Abstract
Notch, a transmembrane protein found in a wide range of organisms, is a component of a pathway that mediates cell-fate decisions that involve intercellular communication. In this paper, we show that in Drosophila melanogaster, Notch (N) is processed in a ligand-dependent fashion to generate phosphorylated, soluble intracellular derivatives. Suppressor of Hairless [Su(H)] is predominantly associated with soluble intracellular N. It has been demonstrated by others that N has access to the nucleus, and we show that when tethered directly to DNA, the cytoplasmic domain of N can activate transcription. Conversely, a viral activator fused to Su(H) can substitute for at least some N functions during embryogenesis. We suggest that one function of soluble forms of N is to bind to Su(H), and in the nucleus, to act directly as a transcriptional transactivator of the latter protein. Although N has functional nuclear localization signals, the N/Su(H) complex accumulates in the cytoplasm and on membranes suggesting that its nuclear entry is regulated. Localization studies in cultured cells and embryos suggest that Su(H) plays a role in this regulation, with the relative levels of Delta, N and Su(H) determining whether a N/Su(H) complex enters the nucleus.
Keywords: Notch, processing, Delta, Suppressor of Hairless, nuclear entry, transcriptional transactivator
Notch (N) is a 2703-amino-acid transmembrane protein that is found in a wide range of organisms from Caenorhabditis elegans to humans. The N gene was originally identified in Drosophila melanogaster in which N is a component of a pathway involved in cell-fate decisions that involve intercellular communication. During Drosophila embryogenesis, this process, which is known as lateral inhibition, operates in all three germ layers and ensures that only one cell of several equipotential cells, termed an equivalence group, will adopt the primary cell fate. Because this pathway was first characterized in the neuroectoderm, in which the absence of any member of the pathway results in overproliferation of neuroblasts, the genes involved in lateral inhibition are known as neurogenic genes. N, and other members of the neurogenic gene family are also conserved during evolution (for review, see Artavanis-Tsakonas et al. 1995; Greenwald 1998).
Genetic mosaic analysis has demonstrated that N is cell autonomous, suggesting that it is a receptor (Hoppe 1986; Heitzler and Simpson 1991). Delta (Dl), also a transmembrane protein, has been identified as the ligand for N in its role in lateral inhibition (for review, see Artavanis-Tsakonas et al. 1995; Nye and Kopan 1995). Activation of N results in transcription of genes of the Enhancer of split [E(spl)]) complex. This transcriptional activation is mediated by Suppressor of Hairless (Su(H)), a DNA binding protein that has been demonstrated to bind to the cytoplasmic domain of N (Fortini and Artavanis-Tsakonas 1994; Jennings et al. 1994; Jarriault et al. 1995; Lecourtois and Schweisguth 1995; Tamura et al. 1995; Kato et al. 1997; Schroeter et al. 1998).
Previous work has shown that expression of N proteins deleted for the extracellular domain results in gain of function phenotypes indicative of ligand-independent activation (for review, see Artavanis-Tsakonas et al. 1995; Greenwald 1998). The cytoplasmic domain of N contains functional nuclear localization signals (Stifani et al. 1992; Lieber et al. 1993), and it has been proposed that on binding Dl, the transmembrane N protein is cleaved releasing the cytoplasmic domain that translocates to the nucleus, where it is tethered to DNA via Su(H) and behaves as a transcriptional transactivator (Lieber et al. 1993; Struhl et al. 1993; Jarriault et al. 1995; Kopan et al. 1996). In tissue culture, Schroeter et al. (1998) have mapped a site in the transmembrane domain of mouse Notch-1 at which it undergoes ligand-dependent proteolytic cleavage. Mutating this site reduces the activity of Notch-1 in cell culture. In addition, it has been found that in vivo the cytoplasmic domain of Drosophila N has access to the nucleus, although the biochemical nature of this access was not determined (Lecourtois and Schweisguth 1998; Struhl and Adachi 1998).
In this paper, we present data indicating that in wild-type Drosophila, N is processed in a ligand-dependent manner to generate a cytoplasmic domain that, on the basis of size and solubility, does not span the membrane. This domain is phosphorylated and Su(H) preferentially associates with this form. The N/Su(H) complex is found associated with membranes and predominantly in the cytoplasm, indicating that there is a mechanism for regulating its subcellular location. Our data suggest that Su(H) can inhibit nuclear entry of the soluble N proteins, and that nuclear entry occurs in a fashion dependent on the relative abundance of soluble N and Su(H). In prior genetic studies of N nuclear activity, it was suggested that nuclear N participates in the transcriptional regulation of downstream target genes (Lecourtois and Schweisguth 1998; Struhl and Adachi 1998). We show that in the nucleus, N behaves as a transcriptional transactivator, and that a heterologous activator fused to Su(H) can substitute for activated N function in embryos.
Results
Su(H) interacts with phosphorylated Notch proteins
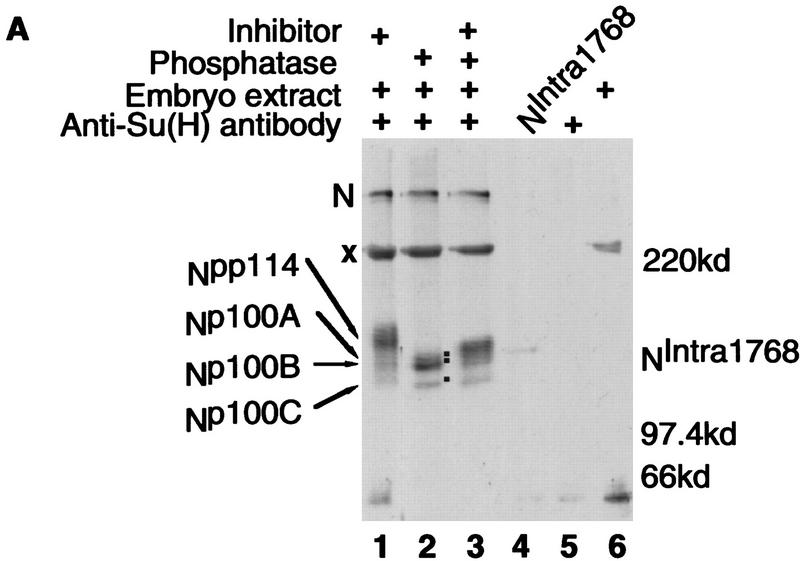
N and Su(H) proteins have been shown to physically interact (Fortini and Artavanis-Tsakonas 1994; Jarriault et al. 1995; Tamura et al. 1995; Kato et al. 1997; Schroeter et al. 1998). To characterize the nature of the associated N and Su(H) proteins in vivo, immunoprecipitations with antibodies against either N or Su(H) were performed on detergent extracts of Drosophila embryos. Following electrophoresis, N proteins in these immunoprecipitates were detected by Western blot. When anti-N and anti-Su(H) immunoprecipitations are probed with anti-N antibodies, only a small subset of the proteins immunoprecipitated by anti-N antibodies are found in the corresponding Su(H) immunoprecipitate [cf. Fig. 2A, below, lanes 1 and 2, anti-Su(H) immunoprecipitation, with Fig. 2C, lanes 1 and 2, anti-N immunoprecipitation]. This suggests that the interactions between N and Su(H) detected in this assay occurred in vivo and not during the course of the immunoprecipitation, as the latter might be expected to result in similar array of N proteins in both immunoprecipitates.
Figure 2.
