Abstract
Objective
To assess the utility of a panviral DNA microarray platform (Virochip) in the detection of viruses associated with pediatric respiratory tract infections (RTIs).
Study design
The Virochip was compared with conventional direct fluorescent antibody (DFA)- and polymerase chain reaction (PCR)-based testing for the detection of respiratory viruses in 278 consecutive nasopharyngeal aspirate samples from 222 children.
Results
The Virochip was superior in performance to DFA, showing a 19% increase in the detection of 7 respiratory viruses included in standard DFA panels, and was similar to virus-specific PCR (sensitivity, 85% to 90%; specificity, ≥99%; positive predictive value, 94% to 96%; negative predictive value, 97% to 98%) in the detection of respiratory syncytial virus, influenza A, and rhinoviruses/enteroviruses. The Virochip also detected viruses not routinely tested for or missed by DFA and PCR, as well as double infections and infections in critically ill patients that DFA failed to detect.
Conclusions
Given its favorable sensitivity and specificity profile and expanded spectrum for detection, microarray-based viral testing holds promise for clinical diagnosis of pediatric RTIs.
Abbreviations: AdV, Adenovirus; CoV, Coronavirus; DFA, Direct fluorescent antibody; EV, Enterovirus; FluA/B, Influenza A/B; HMPV, Human metapneumovirus; HPIV, Human parainfluenza virus; NPA, Nasopharyngeal aspirate; PCR, Polymerase chain reaction; RSV, Respiratory syncytial virus; RT, Reverse-transcriptase; RTI, Respiratory tract infection; RV, Rhinovirus; UCSF, University of California San Francisco
Acute respiratory tract infections (RTIs) are the most common illnesses in humans and are associated with significant morbidity and mortality. In children, viruses are responsible for the majority of RTIs, with bacteria and other pathogens thought to be responsible for fewer than 15% of cases.1 However, even with the best methods for viral detection currently available, a specific pathogen cannot be identified in 20% to 50% of RTIs.1, 2, 3, 4
Existing viral diagnostic methods are limited in sensitivity and scope. Virus isolation by culture takes days to weeks, and many viruses remain fastidious or unculturable. Direct fluorescent antibody (DFA) testing has a turnaround time of 2 hours, but may suffer from low sensitivity and is available for only a limited number of viruses.5 Polymerase chain reaction (PCR) testing is rapid and highly sensitive, and has supplanted culture as the new gold standard for detecting respiratory viruses in research settings.3, 4 However, most PCR tests target only 1 virus at a time, making these assays cumbersome in routine clinical practice. For simultaneous detection of up to 20 viruses, a number of multiplex PCR assays have been proposed.3, 4, 6, 7, 8, 9, 10
Recently, DNA microarray testing has emerged as a promising new technology for broad-spectrum virus detection.11, 12, 13 We previously designed an in-house microarray platform to detect all known viruses, as well as novel viruses related to known viral families (Virochip; University of California San Francisco [UCSF]).13, 14 The Virochip consists of ∼22 000 oligonucleotide probes representing all ∼1800 fully or partially sequenced viruses in GenBank as of Fall 2004. The performance of the Virochip in respiratory virus detection has been tested previously using virally infected tissue culture cells13 and in selected patient cohorts.15 To date, the Virochip has not been compared directly with standard diagnostic tests for viruses in a clinical setting; thus, in the present study, we sought to compare the Virochip with conventional clinical DFA- and PCR-based testing in the detection of respiratory viruses associated with RTIs in children.
Methods
Study Design
This study is a prospective case series of all consecutive samples sent for viral DFA testing from pediatric patients seen at UCSF between December 2003 and June 2004. All patient samples were collected according to protocols approved by UCSF's Institutional Review Board.
Sample Processing
Consecutive nasopharyngeal aspirate (NPA) samples sent to the UCSF clinical laboratory for DFA were analyzed with the Light Diagnostics Respiratory DFA Viral Screening and Identification Kit (Chemicon International, Temecula, CA). This kit detects 7 common respiratory viruses: respiratory syncytial virus (RSV); influenza A and B (FluA and FluB); human parainfluenza virus types 1, 2, and 3 (HPIV-1, 2, and 3); and adenovirus (AdV). Remaining sample material was transferred into a sterile 14-mL conical tube, frozen, and stored at −80°C.
