Using a library of siRNAs, we found that ARHGAP18 was essential for the organization of actin stress fibers and focal adhesion. ARHGAP18 is one of the crucial factors for the regulation of RhoA in order to control cell motility and spreading.
Abstract
Rho GTPases are molecular switches that transmit biochemical signals in response to extracellular stimuli to elicit changes in the actin cytoskeleton. Rho GTPases cycle between an active, GTP-bound state and an inactive, GDP-bound state. These states are regulated by two distinct families of proteins—guanine nucleotide exchange factors and GTPase-activating proteins (GAPs). We studied the role of a previously uncharacterized GAP, ARHGAP18 (MacGAP). Overexpression of ARHGAP18 suppressed the activity of RhoA and disrupted stress fiber formation. Conversely, silencing of ARHGAP18 by small interfering RNA transfection–enhanced stress fiber formation and induced rounding of cells. We examined the role of ARHGAP18 in cell spreading and migration. Immunofluorescence analysis revealed that ARHGAP18 was localized to the leading edge during cell spreading and migration. ARHGAP18-knockdown cells showed impaired spreading, premature formation of stress fibers, and sustained activation of RhoA upon cell attachment. In addition, knockdown and overexpression of ARHGAP18 resulted in the inhibition and promotion of cell migration, respectively. Furthermore, ARHGAP18 was required for the polarization of cells for migration. Our results define ARHGAP18 as one of the crucial factors for the regulation of RhoA for the control of cell shape, spreading, and migration.
INTRODUCTION
Dynamic remodeling of the actin cytoskeleton initiated by cell adhesion to the extracellular matrix (ECM) is essential for fundamental biological processes such as cell migration and spreading. Integrins form a family of heterodimeric transmembrane proteins that functionally link the ECM and actin cytoskeleton (Hynes, 1992; Dedhar and Hannigan, 1996). Integrin–ECM interaction activates a variety of intracellular signaling molecules for the rearrangement of the actin cytoskeleton (Schwartz et al., 1995). One particular family of proteins, the Rho family GTPases, plays a pivotal role in the regulation of signaling pathways that link integrin–ECM interaction to actin cytoskeletal changes (Hall, 1998).
More than 20 members of the Rho family of GTPases have been identified in mammalian cells (Wennerberg and Der, 2004; Hall, 2005). Among these proteins, the best-characterized members of this family are RhoA, Rac1, and Cdc42. Activated Rac1 and Cdc42 are essential for the production of lamellipodia and filopodia, respectively (Ridley et al., 1992; Kozma et al., 1995; Nobes and Hall, 1995). Both structures are commonly found on the periphery of cells in migration or spreading and are required for the cells to extend into the free space (Hall, 1998). In addition, Cdc42 is critically involved in the establishment of cellular polarity (Etienne-Manneville and Hall, 2001; Etienne-Manneville, 2004). Activation of RhoA stimulates the formation of focal adhesions and stress fibers (Ridley and Hall, 1992; Chrzanowska-Wodnicka and Burridge, 1996). Coordinated activation of these GTPases in time and space is essential for the cellular shape changes required for fundamental processes such as cell division and migration (Wennerberg and Der, 2004; Jaffe and Hall, 2005). A number of downstream targets of Rho GTPases have been identified that control actin polymerization or actomyosin contractility. For example, activated RhoA regulates the dynamics of the cytoskeleton through effectors such as Rho kinases/ROCKs and mammalian homologue of diaphanous (mDia; Kimura et al., 1996; Matsui et al., 1996; Narumiya et al., 1997; Watanabe et al., 1997; Kaibuchi et al., 1999; Amano et al., 2010).
