain slightly fewer NES-containing proteins than NetNES (60 versus 64).
Training and prediction pipeline of NESsential
We first applied a pre-filter consensus Φx(2,3)ΦxΦ, where Φ can be substituted by L, I, V, F or M and x is any amino acid, to each protein sequence in the training set. This retrieved 946 matches, including 117 NES sites and 829 spurious matches, according to the annotation of experimentally verified NES regions. These matches constitute our positive and negative training examples. Subsequently, 22 features, such as predicted disorder, were extracted and applied to train SVM models [implemented by LIBSVM 2.9 (18)] to discriminate the real NES sites from the spurious matches. Based on this pipeline, we defined two types of NESsential that differ by a prior classification of matches by disorder prediction: ‘flat’ NESsential contains one SVM model trained by all matches, while ‘split’ NESsential employs different SVM models for disordered and ordered matches as shown in Figure 1. In split NESsential, if every residue is predicted to be ordered by POODLE-L, the pre-filter match is placed in the ordered group; otherwise, the disordered group (which therefore includes matches predicted as partially disordered).
Figure 1.
The training pipelines of two types of NESsential. In split NESsential, pre-filter matches are first divided into disordered or ordered; ordered if POODLE-L predicts ordered for every residue in the match, otherwise disordered. Separate SVMs are used for each group when predicting them as NES’s or spurious matches.
Choice of pre-filter
In our scheme, it is imperative that the pre-filter has high sensitivity. A low precision is tolerable because the SVM classifier has a chance to eliminate false positives. To increase the transparency of the prediction process, it is also desirable for a pre-filter to be a simple pattern. For these reasons, we applied two general patterns with lengths of 6 and 7 residues, ΦxxΦxΦ and ΦxxxΦxΦ, as a pre-filter. The 6-mer pattern matched 491 times, of which 71 were in the experimentally indicated ‘gold standard’ NES regions, while the 7-mer pattern matched 455 times, including 46 in NES regions. This pre-filter achieve the highest sensitivity among all available consensus sequences (Supplementary Figure S1). Moreover, both patterns contain the region bounded by the second and the fourth hydrophobic position of the consensus sequence currently in use. Previous research indicated that the first hydrophobic position in the signal is less conserved (10), which is consistent with some experimental observation indicating that NES activity is more susceptible to mutations of the C-terminal hydrophobic residues within the signal (8,19). To test for statistical significance, we used a binomial test to see if the high frequency of matches within verified NES regions could be explained by chance, obtaining P-values of 1.7e-16 (ΦxxΦxΦ) and 5.6e-7 (ΦxxxΦxΦ), respectively.
Evaluation
To compare our method with NetNES, we collected 70 recently discovered NES-containing proteins. In addition to this new data, we also prepared a set of background proteins and conducted the first evaluation of current methods on the protein-level classification task.
Recently discovered NES-containing proteins
To further our understanding of NESs and evaluate the performance of existing methods, it is important to use as much experimental data as possible; however, unfortunately, many recently discovered NESs are not annotated in UniProt. Starting with the references given by Kosugi et al. (11), we undertook a literature search to collect NES-containing proteins not included in NESbase. In order to allow a fair comparison between our predictor and previous methods, these proteins were used only for evaluation, not training. We checked sequence identity (25%) with NCBI BLASTClust to avoid redundancy between training and test data. As a result, we obtained a test set containing 70 proteins and 85 NESs (some proteins contain multiple NES sites). The addition of these newly collected proteins more than doubles the number of CRM1-dependent NES containing proteins organized in a single data set. This data set is itself an important resource which should contribute to future NES research (Supplementary Table S2).
Background proteins
The 70 NES-containing proteins described in the previous section can serve as positive examples for protein level prediction. Unfortunately, it is difficult to prepare an ideal negative data set, as in general it is difficult to rule out the possibility that a nuclear protein may have a yet undiscovered NES or that a non-nuclear protein might have a cryptic NES which could function if the protein were found in the nucleus. Therefore, we selected 541 yeast proteins whose localization is annotated as either the cytosol (159 proteins) or the nucleus (382 proteins) from the Universal Protein Resource (UniProt) (http://www.uniprot.org/) as background proteins for protein level classification evaluation. A few of these background proteins might contain NESs, but we expect that most do not. Note that we only use these background proteins for evaluation, never for training.