For blinded Virochip analysis, frozen NPA samples were thawed, and 200-μL aliquots were used to extract RNA using the RNeasy Mini Kit (Qiagen Inc, Valencia, California) according to the manufacturer's protocol, including on-column DNase digestion. Microarrays used in this study were identical to those described previously (National Center for Biotechnology Information GEO platform GPL3429).16 RNA samples were amplified and labeled using a round A/B random PCR method and hybridized to the Virochip as reported previously.13 Microarrays were scanned with an Axon 4000B scanner and analyzed using Axon GenePix 6 software (Axon Instruments, Union City, California). All microarray data have been submitted to the National Center for Biotechnology Information GEO database (accession number GSE10294).
Virochip Data Analysis
Virochip data analysis was carried out in 2 stages. First, all microarrays were analyzed with the E-Predict algorithm,17 using a significance value of P < .01 to identify microarrays with statistically significant viral hybridization patterns (176 of 278). To generate an optimized set of oligonucleotide intensity weights for the third-generation Virochip platform, we took the remaining 102 presumably negative microarrays and manually fit a set of functions with the general equation
where w is a weight (value from 0 to 1) for a given oligonucleotide, i is the median of sum-normalized intensities of that oligonucleotide across the 102 negative microarrays, and lower boundary a is the median of medians of the sum-normalized intensities of all oligonucleotides. The upper boundary condition b was expressed as , where c is a constant and σ is the standard deviation of sum-normalized intensities of the oligonucleotide across the 102 negative microarrays. A total of 40 weight sets with c and p as fitting measurements were evaluated using E-Predict profile separation statistics on 6 rhinovirus (RV)-positive, 6 RSV-positive, 4 HPIV-3–positive, and 2 FluA-positive microarrays. The optimal performance was achieved with weights corresponding to c = 0.15 and p = 1.5. These weights were used to generate negative null distributions based on the set of 102 negative microarrays mentioned above, and final microarray virus determinations were made by E-Predict. A microarray was considered E-predict–positive for a given virus if the corresponding energy profile attained a significance value of P < .05.
PCR Analysis
Blinded reverse-transcriptase (RT)-PCR assays for RSV, FluA, RV, and enterovirus (EV) were performed on RNA extracted from the frozen NPA samples at the Viral and Rickettsial Disease Laboratory, California Department of Health Services. For detection of RSV, an RT-PCR assay for RSV targeting the fusion (F) gene was performed using the 1-step Access RT-PCR system as described previously.2 For detection of FluA, primers and fluorescent probes targeting the highly conserved matrix (M) gene of FluA were used with the Roche LightCycler Real-Time PCR System (Roche Diagnostics, Indianapolis, Indiana) as described previously.18 For detection of RV/EV, we first ran an in-house RT-PCR multiplex assay on randomly primed template cDNA using primers targeting the highly conserved 5′ untranslated region of RV15 and EV.19 Positive samples were identified by the presence of amplified PCR bands of the expected size on agarose gel electrophoresis. Follow-up real-time PCR assays for RV/EV detection on discrepant samples between the Virochip and the in-house multiplex RT-PCR were then carried out using the Roche LightCycler Real-Time PCR System as described previously.20
Sequence Confirmation of Virochip-Positive/PCR-Negative Samples
Confirmation of 2 Virochip-positive/PCR-negative samples was done by PCR using alternative primers based on high-intensity microarray oligonucleotides and direct sequencing. The sequence from 1 case of RSV was 97% identical to a 155-bp fragment from the matrix (M) gene for RSV subgroup B strain 9320, and the sequence from 1 case of RV was 96% identical to a 302-bp fragment from the VP4/VP2 region for RV strain QPID03-0003. Another Virochip-positive/PCR-negative case (an EV) was confirmed by a repeat run of the real-time PCR assay.20
Clinical Data Collection
After assay results for the NPA samples were obtained, subjects were identified, and a retrospective review of the medical record was performed. Data were systematically collected using a standard form that documented the following information: age, sex, date and location of sample collection, clinical presentation, presence of immunocompromised state, presence of acute respiratory failure, and DFA result. Location of sample collection was classified as outpatient (clinic or emergency department) or hospital admission. Presenting illness was defined as an upper RTI (ie, cough and/or congestion with or without fever or a clinician's diagnosis), lower RTI (ie, clinician's diagnosis of asthma exacerbation, bronchiolitis, croup, or pneumonia), or no respiratory illness (eg, febrile illness, seizures, or DFA collected on a routine basis, such as before transplantation surgery). Immunocompromised patients were defined as children with solid organ or bone marrow transplants, congenital or acquired immune deficiencies (including those on chemotherapy), or human immunodeficiency virus. Patients with respiratory failure were defined as children who developed acute respiratory decompensation necessitating mechanical ventilation as a result of their respiratory illness. For patients with more than 1 sample, a first-time sample was defined as the earliest sample collected during a single hospitalization or illness.