Rho GTPases are molecular switches that cycle between a GTP-bound, active form and a GDP-bound, inactive form (Etienne-Manneville and Hall, 2002). Activated Rho GTPases associate with a variety of downstream effectors to modulate their activity and localization. Cycling between the active and inactive states is primarily regulated by two distinct families of proteins. Exchange of GDP for GTP is mediated by guanine nucleotide exchange factors (GEFs), whereas hydrolysis of GTP for GDP is stimulated by GTPase-activating proteins (GAPs; Bernards and Settleman, 2004; Bos et al., 2007; Buchsbaum, 2007). A genome-wide analysis revealed that there are a large number of GEFs and GAPs in mammalian cells compared with the number of Rho GTPases (Peck et al., 2002). The finely coordinated activation of Rho GTPases in time and space is mediated by activation of numerous GEFs and GAPs. Therefore it is crucial to elucidate physiological functions of each protein to fully understand the regulatory mechanisms of Rho GTPases.
To search for genes that are essential for actin cytoskeleton organization, we performed a screening using a library of small interfering RNAs (siRNAs) targeting GEFs and GAPs. Because actin remodeling is associated with cellular morphological changes, we transfected each siRNA into HeLa cells and simply examined the cells by microscopy. During this screening, we found that transfection of siRNA targeting ARHGAP18 significantly increased the number of rounded cells. Previous research showed specific expression of ARHGAP18 in the epididymis (Li et al., 2008), but its physiological functions have never been reported. Here we report that ARHGAP18 is a novel RhoA GAP that regulates cell shape, migration, and spreading.
RESULTS
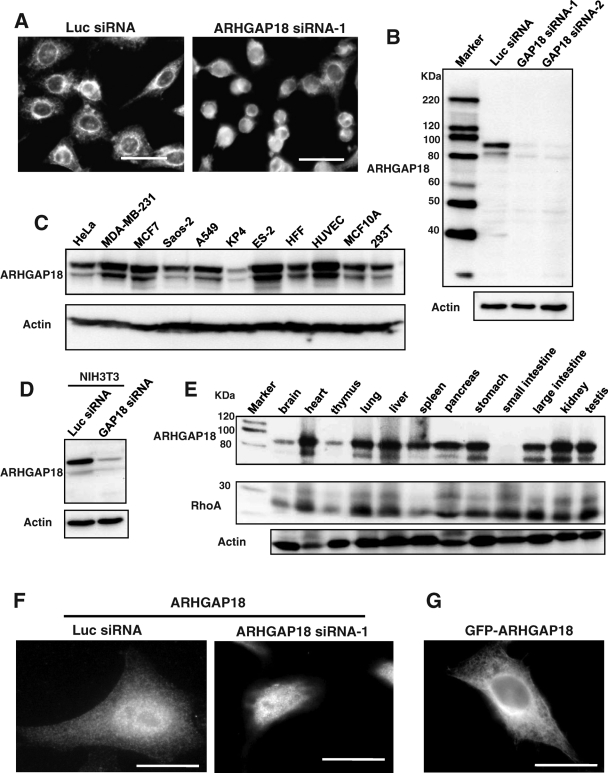
A search for GEFs and GAPs that are required for actin cytoskeleton organization using a library of siRNAs revealed that transfection with siRNA targeting ARHGAP18 increased the number of rounded cells (Figure 1A). ARHGAP18 contains a C-terminal RhoGAP domain without any other specific domains, and its biological function has never been reported. To examine the ability of ARHGAP18 siRNA to knock down the expression of the endogenous protein, we generated affinity-purified polyclonal antibody against amino acids 70–116 of ARHGAP18. Immunoblotting revealed two immunoreactive bands around 90 kDa, both of which were clearly lost with transfection of ARHGAP18 siRNA, indicating that both bands were ARHGAP18 (Figure 1B). Slower-migrating form of proteins are often observed by phosphorylation. We treated immunoprecipitated ARHGAP18 with phosphatase but did not observe any change in the mobility of the slower-migrating band (Supplemental Figure S1A). We found that transfection of both C-terminally Myc-tagged and nontagged ARHGAP18 expressed two bands similar to the endogenous ARHGAP18 (Supplemental Figures S1B and S1C). ARHGAP18 has second ATG at amino acid 55. Transfection of plasmid that encoded the open reading frame of ARHGAP18 from amino acid 55 showed a single band around the molecular size of the fast-migrating form of endogenous ARHGAP18 (Supplemental Figure S1D). These results suggest that the fast-migrating from of ARHGAP18 is a product translated from the secondary start codon. We next examined expression of ARHGAP18 in cell lines and tissues. As shown in Figure 1C, expression of ARHGAP18 was observed in cell lines derived from various tissues. Because the antibody was able to detect mouse ARHGAP18 (Figure 1D), we used mouse tissues to examine the expression. Immunoblotting of tissue lysates revealed expression of ARHGAP18 in most organs except small intestine (Figure 1E). We next investigated the subcellular localization of ARHGAP18. HeLa cells were cultured on fibronectin-coated glass coverslips, transfected with either luciferase (Luc) or ARHGAP18 siRNA, and immunostained with anti-ARHGAP18 antibody (Figure 1F). Signal intensity from the nucleus was similar for Luc and ARHGAP18 siRNA–transfected cells, indicating that the nuclear staining was nonspecific. However, diffuse cytoplasmic localization of ARHGAP18 was visible only in the Luc siRNA–transfected cells. Consistent with this result, green fluorescent protein (GFP)–tagged ARHGAP18 showed localization in the cytoplasm (Figure 1G). We concluded that ARHGAP18 is localized in the cytoplasm.