Performance measurement
To evaluate prediction performance, for each task, we computed the receiver operating characteristic (ROC) curve and its area under the curve (AUC) metric. We also provide precision-recall (PR) curves, which can be more informative than the ROC curve in the case of skewed data sets (20), in the Supplementary Data.
RESULTS
Distribution of predicted disorder scores
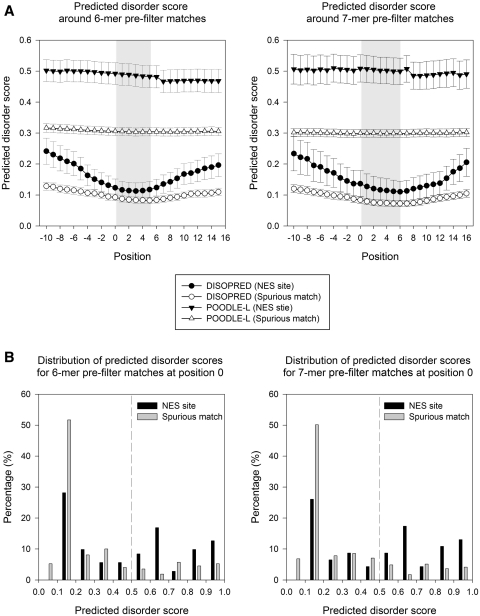
Both DISOPRED and POODLE-L return a probability estimate of each residue being disordered. Figure 2(A) shows the mean and standard error (a measure of the uncertainty of mean estimation) of the predicted disorder scores of NES site and spurious matches in the training data. For both tools, the NES sites display significantly more disorder than spurious matches. Note that DISOPRED uses a window size of 15 aa for prediction, opposed to the 40 aa used by POODLE-L; this may explain why the DISOPRED NES site scores are sensitive to the hydrophobic residues in the pre-filter match, but the POODLE-L curves are almost flat.
Figure 2.
Predicted disorder scores by POODLE-L and DISOPRED2 of NES sites and spurious matches (training data). (A) The mean score and its standard error are shown at each position, where position 0 represents the first residue of 6-mer (ΦxxΦxΦ) or 7-mer (ΦxxxΦxΦ) pre-filter matches. The regions corresponding to ΦxxΦxΦ and ΦxxxΦxΦ are shaded in gray [where Φ denotes (LIVFM) and x denotes any amino acid]. (B) The distribution of POODLE-L disorder scores at the first position of 6-mer and 7-mer are shown. The vertical dashed line indicates the threshold of POODLE-L (0.5).
Distribution of predicted disorder scores for NES sites by POODLE-L
Having established that the average predicted POODLE-L disorder score is higher in NES site matches, we investigated the full distribution of such scores. Figure 2(B) shows the distribution of predicted disorder scores at the first residue of the ΦxxΦxΦ and ΦxxxΦxΦ matches, respectively. The distribution of NES site matches is marked different from that of spurious matches. Indeed, the fraction of NES sites with score >0.5 at this position is significantly higher than that of spurious matches for both 6- and 7-mers (Fisher’s exact test, P-values = 5.3e-07 and 1.1e-06, respectively). Similar results were observed at other positions within the pre-filter region.
Classification of NES-containing proteins versus non-NES-containing proteins
To evaluate current methods, we first applied each predictor to the mixed set of NES-containing test proteins and background proteins, and retrieved the highest predicted score for each protein to generate a ranked list. Based on this list, the performances were evaluated and compared by the ROC curve and the AUC. Meanwhile, the performances of current consensus sequences were also plotted in the ROC space by applying a simple rule that proteins, which have a match to the given consensus are classified as ‘NES-containing’. Surprisingly, Figure 3 shows that the corresponding points of the consensus sequences in ROC space are located below the diagonal, meaning that the performance is worse than random guessing. As for the computational methods, the AUC values of flat NESsential (0.71) and split NESsential (0.63) are higher than that of NetNES (0.60). However, none of them seem high enough in overall performance.
Figure 3.
The ROC curves of two types of NESsential and NetNES. Dots denoting the performance of current consensus sequences are also plotted in the ROC space for comparison.