Results
Demographic and Clinical Data
We analyzed a total of 278 NPA samples collected from 222 children for the presence of viruses using DFA and Virochip. Demographic and clinical data for the 222 patients are given in Table I. Most patients (73%) were hospitalized. Approximately 71% of the enrolled patients (n = 157) had an acute RTI, and the remaining patients (n = 65) had DFA sent for other reasons, most often a nonrespiratory febrile illness (n = 46). Eighteen percent of the patients (n = 39) were immunocompromised, and 8% (n = 17) developed respiratory failure necessitating mechanical ventilation as a result of their illness. Most of the subjects with RSV were younger (mean age, 1.5 years) and required hospitalization (76%), whereas subjects with influenza tended to be older (mean age, 4.1 years) and were treated mainly as outpatients (64%). All cases of human metapneumovirus (HMPV), coronavirus (CoV), and AdV were in hospitalized patients. The majority of viral infections in immunocompromised subjects (60%) were due to picornaviruses.
Table 1.
Demographic and clinical data according to illness and viral pathogen
| Variable | Number (%) of subjects | Age (± standard deviation), years | Male sex | Hospitalization | Respiratory failure | Immunocompromised status |
|---|---|---|---|---|---|---|
| All subjects | 222 (100%) | 3.5 (4.4) | 136 (61%) | 163 (73%) | 17 (8%) | 39 (18%) |
| Respiratory illness | 157 (71%) | 3.2 (4.2) | 92 (59%) | 105 (67%) | 11 (7%) | 17 (11%) |
| Upper RTI | 59 (27%) | 3.5 (4.3) | 39 (66%) | 25 (42%) | 1 (2%) | 10 (17%) |
| Bronchiolitis/croup | 42 (19%) | 1.2 (1.5) | 21 (50%) | 29 (69%) | 1 (2%) | 0 (0%) |
| Pneumonia | 51 (23%) | 4.0 (4.9) | 27 (53%) | 45 (88%) | 9 (18%) | 7 (14%) |
| Asthma exacerbation | 13 (6%) | 3.9 (2.7) | 12 (92%) | 12 (92%) | 0 (0%) | 0 (0%) |
| No respiratory illness | 65 (29%) | 4.0 (4.9) | 44 (68%) | 58 (89%) | 6 (9%) | 22 (34%) |
| RSV | 42 (19%) | 1.5 (1.2) | 23 (55%) | 32 (76%) | 3 (7%) | 1 (2%) |
| FluA/FluB | 25 (11%) | 3.8 (4.1) | 18 (72%) | 9 (36%) | 2 (8%) | 2 (8%) |
| HPIV | 10 (5%) | 2.8 (4.5) | 5 (50%) | 7 (70%) | 1 (10%) | 2 (20%) |
| Picomavirus | 35 (16%) | 3.6 (4.6) | 19 (54%) | 27 (77%) | 4 (11%) | 9 (26%) |
| HMPV | 4 (2%) | 3.1 (2.8) | 2 (50%) | 4 (100%) | 1 (25%) | 0 (0%) |
| CoV | 3 (1%) | 4.2 (2.6) | 1 (33%) | 3 (100%) | 1 (33%) | 1 (33%) |
| AdV | 3 (1%) | 3.8 (1.8) | 3 (100%) | 3 (100%) | 1 (33%) | 0 (0%) |
| No pathogen or other | 106 (48%) | 4.0 (5.0) | 69 (65%) | 83 (78%) | 6 (6%) | 24 (23%) |
Demographic and clinical data were collected for all 222 enrolled subjects in the study and stratified according to type of presenting illness and virus detected by the Virochip. For patients with more than 1 sample collected, only the first-time sample was used in this analysis. The total number of cases summarized in this table (n = 222) differs from the total number of samples reported in Figure 1 (n = 278), because multiple samples were collected for some patients. The sum of the percentages of viruses detected by the Virochip is >100%, because some of the cases are double-virus infections.