FIGURE 1:
Expression and localization of ARHGAP18 in cells. (A) HeLa cells were cultured in 24-well plates and transfected with Luc or ARHGAP18 siRNA. Seventy-two hours later, cells were fixed and stained with FITC-labeled paclitaxel to visualize cells. Scale bar, 50 μm. (B) HeLa cells were transfected with indicated siRNAs, and 72 h later, the cells were lysed and immunoblotted with anti-ARHGAP18 antibody. (C) Expression of ARHGAP18 in cell lines from various tissues. (D) NIH3T3 cells were transfected with either Luc or mouse ARHGAP18 siRNA, and 72 h later, cells were lysed and the expression was examined by immunoblot. (E) Expression of ARHGAP18 and RhoA in mouse tissues was examined by immunoblot. (F) HeLa cells transfected with Luc or ARHGAP18 siRNA were fixed and immunostained with anti-ARHGAP18 antibody. Scale bar, 20 μm. (E) GFP-tagged ARHGAP18 was transiently expressed in HeLa cells. Scale bar, 20 μm.
The actin cytoskeleton is critical for regulating cell shape; therefore we examined actin organization in the absence of ARHGAP18 expression. HeLa cells and MDA-MB-231 breast cancer cells were cultured on fibronectin-coated glass coverslips and transfected with each siRNA. Three days later, cells were fixed and immunostained for the actin cytoskeleton. Silencing of ARHGAP18 did not induce robust cell rounding when cultured on the fibronectin-coated surface, but we observed a clear difference in the formation of actin stress fibers. As shown in Figure 2A, the number of actin stress fibers was clearly increased in the ARHGAP18 siRNA–transfected cells compared with that of Luc siRNA–transfected cells. The organization of focal adhesions is regulated by actin cytoskeleton formation. Thus we immunostained cells with anti-vinculin antibody to visualize focal adhesions. MDA-MB-231 cells have a low number of focal adhesions, but suppression of ARHGAP18 expression increased the number of focal adhesions (Figure 2B). Clear formation of focal adhesions was observed on the edge and ventral surface of Luc siRNA–transfected HeLa cells, whereas ARHGAP18 siRNA–transfected HeLa cells showed a characteristic distribution of focal adhesions clustered along the periphery of the cells (Figure 2B). These results indicate that ARHGAP18 is an essential component that regulates the integrity of the actin cytoskeleton and focal adhesions.
FIGURE 2:
Suppression of ARHGAP18 affects stress fiber and focal adhesion formation. (A) HeLa and MDA-MB-231 cells cultured on the fibronectin-coated glass coverslips were transfected with Luc or ARHGAP18 siRNA and 3 d later, cells were fixed and stained with rhodamine-conjugated phalloidin. Scale bar, 20 μm. (B) HeLa and MDA-MB-231 cells were treated as in A and immunostained with anti-vinculin antibody. Scale bar, 20 μm.