Su(H) is associated with phosphorylated N proteins. (A,B) Western blot analysis of anti-Su(H) immunoprecipitates from Drosophila embryos probed with anti-N (NPCR) antibody. For A, an overnight collection of embryos was used and in B, staged embryos (age indicated in hours at top of lanes; H denotes hatching) were used for detergent extracts. Prior to electrophoresis, some of the samples were treated with alkaline phosphatase with or without inhibitor. (+) Presence of phosphatase and/or inhibitor. (A, Lane 4 and B, lane 9) In vitro-translated NIntra1768, the size of which is not affected by phosphatase treatment; (A, lane 5) Antibody but no extract; (A, lane 6) Embryo extract recovered on protein A–Sepharose beads that were not conjugated to antibody. The locations of N (N), NIntra1768, Npp114, Np100A, Np100B, and Np100C proteins are shown along side each blot, and also by dots on the blot. The × in A indicates a cross-reacting protein nonspecifically bound to the protein A–Sepharose beads; the asterisk (*) in B indicates a hypophosphorylated N protein that comigrates with Np100B. (A, all lanes with extract; B, lanes 3–6) 500 μg of protein was used for each immunoprecipitation; the remaining lanes in B contain 2.5 mg(lanes 1,2) and 1.75 mg (lanes 7,8). (C) Western blot analysis of anti-N (NI) immunoprecipitates from Drosophila embryos probed with anti-N (NPCR) antibody. (Lanes 1, 2) Immunoprecipitates from yw embryos; (lanes 3,4) immunoprecipitates of embryos resulting from a cross of Su(H) hsNintra1790/CyO males to Su(H)SF8 FRT40A/CyO virgins; (lanes 5,6) immunoprecipitates of embryos resulting from a cross of Su(H) hsNintra1790/CyO males to hsflp12/yw; Su(H)SF8 FRT40A/ovoD FRT40A virgins. All NIntra expressing embryos are both zygotically and maternally Su(H) null. For all genotypes, 3- to 5 hr-old embryos were subjected to a 30 min heat shock at 37°C and allowed to recover for 15–30 min prior to collection. (+) Phosphatase treatment. The locations of N (N) and NIntra are indicated next to the blot.
Anti-Su(H) immunoprecipitates contain two major size classes of N proteins, both of which are recognized by antibodies raised against 2 different regions of the intracellular domain of N (see Fig. 1 for antibodies used in this work); one the size of full length N and another, substantially enriched, which migrates as smear of proteins of ∼114 kD (Fig. 2A, lane 1). The existence of a smear of bands at 114 kd suggests that these N proteins might be post-translationally modified, perhaps by phosphorylation. Figure 2A, lane 2, shows that treating the Su(H) immunoprecipitates with alkaline phosphatase results in the smear of proteins being resolved into three proteins of ∼100 kD, which we have termed in order of decreasing molecular weight Np100A, Np100B, and Np100C. Np100A and Np100C differ by ∼4 kD. We have collectively termed the phosphorylated forms of these proteins Npp114. Treatment of the immunoprecipitates with alkaline phosphatase in the presence of phosphatase inhibitors reduces the effect of the phosphatase (Fig. 2A, lane 3). Because Npp114 reacts with two different N antibodies, it is most likely that these proteins are N. As no alternatively spliced N transcripts have been observed, and no appropriately positioned methionine exists at which internal translation could initiate to give rise to a protein with the size and antigenic determinants of Npp114 (Wharton et al. 1985; Kidd et al. 1986; Kopan et al. 1996), it is most likely that these smaller N proteins are the result of proteolytic cleavage.
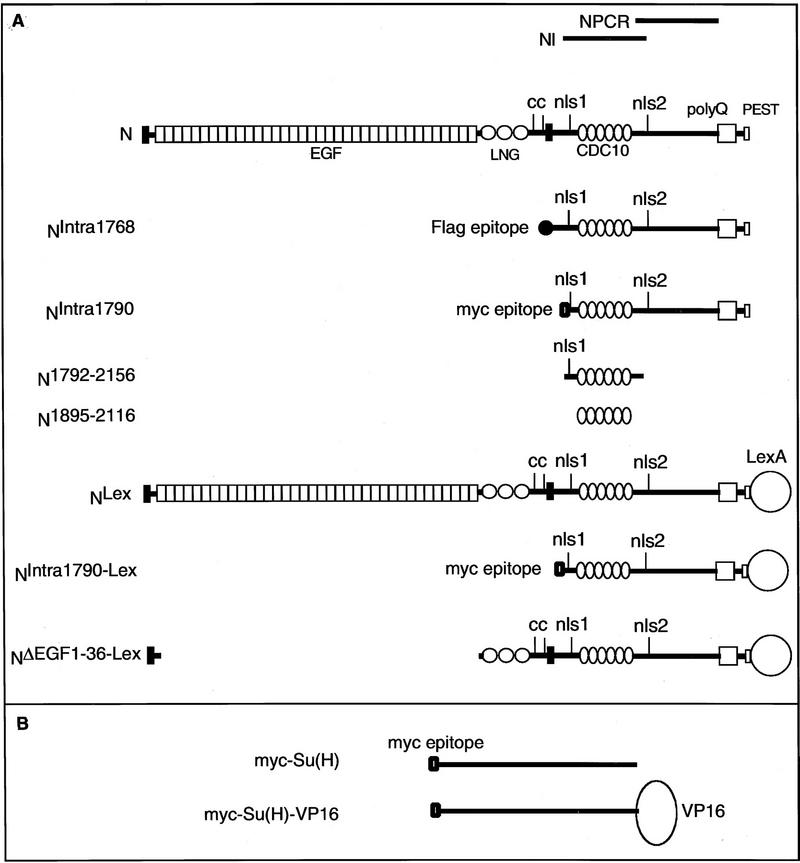
Figure 1.
Constructs and antibodies. (A) A diagram of the N protein (N). (EGF) Epidermal growth factor-like repeats; (LNG) lin-12, Notch, Glp-1 repeats; (cc) evolutionary conserved cysteine residues; (CDC10) CDC10 or ankyrin repeats; (nls1, nls2) putative nuclear localization signals; (polyQ) polymeric glutamines; (PEST) PEST sequence thought to be involved in protein stability (Rogers et al. 1986). The bars above the diagram of N indicate the various regions used as antigens to create anti-N antibodies. Beneath the diagram of N are the N constructs used in this paper. (B) Su(H) constructs used in this paper.
The proteins used for the previous experiment were from an overnight collection of embryos. We wondered if production of the various N proteins associated with Su(H) is developmentally regulated. Figure 2B shows that whereas Npp114 is present throughout embryogenesis with Np100B as its major component, the amount of Np100A appears to increase with age, and Np100C is only found late in embryogenesis. In addition, embryos that are young (0–4 hr) contain significantly more processed N protein that comigrates with hypophosphorylated Np100B (indicated by an asterisk) relative to Npp114. Late in embryogenesis, proteins that comigrate with both hypophosphorylated Np100B and Np100C are found.
The above experiment indicates that phosphorylated, processed N proteins interact with Su(H). To address whether phosphorylation is a consequence of Su(H) binding, we asked whether NIntra1790 (Fig. 1) is phosphorylated in embryos that lack Su(H). As can be seen in Figure 2C, lanes 5 and 6, NIntra1790 expressed in embryos that are both maternally and zygotically Su(H)− is phosphorylated, indicating that phosphorylation of processed N proteins is not dependent on Su(H) binding. Intriguingly, even after phosphatase treatment, NIntra1790 immunoprecipitated from Su(H)− embryos migrates slower than NIntra1790 immunoprecipitated from embryos that contain Su(H) (cf. lanes 4 and 6). This might be because NIntra1790 bound to Su(H) has undergone additional proteolytic processing, or NIntra1790 not bound to Su(H) has been subject to additional post-translational modification. However, in the Su(H)− extracts there is also a smear extending upward from endogenous N. This suggests that in Su(H)− embryos, NIntra1790, as well as being phosphorylated, has undergone additional post-translational modification.
We have also analyzed Su(H) coimmunoprecipitated with anti-N antibodies (data not shown). The Su(H) associated with N comigrates with the Su(H) immunoprecipitated by anti-Su(H) antibodies. In both immunoprecipitates, there is no change in mobility on phosphatase treatment, suggesting that Su(H) is not extensively phosphorylated.
Cleavage of N is ligand dependent
If N signaling is dependent on ligand-induced cleavage of N, then Npp114 might result from the binding of a N ligand. The two experiments shown in Figure 3 demonstrate that the presence of ligand and the ability of N to bind ligand is required for the presence of Npp114. All the known N ligands bind to the extracellular EGF-like repeats, deletion of which results in a nonfunctional protein (Rebay et al. 1991, 1993; Lieber et al. 1993). Extracts from embryos expressing a form of N that spans the membrane but lacks the EGF-like repeats and is tagged at the Carboxyl terminus with the DNA-binding domain of LexA [NΔEGF1-36–LexA (Fig. 1)] were immunoprecipitated with anti-N and anti-Su(H) antibodies, treated with phosphatase, and the Western blot probed with anti-LexA antibody. As can be seen in Figure 3A, lane 4, the anti-Su(H) immunoprecipitates from NΔEGF1-36–LexA embryos do not contain Np100–LexA. Expression of NΔEGF1-36–LexA will not rescue the neurogenic phenotype of a zygotically N− embryo (data not shown). As a control for this experiment, immunoprecipitations from embryos expressing LexA-tagged N (NLexA) were carried out. In these experiments, a LexA-tagged Np100 protein is seen (Fig. 3A, lane 3, Np100–LexA, indicated by an asterisk, presumably the hypophosphorylated form of Npp114–LexA). Expression of NLexA will rescue the neurogenic phenotype of zygotically N− embryos (data not shown).