Detection of Viruses by DFA and the Virochip
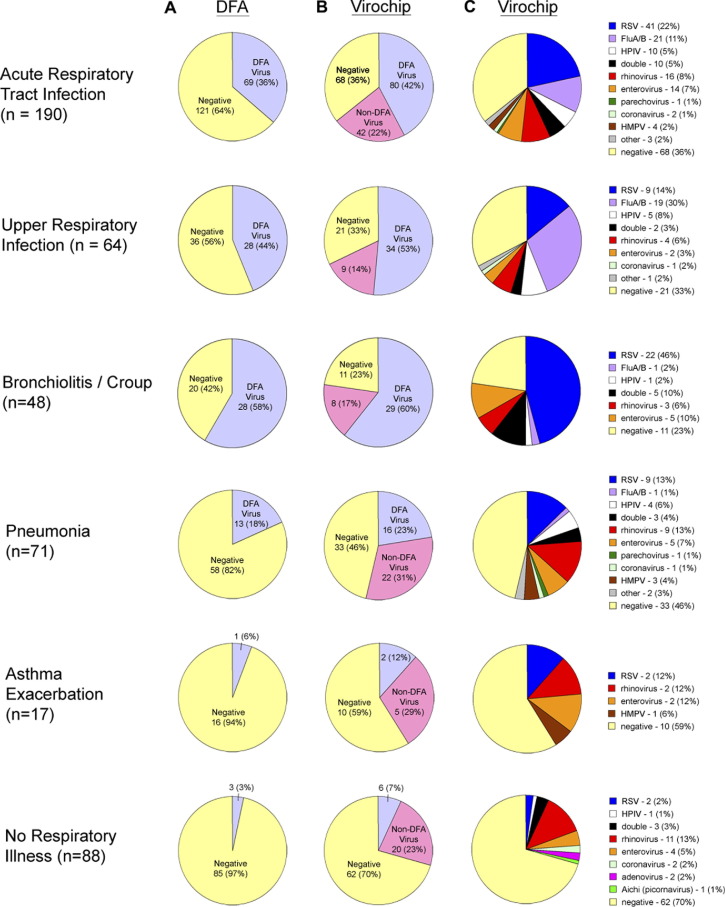
Figure 1 shows the spectrum and frequency of detection of different viruses by the 2 methods. In patients with RTI, DFA detected a virus in only 36% of the samples, whereas the Virochip detected a virus in 64%, for a 75% improvement in the rate of detection. Detection of viruses not included in the DFA panel (non-DFA viruses) accounted for approximately 1/3 of all Virochip-positive identifications. Among these non-DFA viruses, picornaviruses was the largest group (74% of samples positive for a non-DFA virus, or 16% of all samples from patients with RTIs). These picornaviruses included 16 RVs, 14 EVs, and 1 parechovirus. Besides the picornaviruses, the Virochip identified 4 HMPV and 2 CoV infections from patients with RTIs. Human cytomegalovirus–like sequences were detected in 1 sample from 1 febrile patient with pneumonia. In 7 samples, the Virochip detected viruses not typically associated with respiratory disease and of doubtful clinical significance; these viruses included polyomaviruses (SV40 and JC viruses), plant viruses commonly found in the gastrointestinal tract (nanoviruses and geminiviruses),21 and bovine leukemia virus, a virus ubiquitous in cow's milk.22 A virus was detected in a higher proportion of patients with upper RTIs (68%) and bronchiolitis (77%) than in those with pneumonia (54%) or asthma exacerbation (41%). In patients with nonrespiratory illnesses (typically febrile illnesses of uncertain origin), the Virochip detected a virus in 30% of the samples, with the majority accounted for by picornaviruses, whereas DFA detected a virus in only 3%. Overall, at least 1 of the 7 DFA viruses was identified by Virochip in 31% of the RTI samples (n = 86), compared with 25% identified by DFA (n = 72), corresponding to a 19% overall increase in detection by the Virochip compared with DFA (P < .01 by χ2 analysis).
Figure 1.
Viruses detected by DFA and Virochip. A total of 278 nasopharyngeal aspirate samples from 222 patients were analyzed by DFA and Virochip. Number of NPA samples positive for a virus in the DFA panel (“DFA virus,” light blue), positive for a virus not found in the DFA panel (“non-DFA virus,” pink), and negative (negative, yellow) by DFA (A) and by Virochip (B), plotted as a proportion of all samples stratified by presenting illness (rows). C, Number of NPA samples positive for RSV, FluA/B, HPIV, RV, EV, CoV, AdV, and HMPV plotted as a proportion of all samples stratified by presenting illness (rows). The term “double” denotes instances of infections with 2 viruses (see Table II; available at www.jpeds.com), and “other” corresponds to viruses that are not typically considered respiratory pathogens (see text). The respiratory illnesses corresponding to 12 samples from 8 patients (3.6%) are assigned to 2 categories.