We further investigated the higher positions in the ranked lists, corresponding to the performance expected when applying strict thresholds. Among the top ranked positions, flat and split NESsential list two to five times the number of NES-containing test proteins (dark gray stack in Figure 4) than NetNES. This result demonstrates that proteins with high scores predicted by NESsential, split NESsential especially, have a higher chance to contain NESs. The PR curve (Supplementary Figure S2) also gives a relatively strong result for NESsential. For example, at a recall of 20%, NESsential attains a precision of over 50%, while NetNESs precision is only 16%.
Figure 4.
Stacked bar plots of the composition of the top-scoring proteins for the two types of NESsential and NetNES.
Finding NES positions within NES-containing proteins
Unlike the protein-level classification task, there are some complications to make the comparison completely fair and objective: the lack of exact boundaries of gold standard data and the different form of prediction between NESsential and NetNES (explained in detail in Supplementary Figure S3). To evaluate this prediction task previously addressed by NetNES, we converted and assigned the ‘site-level’ predicted scores of NESsential to each residue of the pre-filter match (6 or 7 contiguous residues). As shown in Figure 5, flat NESsential achieves higher AUC values (by 0.01 and 0.09) than NetNES for the disordered and ordered groups, respectively. The PR curves (Supplementary Figure S4) further indicate that flat NESsential achieves better precision than NetNES at low recall level for either group.
Figure 5.
The ROC curves of flat NESsential and NetNES at the residue level. Because many residues are assigned a score of zero (see Supplementary Data), there is a big jump of measurable performance (dashed line) for both methods. Each dashed line connects two specific points in ROC space: one end represents the false positive rate (FP rate) and true positive rate (TP rate) obtained by using the smallest nonzero score as a threshold, while the other end (1, 1) represents the unconditional assignment of all residues as NES positions. The AUC was calculated for each curve including the dashed line.
Analysis of combined data set
To avoid ‘peeking’ at the test data, we intentionally designed NESsential without statistic analysis of the test data. However, our combined data set is roughly twice the size of the training data and warrants statistical analysis. Therefore, we report a post hoc analysis in this section.
Sequence determinants of leucine-rich NESs
Using the verified NESs in NESbase for analysis, la Cour et al. found a preference for negatively charged amino acids around the NES. To test whether this conclusion is still valid, we generated sequence logos for NES sites in the combined data set by aligning three hydrophobic positions within the pre-filter. As shown in Supplementary Figures S5 and S6, the preference for negatively charged residues is generally lower than previously observed. We also note that the 6-mers appear to have a slightly stronger tendency for a fourth leucine to appear upstream, especially in the position four residues upstream from the first hydrophobic position in our consensus filter.
Considering the correlation between the presence of charged residues and intrinsic disorder, we also generated the sequence logos for ordered and disorder groups separately. As expected, the sequence logos for disordered NESs demonstrate higher preference for charged residues (Supplementary Figure S7) than for ordered ones (although this may simply reflect a tendency for charged regions to be predicted as disordered). Meanwhile, the comparison among the three anchor positions shows that leucine is more strongly preferred in disordered NESs than in ordered ones
Although suggestive, sequence logo analysis implicitly compares NESs to what is expected from random sequences, rather than using real sequences as a negative control. Thus, when splitting sequences by disorder prediction, the sequence logo confounds general features of sequences predicted to be disordered with those specific to NESs. Moreover, features evident in sequence logos are not guaranteed to be effective for discrimination between NESs and spurious matches.
Therefore, we also preformed a discriminatory analysis by computing the F-score for each descriptive feature on NES site matches versus spurious matches in our combined dataset. The results shows that some features, such as normalized distance to previous matches, have higher discriminative power than the number of negatively charged residues, for both disordered (Table 1) and ordered (Table 2) NESs. We can also see that, for predicted disordered pre-filter matches, prolines are disfavored compared to the negative controls. This is consistent with the results of Kosugi et al. (11), who used a yeast selection system to screen for sequences from a random peptide library with NES activity and found proline to be underrepresented.
Length of disordered tendency
We investigated the extent to which the mean predicted disorder of NES sites differs from spurious matches. We observed that the disorder score distribution in the NES sites tends to be <0.3 or >0.5—particularly in the flanking regions (Figure 6) and that the mean score for NES versus spurious matches remains different as far as 50 or more residues away (Supplementary Figure S8).
Figure 6.
The distribution of POODLE-L disorder prediction score for NES and spurious pre-filter matches (combined data set) are shown for 6-mer and 7-mer matches and 25 residues upstream and downstream of the first (last) position of the matches.