Because the suppression of ARHGAP18 enhanced stress fiber formation, we speculated that overexpression of ARHGAP18 would disrupt the actin cytoskeleton. We used Saos-2 cells because these cells have clear and thick actin stress fibers. We established Saos-2 cells that constitutively expressed GFP (GFP), GFP-ARHGAP18 (FL), GFP-ARHGAP18 deleted of the RhoGAP domain (ΔGAP), and GFP-ARHGAP18 in which the conserved arginine at 365 in the RhoGAP domain was substituted with alanine (R365A). The substitution of the conserved arginine in the RhoGAP domain to alanine is known to abolish GAP activity (Barrett et al., 1997). As shown in Figure 3A, expression of GFP-ARHGAP18 was considerably higher than that of endogenous protein, and each GFP-tagged protein showed a similar level of expression. We then examined the formation of actin stress fibers and focal adhesions. Although expression of GFP did not affect formation of stress fibers and focal adhesions, FL cells showed a significantly reduced number of stress fibers and focal adhesions (Figure 3, B and C). ΔGAP and R365A cells showed normal organization of actin stress fibers and focal adhesions similar to GFP cells (Figure 3, B and C). These results indicate that ARHGAP18 regulates the formation of the actin cytoskeleton and focal adhesions through GAP activity.
FIGURE 3:
Overexpression of wild-type ARHGAP18, but not GAP-defective ARHGAP18, suppresses formation of stress fibers and focal adhesions. (A) Expression of ARHGAP18 in each cell line was examined with anti-ARHGAP18 and anti-GFP antibodies. FL, full-length ARHGAP18; ΔGAP, ARHGAP18 with the deletion of RhoGAP domain; R365A, full-length ARHGAP18 with the substitution of arginine at 365 to alanine. Arrows indicate endogenous ARHGAP18. (B) Cells cultured on the fibronectin-coated glass coverslips were fixed and stained with rhodamine-conjugated phalloidin. Scale bar, 20 μm. (C) Cells cultured on the fibronectin-coated glass coverslips were fixed and stained with anti-vinculin antibody. Scale bar, 20 μm.
Cellular changes induced by the suppression or overexpression of ARHGAP18 are similar to those induced by activation or inactivation of RhoA, respectively. To determine whether ARHGAP18 regulates the activity of RhoA, we performed a pull-down assay. Lysates of GFP- and GFP-ARHGAP18–overexpressing HeLa cells were incubated with glutathione S-transferase (GST)–Rhotekin–Rho-binding domain (RBD) or GST–p21 activated kinase (PAK)–Rac/Cdc42 (p21) binding domain (PBD) bound to glutathione agarose beads, and affinity precipitated proteins were probed for Rho GTPases by immunoblot analysis. As shown in Figure 4A, the activity of RhoA was diminished in cells that overexpressed ARHGAP18. In contrast, the activities of Rac1 and Cdc42 were not affected by ARHGAP18 expression. Rho GAPs have been reported to bind with a high affinity to activated Rho GTPases (Garcia-Mata et al., 2006). Therefore we examined the association of active RhoA (Q63L) and ARHGAP18. Lysates of HeLa cells expressing GFP-ARHGAP18 were incubated with recombinant GST-fused active RhoA coupled to glutathione beads, and bound proteins were affinity precipitated. As shown in Figure 4B, ARHGAP18 interacted with active RhoA.