Figure 3.
The presence of Notch ligand and the extracellular domain of N are required for the isolation of Np100 bound to Su(H). (A) Detergent extracts from embryos expressing either NLexA (lanes 1,3) or NΔEGF1-36–LexA [lanes 2,4 (see Fig. 1 for structures of NLexA and NΔEGF1-36–LexA] were immunoprecipitated with antibodies against either N (NI) (lanes 1,2) or Su(H) (lanes 3,4) treated with phosphatase, and then probed with antibodies against LexA. Antibodies against N immunoprecipitate NLexA (lane 1) and NΔEGF1-36–LexA (lane 2); the smaller proteins seen in lane 2 are missing from lane 1 and, are most likely nonspecific breakdown products visible because of massive overexpression of NΔEGF1-36–LexA compared with NLexA. NLexA and NΔEGF1-36–LexA are coimmunoprecipitated by anti-Su(H) antibodies as is a smaller, processed form of NLexA (Np100–LEXA, lane 3, *). No such processed protein is seen when the extracellular ligand binding domain of N is deleted (lane 4). (B) Dl temperature-sensitive mutants reduce the level of processed N protein. yw (wt), Dl6B/TM6 (a strong) or DlRF/TM6 (a weak temperature-sensitive Dl allele) males were mated to Dlx/TM6 (an amorphic Dl allele) females. All the embryos resulting from the above crosses were incubated at either room temperature (lanes 1,3,5) or at the nonpermissive temperature, 29°C (lanes 2,4,6). N proteins coimmunoprecipitated from detergent extracts by anti-Su(H) antibody were detected with the NPCR anti-N antibody. Beneath each lane the ratio of processed N to N (termed Npp114/N) is shown. The strong temperature-sensitive Dl allele (lanes 3,4) has a considerably greater effect on the level of processed N than the weaker allele (lanes 5,6)
We then examined the effect of lowering the level of ligand by using temperature-sensitive mutants of the N ligand Delta. Dl6B/TM6 (a strong temperature-sensitive Dl allele) and DlRF/TM6 (a weak temperature-sensitive Dl allele) males were mated to Dlx/TM6 (an amorphic Dl allele) females. All the embryos from this cross were incubated at either the permissive or nonpermissive temperature. Extracts of these embryos were immunoprecipitated with anti-Su(H) antibody. Figure 3B shows that at the nonpermissive temperature, temperature-sensitive Dl alleles reduce the level of processed compared with full-length N bound to Su(H). The ratios of processed to full-length N obtained by scanning this image are shown beneath each lane. The level of reduction produced by each temperature-sensitive Dl allele appears to correlate with the strength of its mutant phenotype. Incubation of the stronger temperature-sensitive allele, Dl6B, at the nonpermissive temperature (Fig. 3, lanes 3,4) reduces the ratio of processed to full-length N from 1.1 to 0.3, whereas the weaker allele, DlRF (lanes 5,6), only reduces the ratio from 1.6 to 0.8. In contrast, in extracts from wild-type embryos, the ratio increases from 2.5 at the permissive to 3.7 at the nonpermissive temperature (lanes 1,2). In extracts of embryos with neurogenic phenotypes produced by expressing antisense m8 RNA or produced by pcx1 parents (Perrimon et al. 1984), there is no decline in the level of processed relative to full-length N (data not shown).
The above experiments indicate that the presence of Npp114 bound to Su(H) is correlated with N function and suggest that ligand binding is required for cleavage of N to generate Npp114 that is bound to Su(H). However, in converse experiments in which the N ligand is overexpressed, a more complex picture emerges. In these experiments, heat shock GAL4 was used to induce expression of UAS Dl in otherwise wild-type embryos. Two hours after induction of Dl, immunoprecipitations with anti-N antibodies reveal that a N protein, the size of Np100B associated with Su(H), has become more abundant than in wild-type extracts or in uninduced UAS Dl embryos (Fig. 4A; cf. the samples of lanes 1, 3, and 5, which are unphosphatased immunoprecipitations). The increase in abundance of this protein is most obvious when comparing wild-type extracts with extracts from heat shock-induced UAS Dl embryos (Fig. 4A, lanes 1, 5). Because of the leakiness of the heat shock promoter, some of this protein can be seen in extracts from uninduced UAS Dl embryos (Fig. 4A, lane 3). The mobility of most of the protein produced by UAS Dl induction does not change significantly on phosphatase treatment, suggesting little or no phosphorylation (Fig. 4A, lanes 5, 6).
Figure 4.
Ligand-induced cleavage of N. (A) Anti-N antibodies immunoprecipitate a N protein, the size of Np100B (*) from embryos exposed to ectopic Dl (lane 5). Embryos aged from 2 to 4 hr were heat-shocked at 37°C for 30 min and allowed to recover at 30°C for 2 hr prior to detergent extraction. Extracts from either heat-shocked yw (lanes 1,2) or heat-shocked GAL4; UAS Dl with (lanes 5,6), or without heat shock (lanes 3,4) were immunoprecipitated with anti-N NI antibody. Lane 7 lacks embryo extract; lane 8 contains NIntra1768 in vitro translation products. After immunoprecipitation, lanes marked with a + were treated with alkaline phosphatase prior to electrophoresis. N proteins were detected with the anti-N antibody NPCR. (Labels) Locations of N (N) and NIntra1768(NIntra). (B) Coexpression of NLexA and Dl results in an increase in the level of Npp114–LexA. Detergent extracts from 4–8 hr UASNlexA × hGAL4 embryos (lanes 1,2,5,6) or UAS Dl30B; UAS NlexA × hGAL4 embryos (lanes 3,4,7,8) were immunoprecipitated with either anti-N antibody (NI) (lanes 1–4) or anti-Su(H) antibody (lanes 5–8). Samples in lanes 2, 4, 6, and 8 were treated with alkaline phosphatase prior to electrophoresis. After blotting, proteins were detected with an anti-LexA monoclonal antibody. Coexpression of Dl and NLexA causes the appearance of a novel hypophosphorylated NLexA protein in the anti-N immunoprecipitations (*, cf. lanes 1 and 3). In this experiment, there was an ∼40%–50% increase in the amount of Npp114–LexA bound to Su(H) (cf. the ratios of Npp114LexA to NLexA in lane 5 with lane 7, and in lane 6 with lane 8). (C) A histogram comparing the relative amounts of processed vs. full-length N associated with Su(H) in the absence or presence of ectopic Dl. The average of the results of four immunoprecipitations are plotted. The fold increase in the amount of processed N compared with full-length N in the presence of UAS Dl for each of the four experiments was as follows: 1.7, 1.5, 2.0, and 1.8.
Although ligand binding is required for production of Npp114 associated with Su(H) (see above), the pool of N protein coimmunoprecipitated by anti-Su(H) on overexpression of the ligand Dl did not appear to increase (data not shown), suggesting that most of this Dl-induced protein has not become associated with Su(H). We reasoned that we were more likely to see an increase in processed N associated with Su(H) if we induced a tagged form of N at the same time as Dl. In this way, rather than looking for a change superimposed on the steady state level N, we would be looking for a change only in the tagged N synthesized at the same time as Dl. Therefore, we compared the amount of processed NLexA bound to Su(H) in extracts of embryos in which NLexA alone was induced by hairy GAL4 with the amount of processed NLexA bound to Su(H) in extracts of embryos in which Dl was induced along with NLexA. The results are shown in Figure 4B. In this experiment, coexpression of NLexA with Dl results in a 42% (the phosphatased samples of Fig. 4B, lanes 6, 8) or 52% (unphosphatased samples of Fig. 4B, lanes 5, 7) increase in the level of Npp114–LexA associated with Su(H) when compared with NLexA expressed in the absence of additional Dl. A histogram summarizing the results of four immunoprecipitations from three protein preparations is shown in Figure 4C. On average, there is a 1.75-fold increase in the amount of processed compared with full-length N associated with Su(H). In addition, as was the case with overexpression of Dl in wild-type flies, overexpression of Dl along with NLexA results in the production of a hypophosphorylated N protein the size of Np100B fused to LexA that is immunoprecipitated with anti-N antibody but does not associate with Su(H) (cf. the unphosphatased samples of Fig. 4B, lanes 1,3).