To see if this observation can be used to improve prediction accuracy, we tried changing the width used for the upstream and downstream average predicted disorder features (Table 3). As we cannot retrain NetNES to do cross-validation, we only evaluate NESsential. Therefore, we did not convert the NESsential output, but simply treated the problem as a binary classification of pre-filter matches. We find that extending the feature width to around 50 does indeed lead to an increase in AUC for flat NESsential and both branches of the split NESsential predictor. Therefore, we also make the SVM model with length 50 disorder features available for download. Interestingly, increasing the flank width did not improve performance for our original separated training/test evaluation (data not shown). This suggests that, as a group, the more newly discovered NESs differ somewhat from the older ones. Indeed, we found that the test proteins contained a significantly (Fisher’s exact test, P-value = 4.4e-05) larger fraction of ordered NESs (73%) than the training proteins (48%). However, disorder is still significantly enriched in the combined data set relative to spurious matches (Fisher’s exact test, P-value = 7.1e-09). We note in passing that the newer data also have a somewhat (but not statistically significantly) higher ratio of 7- to 6-mer matches in gold standard NES regions (68/83 versu 46/71) (Fisher’s exact test, P-value = 0.38).
Table 3.
The AUC values (combined data set, 5-fold cross validation) by using different length of flanking region for averaging the upstream and downstream disorder scores
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | |
|---|---|---|---|---|---|---|---|
| Disordered model of split NESsential | 0.86 | 0.87 | 0.88 | 0.89 | 0.89 | 0.89 | 0.89 |
| Ordered model of split NESsential | 0.79 | 0.81 | 0.82 | 0.82 | 0.82 | 0.82 | 0.82 |
| Combined AUC of split NESsential* | 0.80 | 0.83 | 0.83 | 0.83 | 0.83 | 0.84 | 0.83 |
| Model of flat NESsential | 0.80 | 0.82 | 0.83 | 0.84 | 0.84 | 0.84 | 0.83 |
*We applied the same threshold to the two models of split NESsential, to obtain a single AUC value for comparison with flat NESsential.
DISCUSSION
Performance of protein-level classification task
In this work, we present three measures of performance: protein level, residue level and site level (Supplementary Data). At a conceptual level, site-level prediction is a natural choice, but it is problematic due to the fact that the gold standard does not define sites in a consistent and precise way (Supplementary Figure S3). Residue-level comparison is basically a variant of site-level prediction and suffers from the same fundamental problem. In this sense, protein-level comparison is the most clear-cut, because it is not affected by the gold-standard boundaries. Thus, we discuss the performance on this task in some detail here.
Consensus-based methods
The fact that consensus sequences have negative predictive power indicates the ‘reversed’ classification decision is more effective than the original one. To find a possible explanation, we calculated the expected number of occurrences for both consensus sequences in random sequences with the amino acid composition and length matching that of NES-containing, cytosolic and nuclear proteins respectively (we did not compute this for the Kosugi et al. ‘consensus’, which is actually more complex than a simple regular expression). As the results show (Supplementary Table S3), both simple consensus sequences are more likely to randomly occur in cytosolic and nuclear background proteins than in NES-containing proteins, due to differences in amino acid composition and average length. This can explain the negative correlation between matching the consensus sequences and whether a protein contains an NES.
NESsential and NetNES
The authors of NetNES recognized the importance of the protein-level prediction task and tested it on five proteins, concluding “the performance of the predictor is sufficiently high to allow for identification of new NES-containing proteins.” With the advantage of the large test set, which has become available in the years since their publication, we were able to evaluate this claim quantitatively. Unfortunately, NetNES performed very poorly in our protein-level test. NESsential performed substantially better, but still only its highest scores offer a reasonable probability of being true positives. Therefore, we must be more cautious with our conclusions.
One lingering question is: why does NetNES perform so poorly at the protein-level task, when it clearly has some ability to identify NES sites within NES-containing proteins? It is true that NetNES is not trained to recognize background proteins as negative examples, but that is also true of NESsential.
One hypothesis we considered was that NetNES might tend to be ‘fooled’ by some specific domains containing sequence mimics of NESs and often give high scores to background proteins with such domains. To test this hypothesis, we checked the domain information around the predicted position for the top-40 background proteins ranked by NetNES and NESsential. However, no clear trends were evident (Supplementary Tables S4 and S5). Since NetNES and NESsential differ in many details (feature set, use of pre-filter, machine learning classifier) it is difficult to pin down what exactly causes the difference.