FIGURE 4:
Regulation of RhoA activity by ARHGAP18 controls stress fiber formation. (A) HeLa cells that constitutively expressed either GFP or GFP-tagged ARHGAP18 were lysed and mixed with GST-Rhotekin-RBD (RhoA) or GST-PAK-PBD (Rac1 and Cdc42) bound to glutathione-agarose beads to precipitate the active form of Rho GTPases. The immunoprecipitates were subjected to immunoblot analysis with the indicated antibodies. (B) HeLa cells that constitutively expressed GFP-tagged ARHGAP18 were lysed and mixed with either GST or GST-RhoA (Q63L) coupled to glutathione-agarose beads, and interacting proteins were affinity precipitated. The immunoprecipitates were subjected to immunoblot analysis with anti-GFP antibody to determine the association of GFP-tagged ARHGAP18 and RhoA (Q63L). Bottom, Coomassie blue staining of recombinant proteins. (C) HeLa cells cultured on the fibronectin-coated glass coverslips were transfected with the indicated siRNAs. Three days later, ARHGAP18 siRNA–transfected cells were treated with DMSO or Y27632 for 1 h and immunostained with rhodamine-conjugated phalloidin. Scale bar, 20 μm. (D) HeLa cells were infected with recombinant virus that encoded the shRNA targeting either luciferase (Luc) or ARHGAP18 and selected with puromycin. Expression of ARHGAP18 in each cell line was examined by immunoblot analysis. Cells were transfected with GFP or GFP-tagged dominant-negative RhoA (T19N) and 48 h later, cells were fixed and immunostained with rhodamine-conjugated phalloidin. Scale bar, 20 μm.
To address whether the enhanced organization of actin stress fibers was due to excessive RhoA signaling, we used Y27632, an inhibitor of Rho kinase. As shown in Figure 4C, addition of the inhibitor abolished the enhanced formation of stress fibers by ARHGAP18 knockdown. To further confirm the requirement of RhoA, we expressed dominant-negative RhoA in the absence of ARHGAP18 expression. We established HeLa cells that had reduced expression of ARHGAP18 using recombinant retrovirus that encoded short hairpin RNA (shRNA) against ARHGAP18 (HeLa/shGAP18) and then transfected GFP-tagged dominant-negative RhoA. We observed enhanced formation of actin stress fibers in HeLa/shGAP18 cells compared with control cells, but transient expression of dominant-negative RhoA in HeLa/shGAP18 cells suppressed stress fiber formation (Figure 4D). These results demonstrate that ARHGAP18 regulates stress fiber formation by modulating RhoA activity.
Cell spreading is a process in which cells produce active membrane protrusions upon adhesion to the extracellular matrix (Ito et al., 2010). Activation of Rac1 and inactivation of RhoA are essential for prompt cell spreading (Arthur and Burridge, 2001; Arthur et al., 2002; Sepulveda and Wu, 2006). To address whether ARHGAP18 controls cell spreading, we first examined the localization of ARHGAP18. As shown in Figure 5A, ARHGAP18 was localized on the edge of membrane protrusions during cell spreading. We next examined the spreading of HeLa cells in the absence of ARHGAP18. Cells were transfected with either Luc or ARHGAP18 siRNA. Then, 72 h later, the suspended cells were seeded on fibronectin-coated dishes, and spreading was monitored by time-lapse microscopy (Figure 5B). On attachment to the surface, Luc siRNA transfected cells started to produce active membrane protrusions to facilitate spreading. In contrast, we observed that more than 60% of ARHGAP18 siRNA–transfected cells were impaired in forming protrusions. These cells produced active membrane blebs outward and gradually spread to take a stable morphology (Figure 5B). We fixed cells 1 h after seeding and evaluated the ratio of spread cells. We found that cell spreading was clearly suppressed in ARHGAP18-knockdown cells (Figure 5C). Immunostaining analysis of cells 1 h after plating revealed that ARHGAP18-knockdown cells formed actin stress fibers in the early stage of cell spreading, whereas control cells were still devoid of stress fiber formation (Figure 5D). We measured RhoA activity during cell spreading by precipitating active GTP-bound RhoA with GST-Rhotekin-RBD. Consistent with previous work (Ren et al., 1999; Arthur and Burridge, 2001), RhoA activity was reduced during spreading of Luc siRNA–transfected cells (Figure 5E). In contrast, ARHGAP18 siRNA–transfected cells showed sustained activation of RhoA (Figure 5E), indicating that ARHGAP18 is required for the inactivation of RhoA during cell spreading. To confirm that sustained RhoA activity inhibited cell spreading in ARHGAP18-knockdown cells, we used Rho kinase inhibitor. siRNA-transfected cells were seeded on a fibronectin-coated surface with or without the inhibitor, and cell spreading was examined 1 h later. As shown in Figure 5F, addition of the inhibitor restored spreading of ARHGAP18-knockdown cells. These results indicate that ARHGAP18-mediated suppression of RhoA activity is required for cell spreading. Finally, we examined the effects of silencing ARHGAP18 on cell attachment. As shown in Figure 5G, cell attachment to a fibronectin-coated surface was not affected by ARHGAP18 knockdown.