Intracellular location of the Su(H) bound N proteins
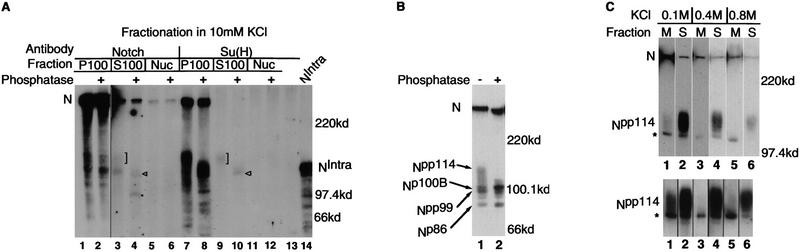
The three dephosphorylated components of Npp114 are small enough to be soluble. To see if this was the case, immunoprecipitations were carried out on subcellular fractions of Drosophila embryos. Equal fractions of each subcellular fraction were immunoprecipitated to allow the relative abundance of the proteins in each fraction to be determined. When the fractionation is carried out under hypotonic conditions (10 mm KCl), the majority of N proteins, full-length as well as Npp114, immunoprecipitated by both N and Su(H) antibodies are in the membrane fraction (Fig. 5A, lanes 1,2,7,8). Some Npp114 is found in the soluble fraction (Fig. 5A, lanes 3,4,9,10) and little or no N is detectable in the nuclear fraction (Fig. 5A, lanes 5, 6, 11, 12). In addition to Npp114, the anti-N immunoprecipitate of the soluble fraction is also enriched for two proteins of ∼99 kD and 86 kD, which are superimposed over an 86–114 kD smear (Fig. 5, A, lane 3, and B, lane 1). Phosphatase treatment reduces the smear to the hypophosphorylated components of Npp114 and a protein of 86 kD (termed Np86) (Fig. 5, A, lane 4 and B, lane 2) suggesting that Npp99 is a phosphorylated form of Np86. Despite being soluble, Npp99 was not found associated with Su(H) (Fig. 5A, cf. lanes 3 and 4 with lanes 9 and 10).
Figure 5.
Npp114 is a soluble protein. (A) Subcellular distribution of Npp114. Subcellular fractions were prepared from Drosophila embryos that had been lysed under hypotonic conditions (10 mm KCl). (Nuc) The nuclear fraction; (P100) the membrane fraction; (S100) the soluble fraction. The position of Npp114 is indicated by a square bracket, Np100B by an arrowhead, and Np86 by a dot. Equal proportions of each fraction were immunoprecipitated with antibodies against either N (NI) (lanes 1–6) or Su(H) (lanes 7–12) and then detected with the anti-NPCR antibody. The autoradiograph has been overexposed to show the presence of processed N in the soluble fraction. Because the amount of N in the membrane fraction is so high, lanes 1 and 2 are from a shorter exposure. (B) Evidence for additional processing of the cytoplasmic domain of N. Soluble proteins extracted in 0.4 m KCl were immunoprecipitated with anti-NI antibody and detected with anti-NPCR antibody. As well as immunoprecipitating the same N proteins as those coimmunoprecipitated by anti-Su(H) antibodies, anti-N antibodies immunoprecipitate two novel proteins of ∼99 kD (Npp99) and 86 kD (Np86) (lane 1), treatment with alkaline phosphatase (lane 2) reduces the amount of Npp99 and increases the amount of Np86, suggesting that Npp99 is a phosphorylated form of Np86. (C) Some Npp114 is associated, but not stably, with membranes. Postnuclear supernatants were incubated with increasing amounts of KCl prior to fractionation by centrifugation into membrane bound (lanes marked M) or soluble proteins (lanes marked S). Fractionated proteins were then immunoprecipitated with anti-Su(H) antibody and detected with the anti-N NPCR antibody, a longer exposure of the relevant region of the resulting autoradiograph is shown at bottom. Some Npp114 can be seen to be associated with membranes when extracted at 0.1 m KCl but is removed at higher salt concentrations. The protein that comigrates with Np100B and appears to be stably associated with membranes is marked by an asterisk.
In contrast, when subcellular fractionations are carried out under physiological salt conditions (100 mm KCl) the majority of Npp114 associated with Su(H) is found in the soluble fraction (Fig. 5C, lane 2). Although there is a significant amount of Npp114 still found associated with the membrane fraction (Fig. 5C, lane 1), incubation of the post-nuclear supernatant in increasing concentrations of KCl prior to centrifugation disrupts the association of Npp114 with membranes (Fig. 5C, lanes 3,5). These results support the notion that Npp114 is the soluble, phosphorylated intracellular domain of N. In conjunction with the results we have presented above, this suggests that on binding its ligand Dl, transmembrane N is cleaved, generating a soluble form that encompasses its cytoplasmic domain, is phosphorylated, and associates with Su(H).
As well as coimmunoprecipitating Npp114, anti-Su(H) antibodies also precipitate a N protein the size of hypophosphorylated Np100B from the membrane fraction (indicated by an asterisk in Fig. 5C). The same salt conditions which disrupted the association of Npp114 with membranes do not do so with this protein, suggesting a stronger association with the membrane.
Su(H) is capable of retaining the cytoplasmic domain of N in the cytoplasm
The biochemical data presented above suggest that some processed N complexed with Su(H) protein is still associated with membranes even though, on the basis of size, it probably lacks a transmembrane domain. In addition, there are larger amounts of N, principally Npp114, in the soluble fraction. This is surprising, as we and others have shown previously that the intracellular domain of N has functional nuclear localization signals and can localize to nuclei (Stifani et al. 1992; Lieber et al. 1993).
Given our fractionation studies of Npp114, subcellular localizations of NIntra were further examined. Whereas in S2 cells NIntra1768 (Fig. 1) is totally nuclear (Fig. 6A), in embryos, a substantial fraction of NIntra1768 is retained in the cytoplasm. This is illustrated in Figure 6B by use of an anti-Flag-antibody to recognize Flag-tagged NIntra1768. Using an anti-N antibody, we found that it is in cells in which NIntra1768 is expressed at higher levels that it is found in nuclei. In cells in which NIntra1768 is expressed at lower levels, it is primarily cytoplasmic (Fig. 6C). This suggests that there is something in embryos that is retaining NIntra1768 in the cytoplasm, and that this retention mechanism can be saturated by high levels of NIntra1768. It has been shown that the cdc10 repeats of N can mediate homotypic N interactions (Roehl et al. 1996; Matsuno et al. 1997). However, in embryos that are both maternally and zygotically N null, there is still substantial cytoplasmic localization of NIntra1768 (Fig. 6D).
Figure 6.

NIntra is retained in the cytoplasm in embryos. Confocal images showing the localization of NIntra in embryos and Drosophila S2 cells as detected by immunofluorescence. With the exception of the left panel in C, NIntra protein is represented in green and the nuclei in red. This panel is a pseudocolored representation to illustrate the relative amounts of NIntra1768. The correspondence of the colors with the intensity of the signal is indicated by the pseudocolor bar, with the more intense signals being depicted by colors higher up the bar. (A) Heat shock-induced NIntra1768 appears to be totally nuclear in S2 cells. UAS NIntra1768 was cotransfected along with HS GAL4 and detected with a rabbit anti-N (NI) antibody. The nucleus was detected with SYTOX Green. (B,C) In embryos (hGAL4; UAS Nintra1768) there is retention of NIntra1768 in the cytoplasm. (B,C, right) The merged images of the N signal in green and the nuclei in red. (C, left) NIntra1768 is primarily nuclear in those cells in which it is most highly expressed. Mouse anti-NPCR antibody was used to detect NIntra1768. (B) Mouse anti-Flag antibody was used to detect Flag-tagged NIntra1768. The nuclei were detected by propidium iodide. (D) NIntra1768 is still retained in the cytoplasm in embryos that are maternally and zygotically N null (N264–47 FRT/ovoD FRT; hGAL4/hsflp X FM7/Y; Nintra1768. (D, left) Mouse NPCR antibody was used to detect NIntra1768; (D, right) a merged image of N (green) and nuclei (red). (E) NIntra1790 is predominantly nuclear in embryos with reduced levels of Su(H). Anti- myc antibody was used to detect myc-tagged Nintra1790 in embryos that are maternally Su(H)− (hsflp/yw;Su(H)SF8 FRT/ovoD FRT;hGAL4 X Su(H)/CyO; UASNIntra1790). In embryos that are maternally Su(H)+ (hGAL4 X Su(H)/CyO;UASNIntra1790) there is retention of NIntra1790 in the cytoplasm (data not shown).