Searching for potential novel NES-containing proteins
Split NESsential is capable of retrieving 20% of NES-containing proteins with a precision of over 50%. Moreover, 6 out of the 11 NES-containing proteins in the top-20 positions are correctly predicted not only at the protein level but also at the site level. These results demonstrate that proteins attaining a high split NESsential score have a reasonably high probability of containing NESs, and should be useful when searching for potential candidates. We therefore retrained split NESsential on all of the data (training and test set) and computed the scores for a set of nucleocytoplasmic dually localized yeast proteins downloaded from UniProt (Supplementary Table S6). Interestingly, one of the top-ranked proteins, the yeast nucleosome assembly protein (NAP1), was previously suggested to be exported by multiple proteins and CRM1 might be one of its nuclear exporters (21,22). However, it should be mentioned that the current annotation of subcellular localization is not completely perfect, which means some of the cytosolic and nuclear proteins may contain undiscovered NESs, although the ratio is probably lower than for proteins annotated as dually localized. Therefore, we also provide lists of nuclear (Supplementary Table S7) and cytosolic (Supplementary Table S8) proteins ranked by their scores given by split NESsential (trained on all data). The top-ranked nuclear protein, the nitrogen regulatory protein GLN3 for instance, has been reported to contain a CRM1-mediated NES (23) (although not verified with leptomycin B).
Sequence conservation as a relevant feature
Sequence conservation among orthologous proteins might be expected to provide useful information to improve NES recognition since the CRM1-mediated export pathway and the leucine-rich NES are found in all major branches of the eukaryotes. However, spurious consensus matches are often located in the hydrophobic core where the sequence is also conserved among orthologs. In fact, one should be careful when trying to apply sequence conservation to NES prediction, since NESs are not necessarily conserved among all orthologs. For example, the NES of the Snail transcription factor were found to be conserved only in mammalian orthologs, while the NES is not present in other family members (24). As another example, although the real NES of Human TPP1 is conserved among human, mice and frogs, spurious pre-filter matches in this protein also show high sequence conservation (25). Although these examples might be special cases, they suggest that it may be challenging to significantly improve prediction by the use of sequence conservation.
Pros and cons of pre-filtering
One of many differences between NESsential and NetNES is the use of a pre-filter match. This has the obvious disadvantage of making it impossible to correctly predict nonclassical NESs. On the other hand, it has advantages, albeit technical, as well. While NetNES must learn to discriminate any non-NES region from NES regions, NESsential can focus on what separates NES site pre-filter matches from spurious matches. The more balanced number of negative versus positive examples eases the learning task for the SVM classifier and allows simple statistical measures, like the F-score, to give meaningful results from limited data. In contrast, without the pre-filter, the vast majority of negative examples are completely unlike NESs and, from the standpoint of understanding NESs, their derived feature values (e.g. disorder) are essentially noise.
Directions for future improvement
As mentioned earlier, by employing a pre-filter, NESsential completely forfeits any chance to predict noncanonically spaced NESs. One possible approach would be to attempt to improve overall prediction accuracy by removing the pre-filter, or using a less stringent one. Unfortunately, with the current data set this would be very challenging. Among all the verified NESs we were able to collect, only 25 out of 170 failed to match the pre-filter. Given the generally low performance of all NES predictors (including NESsential), it seems unlikely that the benefit in terms of increased sensitivity would overcome the cost in increased false positives.
Since the same feature set was used in training all SVM models in this study, it is interesting to discuss what caused the different performance in 5-fold cross-validation between the disordered and ordered models of split NESsential. One might speculate that the unequal performance is a result of the different ratio between positive and negative data for the ordered and disordered pre-filter matches (Figure 1). However, we tested this hypothesis by training and evaluating models for the ordered group using randomly selected negative data to mimic the ratio found in the disordered group, but no significant AUC improvement was observed (data not shown). Thus, it appears that the effect of unbalanced data sets cannot explain the difference in AUC, but rather the ordered NESs are less well described by our feature set. Our features mainly focus on the local information surrounding the NES site. However, the ordered NESs might be located in more buried regions and therefore require more complicated conformational changes to expose themselves to CRM1. Previous research has demonstrated some specific regulation, such as nearby phosphorylation sites (26) or the oligomeric state (27) of proteins with buried NES. Although these features seem to be required for specific proteins, we cannot exclude the possibility that these features will be found in other NES-containing proteins and be helpful for future improvement, especially for the ordered group.