Virochip Detection of Double and Critical Viral Infections
The Virochip detected 13 cases of simultaneous infection by 2 viruses (9% of Virochip-positive samples, 5% of all samples; Table II; available at www.jpeds.com). In contrast, DFA detected no cases of double infection. The Virochip also detected a viral pathogen in 12 of 17 critically ill patients who developed respiratory failure necessitating mechanical ventilation, whereas a virus was detected by DFA in only 5 such patients. Two double infections (1 case of FluA/AdV and 1 case of RSV/RV) were identified among these life-threatening cases.
Table II.
Detection of double and critical infections by Virochip and DFA†
| DFA⁎ | Virochip (virus 1) | Virochip (virus 2) | Presenting illness |
|---|---|---|---|
| Double infections | |||
| RSV | FluA | RSV | Bronchiolitis |
| FluA | FluA | AdV | Pneumonia |
| RSV | EV | RSV | Bronchiolitis |
| RSV | Parechovirus | RSV | Bronchiolitis |
| RSV | EV | RSV | Bronchiolitis |
| RSV | RV | RSV | Bronchiolitis |
| FluA | FluA | Nanovirus | Upper RTI |
| Negative | HMPV | Polyomavirus (JC virus) | Pneumonia |
| Negative | FluA | Polyomavirus (SV40) | Pneumonia |
| Negative | Parvovirus (AAV-5) | RV | Upper RTI |
| Negative | HPIV-1 | Bovine leukemia virus | Fever |
| Negative | Picomavirus (Aichi) | Polyomavirus (JC virus) | Fever |
| Critical infections | |||
| FluA | FluA, AdV | Community-acquired pneumonia | |
| FluA | FluA | Community-acquired pneumonia | |
| RSV | RSV, RV | Bronchiolitis | |
| RSV | RSV | Community-acquired pneumonia | |
| HPIV-1 | HPIV-1 | Community-acquired pneumonia | |
| Negative | RSV | Upper RTI progressing to respiratory failure | |
| Negative | RV | Pneumonia in immunocompromised patient | |
| Negative | HMPV, polyomavirus | Apnea with pneumonia | |
| Negative | CoV | Fever with seizures | |
| Negative | RV | Fever and respiratory distress | |
| Negative | EV | Aspiration pneumonitis | |
| Negative | Streptococcus pyogenes bacteriophage | Aspiration pneumonia | |
| Negative (5) | Negative (5) | Aspiration pneumonia (2), apnea (1), pulmonary hypertension (1), gastroenteritis (1) | |
HPIV-1, human parainfluenza virus type 1; AAV-5, adeno-associated virus type 5.
DFA and Virochip results corresponding to cases of double infection (n = 12) and critical respiratory illness requiring mechanical ventilation (n = 17) are shown. Virus 1 and Virus 2 correspond to the top and second statistically significant Virochip predictions, respectively.
No cases of double infection were detected by DFA.
Viruses that may be commensal or food-associated.
Comparison of the Virochip with PCR
To assess Virochip results that were discordant with DFA and overall sensitivity and specificity, we carried out RT-PCR assays for RSV, FluA, and RV/EV, the 3 major groups of viruses detected by the Virochip (Figure 2). For detection of RSV and FluA, the overall sensitivity of the Virochip (86%) relative to PCR was significantly better than that of DFA (71%; P < .05 by χ2 analysis). The corresponding specificities were ≥ 99%. There was also a greater overlap in positives between Virochip and PCR than between DFA and PCR for these 2 viruses; 16 of of 18 (89%) of Virochip-positive/DFA-negative RSV and FluA samples were confirmed to be true positives by PCR (Figure 2; Venn diagram).
Figure 2.
Comparison of the performance of DFA and Virochip relative to PCR. DFA and Virochip were compared with specific PCR using 2 × 2 contingency tables and Venn diagram analysis for detection of RSV (A) and FluA (B). In addition, Virochip was compared with specific PCR using a 2 × 2 contingency table for detection of RV and EV, 2 picornaviruses not tested for by DFA (C). Because the specific PCR assays for RV/EV used here do not adequately distinguish RV from EV, results are reported as the total number of RV/EV-positive cases.
For detection of RV/EV, we first used an in-house RT-PCR multiplex assay based on methods reported previously.15, 19 Using this assay, 31 NPA samples were positive by both Virochip and PCR, 18 NPA samples were Virochip-positive/PCR-negative, and 5 NPA samples were Virochip-negative/PCR-positive. Because these results were inconsistent with the rates of detection using clinically validated PCR assays for the other viruses (RSV and FluA), we further analyzed the 23 discrepant samples in a blinded fashion using a more sensitive clinically validated real-time PCR assay for RV/EV.20 Results obtained by combining these 2 assays show that the Virochip had overall high sensitivity (90%) and specificity (99%) for detection of RV/EV (Figure 2C).