An obvious candidate for a factor influencing subcellular localization of NIntra in the absence of transmembrane N is Su(H). In early embryos, Su(H) is present ubiquitously and localizes to both cytoplasm and nuclei (data not shown). It has been shown in wing discs that Su(H) is present in the cytoplasm, and that when NIntra is expressed to high levels, it is capable of dragging endogenous Su(H) into nuclei (Gho et al. 1996). Although this is true for high levels of NIntra1768 (Fig. 7, cf. A with B and C), when low levels of NIntra1768 are coexpressed with Su(H) in S2 cells, NIntra1768 is retained in the cytoplasm (Fig. 7D–G). Thus, raising the relative level of Su(H) favors cytoplasmic localization of NIntra. In accord with this observation, NIntra1790 is predominantly nuclear in embryos with reduced levels of Su(H) (Fig. 6E).
Figure 7.
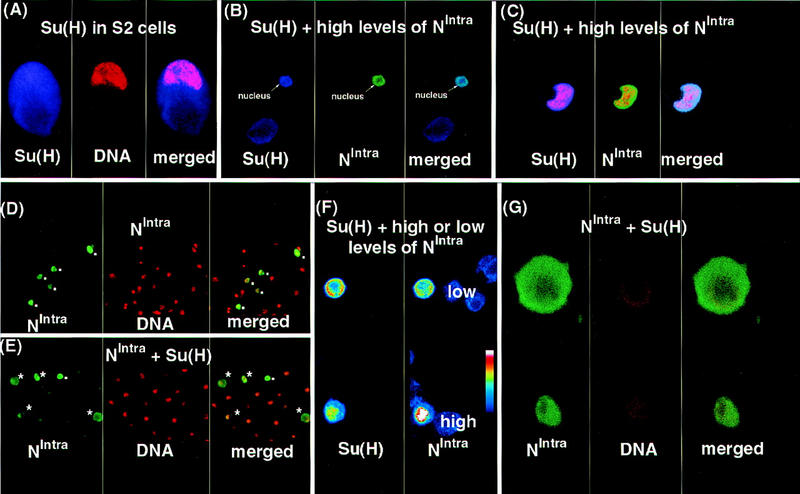
Su(H) can retain NIntra in the cytoplasm. Confocal images showing the localization of Su(H) and NIntra1768. With the exception of F, which is a pseudocolored image, Su(H) is represented in blue, NIntra in green, and the nuclei in red. (A) Su(H) is expressed in both the cytoplasm and nuclei of transfected S2 cells. S2 cells were transfected with a construct expressing Su(H) under the control of an actin promoter. Su(H) is detected with rat anti-Su(H) antibody and the DNA with SYTOX Green. (B,C) Coexpression of high levels of NIntra1768 along with Su(H) results in both being found in nuclei of S2 cells. (B) Two cells are depicted showing Su(H) expression in blue and N expression in green. In the lower cell, no NIntra1768 is present and Su(H) is localized ubiquitously. In contrast, the upper cell expresses high levels of NIntra1768 along with Su(H) and both are localized in the nucleus. A merged image of the first two panels is shown at right. The cell shown in C was probed with a DNA marker as well as with anti-N and Su(H) antibodies. Actin driven Su(H) was cotransfected with UAS NIntra1768 and HS GAL4. Su(H) protein was detected by rat anti-Su(H) antibody, N protein was detected by a mouse anti-Flag antibody, and the DNA with SYTOX Green. (D,E,G) The difference in localization of NIntra1768 promoted by expression of an excess of Su(H). NIntra is represented in green and the nuclei in red. (D) UAS NIntra1768 and HS GAL4 were transfected into S2 cells. (E,G) A 20× mass excess of actin Su(H) was cotransfected along with UAS NIntra1768 and HS GAL4. (D,E) Localization of NIntra1768 in nuclei is indicated by dots and retention in the cytoplasm by asterisks. Two cells from those shown in E are depicted at higher magnification in G. As cytoplasmic localization of NIntra1768 is never seen in cells lacking ectopic Su(H) (D), the upper cell in G must have received more Su(H) relative to NIntra1768 than the lower one, resulting in NIntra1768 being retained in the cytoplasm. NIntra1768 was detected with a rabbit anti-N (NI) antibody and the nuclei with SYTOX Green. (F) When low levels of NIntra1768 are expressed along with Su(H), NIntra1768 is retained in the cytoplasm. A pseudocolored confocal image showing the relative levels on N at right and Su(H) at left is portrayed. The intensity of staining is depicted by the pseudocolor bar with the colors representing the more intensely stained regions being higher up the bar. In the lower cell, NIntra1768 is expressed at relatively high levels and both NIntra1768 and Su(H) are found in the nucleus. In the upper cell, NIntra1768 is expressed at relatively low levels and both NIntra1768 and Su(H) are found in the cytoplasm. When cells are doubly stained for DNA and NIntra1768, the two stains converge only when levels of NIntra1768 are high compared with Su(H) (data not shown). Su(H) was detected with a rat anti-Su(H) antibody and NIntra1768 with a mouse anti-Flag antibody.
The cytoplasmic domain of N behaves as an activator when bound to DNA
The data we have presented above indicate that N is processed and associates with Su(H), and the entry of this complex into the nucleus appears to be dependent on the relative levels of processed N and Su(H). During the course of yeast two-hybrid experiments, it was found that the cytoplasmic domain of N was a strong activator. Figure 8A shows a comparison in yeast of the activating ability of the N cytoplasmic domain with that of the well-characterized transcriptional activator GAL4. It can be seen that the cytoplasmic domain of N has almost as much activator activity (85%) as GAL4. Smaller derivatives of the N cytoplasmic domain activate to a lesser degree. Thus, in a heterologous system, the cytoplasmic domain of N strongly activates transcription from a heterologous promoter. This suggests that at least one aspect of N function could be mediated by its ability to act as a transcriptional transactivator for Su(H). We tested this in two ways. First, we fused the DNA-binding domain of the bacterial repressor LexA to the cytoplasmic domain of N. In S2 cells, this NLexA fusion protein (NIntra–LexA; Fig. 1), but not NIntra1790, activates transcription from a LexA reporter (data not shown). In Figure 8D, we show that in embryos, NIntra–LexA can activate transcription from a LexA–β-galactosidase reporter. The pattern of expression of NIntra–LexA is presented in Figure 8C, and coincides well with the pattern of induced β-galactosidase reporter. Figure 8B shows that NIntra1790 alone, although expressed in the same pattern as NIntra–LexA (data not shown), cannot activate transcription of the LexA reporter. This experiment indicates that in vivo, when N is directly tethered to DNA, it behaves as a transcriptional activator and suggests that the role of Su(H) is to guide a transcriptional activator to DNA.
Figure 8.