Güttler et al. (28) recently provided detailed structural information of NES–RanGTP–CRM1 complex for PKI and HIV-1 Rev NESs by using NES-SPN1 chimera proteins. Their data confirm the wide range of local structure possible for NESs and suggest the existence of a fifth hydrophobic position which can potentially contribute to NES recognition. We expect the concerted efforts of experimental and computational groups will continue to gradually reveal the nature of this elusive signal.
Availability
We have made our source code publicly available at http://seq.cbrc.jp/NESsential/. NESsential depends on the third-party software tools LIBSVM, SABLE and POODLE-L. Unfortunately, POODLE-L is not open source, but a free web server is available and we provide a script to automatically query it and forward the results to NESsential. We have also placed the training (from NESbase) and testing (Supplementary Table S2) data sets on the same website.
CONCLUSION
We present NESsential. NESsential uses long disorder region prediction and other derived features along with more direct primary sequence features, such as the frequency of specific amino acids within particular regions, to predict leucine-rich NESs from amino acid sequence. Our results show that NESsential is much more effective than the other available tool, NetNES, at detecting NES containing proteins. The two tools perform comparably at the task of finding the correct NES sites within NES-containing proteins, except that NESsential is more flexible in the trade-off between sensitivity and precision. In addition, we provide a test data set of 85 verified NES sites (in 70 proteins) as an up-to-date resource for the community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Interchange association Japan (IAJ; to S.C.F.); and a research fellowship of the Japan Society for the Promotion of Science (JSPS; to K.I.). Funding for open access charge: research institute (AIST, Computational Biology Research Center, Tokyo, Japan).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Dr Shunichi Kosugi provided further supporting information of their original paper. Dr Raymond Wan advised in machine learning and evaluation. Dr Martin Frith advised in motif finding. Dr Noriyuki Sakiyama and Dr Shuichi Hirose advised in disorder prediction.
REFERENCES
- 1.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 2.Turner JG, Sullivan DM. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr. Med. Chem. 2008;15:2648–2655. doi: 10.2174/092986708786242859. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen T, Rosendal KR, Sørensen V, Wesche J, Olsnes S, Wiedłocha A. A nuclear export sequence located on a beta-strand in fibroblast growth factor-1. J. Biol. Chem. 2007;282:26245–26256. doi: 10.1074/jbc.M611234200. [DOI] [PubMed] [Google Scholar]
- 4.Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 5.Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 8.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 9.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 11.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic. 2008;9:2053–2062. doi: 10.1111/j.1600-0854.2008.00825.x. [DOI] [PubMed] [Google Scholar]
- 12.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 13.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 14.Kutay U, Güttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Adamczak R, Porollo A, Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 16.Hirose S, Shimizu K, Kanai S, Kuroda Y, Noguchi T. POODLE-L: a two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics. 2007;23:2046–2053. doi: 10.1093/bioinformatics/btm302. [DOI] [PubMed] [Google Scholar]
- 17.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 18.Chang C-c, Lin C-J. LIBSVM: a Library for Support Vector Machines. 2001. [Google Scholar]
- 19.Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 20.He H, Garcia EA. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009;21:1263–1284. [Google Scholar]
- 21.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park Y-J, Luger K. The structure of nucleosome assembly protein 1. Proc. Natl Acad. Sci. USA. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho J, Zheng XFS. Domains of Gln3p interacting with karyopherins, Ure2p, and the target of rapamycin protein. J. Biol. Chem. 2003;278:16878–16886. doi: 10.1074/jbc.M300429200. [DOI] [PubMed] [Google Scholar]
- 24.Garcia de Herreros A, Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol. Cell. Biol. 2003;23:5078. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L-Y, Liu D, Songyang Z. Telomere maintenance through spatial control of telomeric proteins. Mol. Cell. Biol. 2007;27:5898–5909. doi: 10.1128/MCB.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng W, Swenson LL, Fitzgibbon MJ, Hayakawa K, Ter Haar E, Behrens AE, Fulghum JR, Lippke JA. Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J. Biol. Chem. 2002;277:37401–37405. doi: 10.1074/jbc.C200418200. [DOI] [PubMed] [Google Scholar]
- 27.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 2010;17:1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.