Virochip Detection of PCR-Negative Cases of Respiratory Virus Infection
Six cases (3 picornavirus, 2 RSV, and 1 FluA) detected by the Virochip tested negative in the corresponding PCR assays (Figure 2). We hypothesized that most of these cases were viral strains that had failed detection with standard PCR primers. To investigate this possibility, we recovered viral sequences from 2 Virochip-positive/PCR-negative cases (1 RV and 1 RSV) and separately confirmed a case of EV that was Virochip-positive and previously PCR-negative by repeating the real-time PCR assay. Similar attempts to confirm the remaining 3 Virochip-positive/PCR-negative cases with the limited amount of sample available were unsuccessful. Thus, at least 3 of the 6 Virochip-positive/PCR-negative represent PCR false-negatives, and, consequently, the reported sensitivity of the Virochip may be underestimated.
Discussion
This study of pediatric RTIs used a DNA microarray that aims to detect all known viral species simultaneously. The most common viral pathogens identified were RSV (19%), picornaviruses (16%), and FluA/B (11%). Notably, most viruses detected in the respiratory tracts of the patients with nonrespiratory illnesses and in the immunocompromised patients were picornaviruses. These findings are consistent with the observation that asymptomatic RV infections can be seen in 4% to 12% of healthy individuals and suggest that immunocompromised hosts may be more susceptible to colonization or overt respiratory illness by picornaviruses.23 Our frequency of detection of 16% for picornaviruses is consistent with that of a previously published report (18% RVs, 2.9% EVs) that also used consecutive NPA samples.24
Our comparison of the Virochip with DFA demonstrates that the former has superior sensitivity for respiratory virus detection. Importantly, about 50% of the overall increase in detection rate corresponds to samples with inconclusive DFA results due to low cellular content. Unlike DFA, nucleic acid detection methods, such as microarray and PCR, are capable of detecting free viral particles in addition to virus-infected cells. Another significant advantage of a panviral microarray over DFA is the ability to screen for all known viruses simultaneously. DFA panels in current clinical use do not test for RV/EV, HMPV, or CoV, which in our study composed more than 1/3 of the detected viruses in the patients with RTIs.
The 17 patients presenting with illness severe enough to require mechanical ventilation included 3 cases of critical RV infection, including 1 immunocompromised individual. RV infection is thought to comprise a spectrum of disease ranging from asymptomatic infection to life-threatening childhood pneumonia.25 Our findings of cases of critical respiratory tract illness associated with RV infection is consistent with growing evidence linking RV with hospitalizations in young children.26 The Virochip was also superior to DFA for detecting double-virus infections. Two cases of double infection, in which both viruses could be detected by DFA in principle, were characterized as single-virus infections by DFA. Previous studies have suggested that double-virus infections are associated with greater severity of RTI.27 Higher efficiency of the microarray in detecting critical as well as double-virus infections is an important advantage of the method; timely detection of such infections may allow clinicians to avoid unnecessary antibiotics and invasive procedures and begin appropriate antiviral treatment, if available.
Besides being a highly parallel methodology that uses thousands of oligonucleotide probes for simultaneous detection, DNA microarray platforms like the Virochip are expandable and adaptable. The automated oligonucleotide design allows straightforward addition of new probes for better detection of known viruses, as well as expanded coverage of novel or evolving viral species.16, 28, 29 DNA microarrays also can be used for detection of nonviral pathogens, including bacteria and fungi.30 Interestingly, in this study, the Virochip detected Streptococcus pyogenes bacteriophages in a sample from a patient with aspiration pneumonia who also had a positive sputum culture for Streptococcus pyogenes. This result suggests a possible strategy of using phage sequences as an indirect means of detecting bacterial pathogens.