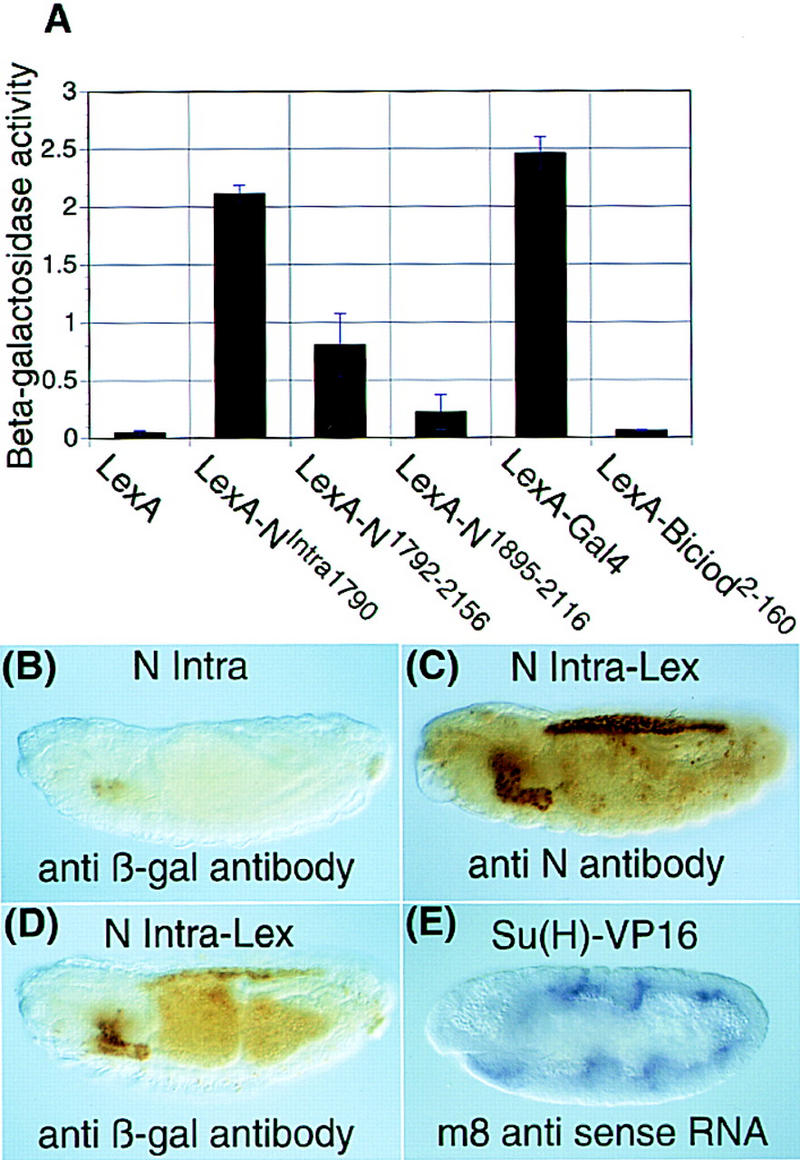
The cytoplasmic domain of N can behave as a transcriptional activator. (A) The cytoplasmic domain of N is a transcriptional activator in yeast. The transcriptional activation activity of various N constructs fused to the DNA-binding domain of LexA (Fig. 1) was determined by their ability to drive expression from a LexA–β-galactosidase reporter. The bar marked LexA is the β-galactosidase activity of yeast expressing the vector alone, LexA–Bicoid2-160 is a negative control. The height of the bar is the average of three samples, standard deviations are shown by the error bars. (B) NIntra1790 cannot activate transcription from LexA–β-galactosidase reporter. A lexA–β-gal; UAS Nintra1790; HS GAL4 embryo is stained with anti β-galactosidase antibody. There is anti β-galactosidase reactivity in the secretory cells and the anal pads that results from leakiness of the reporter. (C) NIntra1790–LexA accumulates to its highest levels in the salivary glands, amnioserosa, and midgut. A lexA–β-gal; UAS Nintra1790–lexA; HS GAL4 embryo is stained with anti-NPCR antibody. (D) NIntra1790–LexA induces expression of β-gal from the LexA–β-gal reporter. A lexA–β-gal; UAS Nintra1790–LexA; HS GAL4 embryo is stained with anti β-galactosidase antibody. β-Galactosidase accumulates in the salivary glands, amnioserosa and midgut, which correspond with regions that accumulate the highest levels of NIntra1790–LexA in C. (B,C,D) Embryos were fixed 2 hr after a 30-min heat shock. (E) A heterologous activator fused to Su(H) can substitute for N function and activate transcription of m8. N264-47 FRT/ovoD FRT; h GAL4/hsflp × FM7 lac-Z/Y; myc Su(H)–VP16 embryos are stained with an m8 probe. m8 expression is induced in seven stripes, in which h is expressed. Overexpression of Su(H) alone does not result in induction of m8 expression (data not shown). Anti β-galactosidase antibody was used to distinguish the N null embryos.
If N is functioning as a transcriptional transactivator, one would predict that a transcriptional activator directly coupled to Su(H) could substitute for at least some aspects of N function. To test this, we fused the viral activator VP16 to Su(H) (Fig. 1). In S2 cells this Su(H)–VP16 fusion, but not Su(H), activates transcription from an m8 reporter (data not shown). In Figure 8E we show that this Su(H)–VP16 fusion but not Su(H) (data not shown) can activate m8 transcription in an embryo that is both maternally and zygotically N−. This experiment demonstrates that a role of N is to either directly or indirectly provide activator function to Su(H).
Discussion
Previous work has led to the model that on ligand binding, N is cleaved, and the cytoplasmic domain enters the nucleus where, in concert with Su(H), it activates transcription of genes such as m8, a member of the E(spl) complex (Lieber et al. 1993; Jarriault et al. 1995; Kopan et al. 1996; Lecourtois and Schweisguth 1998; Schroeter et al. 1998; Struhl and Adachi 1998). In this study, it has been shown that (1) soluble cytoplasmic N proteins are produced in vivo in response to the N ligand, Dl (2) Su(H) is recovered in association with these soluble forms of N, and (3) intracellular forms of N appear to function as transcriptional activators in embryos when physically associated with Su(H).
We have used antibodies against Su(H) and N to examine the structure of the N proteins associated with Su(H). During most of Drosophila embryogenesis, two size classes of N proteins are coimmunoprecipitated by antibodies against Su(H). These include full-length N proteins and, to a greater extent, phosphoproteins of ∼114-kD, Npp114. Unlike mammalian systems in which N exists predominantly as a heterodimer, during Drosophila embryogenesis, the bulk of N exists as the full-length form (Results; Kidd et al. 1989; Blaumueller et al. 1997). When dephosphorylated, Npp114 resolves into three proteins, Np100A, Np100B, and Np100C of ∼100 kD. Through most of embryogenesis, the most abundant of these proteins is Np100B, Np100C being found only late in development. The size difference between the two proteins might be because Np100C has been cleaved further into the intracellular domain than Np100B, or the two proteins may both have the same amino termini, but Np100C might have been additionally cleaved at the carboxyl terminus. It is also possible that there is a precursor product relationship between the two. In any case, the occurrence of Np100C only late in embryogenesis suggests that production of these forms of N is under developmental control.
Throughout most of embryogenesis, the majority of processed N proteins that are associated with Su(H) show some level of phosphorylation. Full-length N has been shown previously to be phosphorylated on serines (Kidd et al. 1989). We do not know how the latter relates to the phosphorylation described here, although the presence of hypophosphorylated forms of N bound to Su(H) suggests that the two events are unrelated. How this phosphorylation is effected and how it influences N function is not known. There are two lines of evidence that suggest that phosphorylation is not an immediate consequence of ligand binding and cleavage. First, most if not all of NIntra1790, none of which has been produced as a result of ligand binding and cleavage of N, is phosphorylated (Fig. 2C). Second, overexpression of Dl induces at least one processed form of N which is hypophosphorylated (Fig. 4). In addition, we have shown that phosphorylation of NIntra1790 is not dependent on the presence of Su(H) (Fig. 2C). Because most, if not all, of NIntra1790 is phosphorylated and there is an enrichment of Npp114 in the soluble fraction (Fig. 5), perhaps phosphorylation is related to the release of cleaved intracellular N from the membrane. Alternatively, phosphorylation may promote nuclear translocation or association with Su(H), or both.
There is some salt extractable Npp114 associated with Su(H) in the membrane fraction. Finding the intracellular domain of N, which contains functional nuclear localization signals either in the membrane or cytoplasmic fractions, indicates that the cell contains mechanisms to restrain the nuclear entry of N cleavage products. Because it has been demonstrated that the cdc10 repeats of N mediate homodimerization (Matsuno et al. 1995; Roehl et al. 1996), newly produced intracellular forms of N may be retained by full-length forms of N at the membrane. This association might be particularly favored if, as believed, the receptor is presented at the cell surface as a dimer (Foster 1975; Portin 1975; Kelley et al. 1987; de Celis and Garcia-Bellido 1994). It is also conceivable that Npp114 is retained on the membrane by a complex of Su(H) and full-length N.