Despite its panviral scope, the Virochip detected a virus in only 64% of RTI cases overall. Although comparable with findings from other detection methods,1, 2, 3, 4, 8, 9 this result is likely an underestimation of the true number of positives arising from several factors. First, some samples may have been missed by the Virochip due to low virus titers at the detection limits of PCR. Second, the use of RNA (rather than DNA) as the source material in this study may have given rise to lower-than-expected rates of detection of DNA viruses such as AdVs, herpesviruses, and parvoviruses. The Virochip may not have detected any cases of human bocavirus, a recently described parvovirus,31 because this virus shares low sequence identity to parvovirus sequences currently represented on the Virochip. Finally, 32% of patients with RTIs in this study were diagnosed with pneumonia, 35% of which were hospital-associated. Many of these cases of pneumonia may be nonviral in origin, as suggested by previous epidemiologic studies of pneumonia in hospitalized children.32, 33
Using custom PCR primers, we confirmed at least 3 of the 6 microarray-positive/PCR-negative cases (1 RSV, 1 RV, and 1 EV) as true positive tests for viruses. Detection of such cases by microarray is not unexpected, because the Virochip uses multiple probes that are derived from different sites in the viral genome and have a higher tolerance for sequence mismatches than the primers used in specific PCR. This results in improved detection of divergent viruses.14, 34 We suspect that the microarray-positive/PCR-negative cases of virus infection were missed by clinical PCR likely due to mismatches between primer and target sequences,35 although limited specimen availability affected our ability to show this definitively.
Although the prospect of comprehensive viral detection with a single microarray assay is appealing, several challenges must be addressed before the use of this technology can become practical in clinical diagnosis. First, setting up the technology entails significant initial startup costs, including the cost of the microarray printer and viral oligonucleotides, as well as ongoing material and labor costs. Second, the turnaround time for the assay is currently ∼24 hours; it may be possible to decrease the processing duration by using ultra-rapid polymerases for amplification or controlled agitation techniques during hybridization. Third, reproducibility and consistent array quality are of concern. In our study, none of the microarray assays had to be repeated, and the inherent probe redundancy built into the method makes the assay robust for the purpose of virus detection. For example, of the 300 oligonucleotide probes on the Virochip designed to hybridize to rhinoviruses, as few as 4 high-intensity oligonucleotides are sufficient to make a successful virus identification. Statistical methods for interpreting the microarray data, as exemplified by E-Predict, can be completely automated, allowing ease of use and freedom from operator bias. Smaller microarrays aimed at detecting targeted virus sets (eg, respiratory viruses only) would reduce costs and simplify issues of reproducibility, quality control, and data analysis.
Looking to the future, we can envision 2 possible approaches to using viral detection microarrays in clinical practice. One approach would be to use the platform for direct diagnosis of respiratory infections, as reported here. An alternative approach could be to deploy a viral microarray as an instrument of discovery rather than of routine detection, with the goal of identifying divergent or unexpected viruses that elude diagnosis using conventional methods. Once a new candidate pathogen was identified, a specific PCR-based or DFA-based assay could then be developed to detect the virus with a high degree of sensitivity in clinical samples. In this scenario, the microarray assay would complement rather than replace existing diagnostic techniques such as PCR.
Footnotes
Supported by a Genentech Graduate Fellowship (A.U.) and grants from the Glaser Pediatric Network (T.G.), the Sandler Program for Asthma Research (J.D.), the Howard Hughes Medical Institute (J.D. and D.G.), and the Doris Duke Charitable Foundation (C.C., J.D., and D.G.).
Supplemental information on the output of the E-Predict analysis is available from the authors upon request.
References
- 1.van Gageldonk-Lafeber A.B., Heijnen M.L., Bartelds A.I., Peters M.F., van der Plas S.M., Wilbrink B. A case-control study of acute respiratory tract infection in general practice patients in The Netherlands. Clin Infect Dis. 2005;41:490–497. doi: 10.1086/431982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erdman D.D., Weinberg G.A., Edwards K.M., Walker F.J., Anderson B.C., Winter J. GeneScan reverse- transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J Clin Microbiol. 2003;41:4298–4303. doi: 10.1128/JCM.41.9.4298-4303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruteke P., Glas A.S., Dierdorp M., Vreede W.B., Pilon J.W., Bruisten S.M. Practical implementation of a multiplex PCR for acute respiratory tract infections in children. J Clin Microbiol. 2004;42:5596–5603. doi: 10.1128/JCM.42.12.5596-5603.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syrmis M.W., Whiley D.M., Thomas M., Mackay I.M., Williamson J., Siebert D.J. A sensitive, specific, and cost-effective multiplex reverse-transcriptase–PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004;6:125–131. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casiano-Colon A.E., Hulbert B.B., Mayer T.K., Walsh E.E., Falsey A.R. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol. 2003;28:169–174. doi: 10.1016/s1386-6532(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 6.Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coiras M.T., Aguilar J.C., Garcia M.L., Casas I., Perez-Brena P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse-transcription nested-PCR assays. J Med Virol. 2004;72:484–495. doi: 10.1002/jmv.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freymuth F., Vabret A., Cuvillon-Nimal D., Simon S., Dina J., Legrand L. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006;78:1498–1504. doi: 10.1002/jmv.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kehl S.C., Henrickson K.J., Hua W., Fan J. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J Clin Microbiol. 2001;39:1696–1701. doi: 10.1128/JCM.39.5.1696-1701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee W.M., Grindle K., Pappas T., Marshall D.J., Moser M.J., Beaty E.L. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol. 2007;45:2626–2634. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin B., Blaney K.M., Malanoski A.P., Ligler A.G., Schnur J.M., Metzgar D. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J Clin Microbiol. 2007;45:443–452. doi: 10.1128/JCM.01870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palacios G., Quan P.L., Jabado O.J., Conlan S., Hirschberg D.L., Liu Y. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D., Coscoy L., Zylberberg M., Avila P.C., Boushey H.A., Ganem D. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D., Urisman A., Liu Y.T., Springer M., Ksiazek T.G., Erdman D.D. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu C.Y., Rouskin S., Koshy A., Urisman A., Fischer K., Yagi S. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43:e71–e76. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urisman A., Fischer K.F., Chiu C.Y., Kistler A.L., Beck S., Wang D. E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol. 2005;6:R78. doi: 10.1186/gb-2005-6-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louie J.K., Hacker J.K., Gonzales R., Mark J., Maselli J.H., Yagi S. Characterization of viral agents causing acute respiratory infection in a San Francisco University Medical Center Clinic during the influenza season. Clin Infect Dis. 2005;41:822–828. doi: 10.1086/432800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rotbart H.A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990;28:438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kares S., Lonnrot M., Vuorinen P., Oikarinen S., Taurianen S., Hyoty H. Real-time PCR for rapid diagnosis of entero- and rhinovirus infections using LightCycler. J Clin Virol. 2004;29:99–104. doi: 10.1016/s1386-6532(03)00093-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhang T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuckleburg C.J., Chase C.C., Nelson E.A., Marras S.A., Dammen M.A., Christopher-Hennings J. Detection of bovine leukemia virus in blood and milk by nested and real-time polymerase chain reactions. J Vet Diagn Invest. 2003;15:72–76. doi: 10.1177/104063870301500117. [DOI] [PubMed] [Google Scholar]
- 23.Hayden F.G. Rhinovirus and the lower respiratory tract. Rev Med Virol. 2004;14:17–31. doi: 10.1002/rmv.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loens K., Goossens H., de Laat C., Foolen H., Oudshoorn P., Pattyn S. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse-transcription–PCR, in children with acute respiratory infections during a winter season. J Clin Microbiol. 2006;44:166–171. doi: 10.1128/JCM.44.1.166-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsolia M.N., Psarras S., Bossios A., Audi H., Paldanius M., Gourgiotis D. Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis. 2004;39:681–686. doi: 10.1086/422996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller E.K., Lu X., Erdman D.D., Poehling K.A., Zhu Y., Griffin M.R. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 2007;195:773–781. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Templeton K.E., Scheltinga S.A., van den Eeden W.C., Graffelman A.W., van den Broek P.J., Claas E.C. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41:345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehlmann M., Dawson E.D., Townsend M.B., Smagala J.A., Moore C.L., Smith C.B. Robust sequence selection method used to develop the FluChip diagnostic microarray for influenza virus. J Clin Microbiol. 2006;44:2857–2862. doi: 10.1128/JCM.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell R. Designing microarray oligonucleotide probes. Brief Bioinform. 2003;4:361–367. doi: 10.1093/bib/4.4.361. [DOI] [PubMed] [Google Scholar]
- 30.Palacios G., Quan P.-L., Jabado O.J., Conlan S., Hirschberg D.L., Liu Y. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juven T., Mertsola J., Waris M., Leinonen M., Meurman O., Roivainen M. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J. 2000;19:293–298. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 33.Michelow I.C., Olsen K., Lozano J., Rollins N.K., Duffy L.B., Ziegler T. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics. 2004;113:701–707. doi: 10.1542/peds.113.4.701. [DOI] [PubMed] [Google Scholar]
- 34.Urisman A., Molinaro R.J., Fischer N., Plummer S.J., Casey G., Klein E.A. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Chiu C.Y., Alizadeh A.A., Rouskin S., Merker J.D., Yeh E., Yagi S. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J Clin Microbiol. 2007;45:2340–2343. doi: 10.1128/JCM.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]