Su(H) may regulate nuclear entry of N
With respect to cytoplasmic retention of Su(H)/Npp114 complexes, regulation may come from Su(H) itself. We have shown that whereas coexpressing high levels of NIntra along with Su(H) in S2 cells results in both proteins translocating to nuclei, when low levels of NIntra are coexpressed along with Su(H) in S2 cells, there is retention of NIntra in the cytoplasm. This suggests that excess Su(H) can promote cytoplasmic localization of soluble, intracellular forms of N. Given that there are multiple binding sites for Su(H) in the cytoplasmic domain of N (Kato et al. 1997; Wettstein et al. 1997; S. Kidd, unpubl.), differences in subcellular localization could reflect the number of Su(H) molecules bound to N, with changes in stoichiometry resulting from increased levels of intracellular N in response to ligand. Because in vivo levels of Su(H) appear to be in excess to soluble N product, as there is sufficient Su(H) to bind to ectopically expressed NIntra and generate gain of function phenotypes (Lieber et al. 1993; Rebay et al. 1993; Struhl et al. 1993), the cytoplasmic retention we observe in Su(H)+ embryos is expected from the S2 cell studies. Further supporting our view that Su(H) can retain soluble N in the cytoplasm in vivo, we found that lowering the dose of Su(H) promotes nuclear localization of NIntra in embryos (Fig. 6E). We also find that lowering the dose of Su(H) increases the severity of the phenotype produced by ectopic expression of gain-of-function N proteins in transgenic flies: Whereas complete loss of Su(H) abolishes the ability of the E(spl) complex to respond to activated N (Bailey and Posakony 1995), lowering the Su(H) dose by one-half increases the lethality as well as the bristle loss observed in transgenic flies carrying NΔLNrpts under control of a heat shock promoter (T. Lieber, unpubl.). A priori, one would have predicted that lowering the dose of a downstream component in the N pathway would decrease the severity of gain-of-function N mutations. Lastly, it is possible that the subcellular distribution of Su(H)/Npp114 complexes is regulated by interaction with additional factors. For example, it has been shown that numb, a membrane-associated protein that is asymmetrically localized during division of sensory organ precursor cells in the peripheral nervous system, is able to retain NIntra at the membrane and in the cytoplasm of S2 cells (Frise et al. 1996).
In the absence of Su(H), both NIntra and N appear to have undergone additional modification. In addition, many intermediately sized N proteins are missing (Fig. 2C). Lecourtois and Schweisguth, (1998) and Schroeter et al. (1998) have suggested that the processed form of N is less stable in the absence of Su(H). Many proteins are targeted to the proteosome by ubiquitinylation. Perhaps the modification of NIntra and N we see in the absence of Su(H) is ubiquitinylation. Interestingly, phosphorylation has also been shown to target proteins to the ubiquitinylation machinery (King et al. 1996).
In addition to being required for the production of Npp114, the N ligand Dl, when overexpressed, promotes accumulation of a hypophosphorylated N protein that has approximately the same mobility as Np100B (Fig. 4A). Our fractionation studies also showed the presence of a hypophosphorylated protein of approximately the same size as Np100B, in this case associated with Su(H). This protein is retained in the membrane fraction under salt conditions that remove Npp114, suggesting that it is tightly associated with the membrane and may well span it (Fig. 5C). The extracellular domain of N has been shown previously to be cleaved at several positions (Blaumueller and Artavanis-Tsakonas 1997; Pan and Rubin 1997; Logeat et al. 1998). It has been proposed that the cleavage closest to the membrane is ligand dependent (Logeat et al. 1998). Such a cleavage product may correspond to the protein we described above.
Soluble N as a transcriptional transactivator
The work of Lecourtois and Schweisguth (1997) and Struhl and Adachi (1998) has shown genetically that the cytoplasmic domain of N has access to the nucleus. The most likely explanation for their results is that Drosophila N is proteolytically cleaved at the site described by Schroeter et al. (1998) to produce the fragment of N, Npp114, that we have described in this paper. We have shown that when tethered directly to DNA via a bacterial DNA-binding domain, the cytoplasmic domain of N can activate transcription both in yeast and in vivo. Conversely, a viral activator fused to Su(H) can substitute for the functions of N mediated by its ability to activate transcription of m8, a natural target of N signaling, in embryos. Whereas maximal activation in yeast is seen with the entire cytoplasmic domain, in agreement with the results of Roehl et al. (1996) a truncated form of the cytoplasmic domain (N1792–2156; Fig. 1) encompassing the cdc10 repeats does weakly activate and has a gain-of-function phenotype in embryos (T. Lieber, unpubl.). Smaller versions of the cytoplasmic domain (N1895–2156) spanning just the cdc10 repeats are even weaker activators and when expressed in wild-type embryos do not have a gain-of-function phenotype (T. Lieber, unpubl.). Our data suggest that the prime function of the sequences downstream of the cdc10 repeats is to provide transactivator activity. In accord with this, the cytoplasmic domain of N has many features that are found in transcriptional activators (Lieber et al. 1993). Although it is possible that N indirectly confers activating ability on Su(H), given the finding of appropriately processed N proteins, which contain functional nuclear localization signals preferentially associated with Su(H), the simplest interpretation of our results is that one function of N is to bind to Su(H) and in the nucleus to directly act as its transcriptional transactivator. Recently it has been suggested that N activates transcription by disrupting the formation of a repressor complex between Su(H) and a histone deacetylase complex (SMRT/HDAC-1) (Kao et al. 1998). Our data suggest that rather than simply disrupting the Su(H)/SMRT/HDAC-1 complex, Npp114 plays a more active role of providing transactivator activity to Su(H).
One other class of membrane-bound transcription factors has been identified previously. The proteolysis of sterol regulatory element binding proteins (SREBPs) (for review, see Brown and Goldstein 1997) is regulated by sterols that accumulate in membranes. As N like molecules have been found in all multicellular organisms where they have been sought, N is an evolutionarily old protein. The existence of a transcription factor that spans the membrane with an extracellular domain capable of interacting with ligands and an intracellular domain that can enter nuclei and activate transcription would provide a simple means for transducing information from neighboring cells. Possibly, the only additional components required would be a protease capable of recognizing a conformational change induced in N on ligand binding resulting in its cleavage, and a second protease that would degrade the cytoplasmic domain in nuclei so that the signaling could be terminated.
Establishing a threshold for Notch signaling
The binary epidermal versus neural cell fate choice mediated by the N signaling pathway involves regulating groups of initially equivalent cells that express both ligand and receptor. Schroeter et al. (1998) have shown that in vertebrate cell culture, extremely low levels of nuclear N are sufficient for function, and our studies of the wild-type Drosophila embryo are consistent with this finding in that no N is detected in the nucleus either biochemically or by immunofluorescence. However, relatively abundant cleaved N associated with Su(H) is detected in the cytoplasm. Why should there be such a disparity between levels of soluble N in the cytoplasm and nucleus, and why shouldn’t such a potent nuclear N signal favor saltatory cell fate decisions, with all cells composing an equivalence group assuming the same secondary cell fate? Uniform expression of ligand and receptor among interacting cells might also be expected to favor a saltatory response.
Some of the puzzling aspects of N signaling are reminiscent of ultrasensitive systems such as the Xenopus oocyte system described by Ferrell and Machleder (1998), in which a continuously variable signal, progesterone, is converted into an all-or-none response, oocyte maturation. An ultrasensitive system exhibits little response to low levels of stimulus but switches from off to on over a narrow range of stimulus concentration. We suggest that the cytoplasmic retention of Npp114/Su(H) complexes described in the present study may similarly reflect a mechanism in which the response to low levels of signal is damped. In such a model for N signaling, only high levels of signal result in sufficient cytoplasmic accumulation of Npp114/Su(H) complex to permit its nuclear entry.
In the Xenopus oocyte system, added ultrasensitivity is provided by a positive feedback loop. Earlier genetic studies have suggested that small differences in the expression of N and Dl may also be amplified by positive feedback to generate robust intercellular differences in the expression of these proteins among cells derived from an equivalence group (Seydoux and Greenwald 1989; Heitzler and Simpson 1991; for review, see Greenwald 1998). By affecting the nuclear entry of N/Su(H) complex, which functions as a transcriptional activator, Su(H) would also be an element of such a feedback mechanism. We suggest that the N signal is initiated and maintained according to the relative amounts of N, Dl, and Su(H). Together, these would determine the rate and duration of accumulation of N/Su(H) complex and the threshold at which it enters the nucleus.
Materials and methods
Constructs
NIntra1768 was expressed by cloning into a derivative of pUAST (Brand and Perrimon 1993) which contains the cactus initiation codon fused to a Flag epitope (Kidd 1992). To express the cytoplasmic domain of N by coupled in vitro transcription/translation (Promega), a derivative of NIntra1768 was made in which the region containing the last three introns of N was replaced with the corresponding segment of cDNA. NIntra1790 contains the first two amino acids of cactus followed by a 14 amino acid myc epitope (Xu and Rubin 1993), which was then fused to amino acid 1790 of N. The hsNIntra 1790 used in Fig. 2C is NIntracellular domain (Lieber et al. 1993).
NLexA has amino acids 1–87 of LexA fused to the carboxyl terminus of N. NΔEGF1-36–LexA is a derivative of the above lacking the 36 EGF repeats. NIntra1790–LexA has amino acids 1–87 of LexA fused to the carboxyl terminus of NIntra1790. The LexA–β-galactosidase, reporter has eight LexA operator sites (Ebina et al. 1983) upstream of a heat shock minimal promoter. This was then inserted in place of the GAL4–UAS region of pUAST (Brand and Perrimon 1993).
Myc-tagged Su(H) contains the first two amino acids of cactus followed by a 14 amino acid myc epitope that was then fused to amino acid 10 of Su(H) (Schweisguth and Posakony 1992). Myc-tagged Su(H)–VP16 has amino acids 19–105 of VP16 (Campbell et al. 1984) fused to the carboxyl terminus of myc Su(H).
With the exception of LexA–β-gal, all constructs were subcloned into pUAST (Brand and Perrimon 1993) for transformation into flies.
Yeast expression experiments were carried out as described by (Gyuris et al. 1993).
Fly stocks
The following fly stocks, w ovoD1 FRT101; hsFLP, yw FRT101 (Chou et al. 1993), hsFLP12; Sco/Cyo, ovoD1 FRT40A/Cyo (Chou and Perrimon 1996), h-GAL4, hs-GAL4 (Brand and Perrimon 1993) were obtained from A. Brand and N. Perrimon; Su(H) (FlyBase 1998) was obtained from the Bloomington Stock Center; Su(H)SF8 FRT40A/CyO (Schweisguth and Posakony 1994) was obtained from F. Schweisguth; Dl6B and DlRF (Parks and Muskavitch 1993) were obtained from M. Muskavitch, UAS–Dl, (Doherty et al. 1996) was obtained from Y. Jan. N264–47 and Dlx are described by (FlyBase 1998).
Antibodies
Antibodies were raised against histidine-tagged Su(H) as previously described (Kidd et al. 1986; Lieber et al. 1993). The remaining N antibodies (shown in Fig. 1) have been described previously (Lieber et al. 1993). Anti-LexA monoclonal antibody was from Clonetech. M5 anti-Flag antibody was from Kodak. c-Myc antibody was from Calbiochem. SYTOX Green used to label S2 cell nuclei was from Molecular Probes. Immunocytochemistry and immunofluorescence was as described previously (Lieber et al. 1993). Double labeling with RNA and antibody was as described by Azpiazu and Frasch (1993).
Immunoprecipitations
Embryo extractions and immunopreciptations were essentially as described by Kidd (1992). Between 300 μg and 1 mg of protein were used for immunoprecipitation with anti-Su(H) antibodies, one-fifth of this amount was used with anti-N antibodies. Immunoprecipitions were carried out overnight with protein A–Sepharose and Gamma Bind (Pharmacia) to collect rabbit and rat and mouse antibodies, respectively. After washing, the immunoprecipitates were treated with alkaline phosphatase (Boehringer Mannheim) as described previously (Kidd 1992) and electrophoresed without further washes. After blotting, N in rabbit anti-N and rat anti-Su(H) immunoprecipitates was detected with mouse anti-N, Su(H) in mouse anti-N immunoprecipitates was detected with rat anti-Su(H). Horseradish peroxidase (HRP) conjugated secondary antibodies were from Jackson. HRP activity was detected by the ECL system (Amersham).
Scanned autoradiographs were quantitated on a Macintosh computer with the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).
Two procedures were used to produce subcellular fractions of embryos. The first, used for Figure 5A, was based on procedures for producing extracts for gel shifts (Andrews and Faller 1991). The second procedure, used for Figure 5, B and C, was as follows: Dechorionated embryos were extensively homogenized in 10 mm HEPES (pH7.6), 100 mm KCl, 50 mm NaF, 2 mm EDTA, 2 mm EGTA, and 2 mm ammonium molybdate with protease inhibitors. After centrifuging the homogenate at 900g for 5 min, the resulting postnuclear supernatant was incubated on ice for 20 min either with no additional salt or an additional 0.4 or 0.8 m KCl, and then centrifuged for 2 hr at 100,000g. The supernatants were adjusted to 0.5% Triton X-100 and to ∼400 mm KCl, and the pellets resuspended in the Triton lysis solution prior to immunoprecipitations.
Acknowledgments
We thank A. Brand, N. Perrimon, M. Muskavitch, F. Schweisguth, and Y. Jan for fly stocks, F. Schweisguth, and K. Nakao for DNAs, Krishna Patel, Marla Abodeely, and Yvonne DeLotto for excellent technical assistance, and reviewers for insightful comments. This work was supported by National Institutes of Health grant GM25103 to M.W.Y.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL young@rockvax.rockefeller.edu; FAX 212-327-8695.
References
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. Tinman and bagpipe: Two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes & Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Campbell ME, Palfreyman JW, Preston CM. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Garcia-Bellido A. Modifications of the Notch function by Abruptex mutations in Drosophila melanogaster. Genetics. 1994;136:183–194. doi: 10.1093/genetics/136.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes & Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- Ebina Y, Takahara Y, Kishi F, Nakazawa A, Brent R. LexA protein is a repressor of the colicin E1 gene. J Biol Chem. 1983;258:13258–13261. [PubMed] [Google Scholar]
- Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- FlyBase. FlyBase: A Drosophila database. Nucleic Acid Res. 1998;26:85–88. doi: 10.1093/nar/26.1.85. http://flybase.bio.indiana.edu/ http://flybase.bio.indiana.edu/. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The Suppressor of Hairless protein participates in Notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Foster GG. Negative complementation at the Notch locus of Drosophila melanogaster. Genetics. 1975;81:99–120. doi: 10.1093/genetics/81.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M, Lecourtois M, Geraud G, Posakony JW, Schweisguth F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development. 1996;122:1673–1682. doi: 10.1242/dev.122.6.1673. [DOI] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: Lessons from worms and flies. Genes & Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Hoppe PE, Greenspan RJ. Local function of the Notch gene for embryonic ectodermal pathway choice in Drosophila. Cell. 1986;46:773–783. doi: 10.1016/0092-8674(86)90353-3. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Kao H-Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch 1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kelley MR, Kidd S, Deutsch WA, Young MW. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell. 1987;51:539–548. doi: 10.1016/0092-8674(87)90123-1. [DOI] [PubMed] [Google Scholar]
- Kidd S. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell. 1992;71:623–635. doi: 10.1016/0092-8674(92)90596-5. [DOI] [PubMed] [Google Scholar]
- Kidd S, Kelley MR, Young MW. Sequence of the Notch locus of Drosophila: Relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986;6:3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Baylies MK, Gasic GP, Young MW. Structure and distribution of the Notch protein in Developing Drosophila. Genes & Dev. 1989;3:1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes & Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of Notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes & Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Go MJ, Sun X, Eastman DS, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- Nye JS, Kopan R. Developmental signaling. Vertebrate ligands for Notch. Curr Biol. 1995;5:966–969. doi: 10.1016/s0960-9822(95)00189-8. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Parks AL, Muskavitch MAT. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev Biol. 1993;157:484–496. doi: 10.1006/dbio.1993.1151. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. Developmental genetics of the 2E-F region the Drosophila X chromosome: A region rich in ‘Developmentally important’ genes. Genetics. 1984;108:559–572. doi: 10.1093/genetics/108.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portin P. Allelic negative complementation at the Abruptex locus of Drosophila melanogaster. Genetics. 1975;81:121–133. doi: 10.1093/genetics/81.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- Roehl H, Bosenberg M, Blelloch R, Kimble J. Roles of the RAM and ANK domains in signalling by the C. elegans GLP-1 receptor. EMBO J. 1996;15:7002–7012. [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Schweisguth F, Posakony JW. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- ————— Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989;57:1237–1245. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of Split gene product define a novel family of nuclear proteins. Nature Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with protein containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]