FIGURE 5:
Inactivation of RhoA by ARHGAP18 is necessary for prompt cell spreading. (A) HeLa cells were seeded on fibronectin-coated surfaces and fixed 1 h later. Cells were immunostained with anti-ARHGAP18 or anti-GST antibody. Scale bar, 20 μm. (B) HeLa cells were transfected with either Luc or ARHGAP18 siRNA, and 72 h later, suspended cells were seeded onto fibronectin-coated dishes. Cell spreading was monitored by time-lapse microscopy. Representative images are shown. Scale bar, 20 μm. (C) siRNA-transfected HeLa cells were seeded on the fibronectin-coated dishes, and 1 h later, cells were fixed and evaluated for the ratio of spread cells. The graph shows ratios of spread cells counted from five random fields of three independent experiments (means ± SD; *p < 0.01). (D) siRNA-transfected HeLa cells were plated on fibronectin-coated glass coverslips, and 1 h later, cells were fixed and immunostained with rhodamine-conjugated phalloidin. scale bar, 20 μm. (E) HeLa/shLuc or HeLa/shGAP18 cells were seeded on fibronectin-coated dishes and lysed at the indicated time points. Cell lysates were mixed with GST-Rhotekin-RBD coupled to glutathione-agarose beads to precipitate active RhoA. The immunoprecipitates were subjected to immunoblot analysis with anti-RhoA antibody. (F) siRNA-transfected HeLa cells were seeded on the fibronectin-coated surface, and 15 min later either DMSO or Y27632 was added. One hour after seeding, the cells were fixed and counted for the ratio of spread cells. The graphs indicate the ratio of spread cells counted in five randomly selected fields from three independent experiments (means ± SD; *p < 0.01). (G) Cell attachment assays of Luc and GAP18 siRNA–transfected HeLa cells. Cells (1 × 105) were seeded onto fibronectin-coated 24-well plates, and 20 min later, unattached cells were washed out and attached cells in five randomly selected fields were counted. The graph shows the relative ratio of attached cells from three independent experiments (means ± SD; *p < 0.01).
We next tested whether ARHGAP18 is involved in cell migration by using a wound-healing assay. We first examined localization of ARHGAP18 during cell migration. A confluent monolayer of MDA-MB-231 cells was scratched, and 4 h later, cells were fixed and immunostained for ARHGAP18. Similar to the localization during spreading, ARHGAP18 was localized to the leading edge of migrating cells (Figure 6A). To determine the effects of ARHGAP18 knockdown on migration, a confluent monolayer of siRNA-transfected cells was scratched, and migration was examined. We found that wound closure was delayed in ARHGAP18 siRNA-treated cells relative to Luc siRNA-treated cells (Figure 6B). To further confirm the promigratory role of ARHGAP18, we examined cell migration using a Boyden chamber. Consistent with the results obtained from the wound-healing assay, delayed migration was observed in ARHGAP18-knockdown cells (Figure 6C). In addition, overexpression of ARHGAP18 enhanced cell migration (Figure 6D). These results clearly demonstrate that ARHGAP18 is essential for the promotion of cell migration.
FIGURE 6:
ARHGAP18 regulates cell migration. (A) Confluent monolayers of MDA-MB-231 cells were scratched and fixed 4 h later. Cells were immunostained with anti-ARHGAP or anti-GST antibody. (B) MDA-MB-231 cells were transfected with Luc or ARHGAP18 siRNA, and 3 d later, a scratch was made and migration was examined for 18 h. The graph shows the distance of the wound measured in five randomly selected points from three independent experiments (means ± SD; *p < 0.01). Right, representative images of cells in the assay. Scale bar, 500 μm. (C) siRNA-transfected MDA-MB-231 cells were loaded onto the upper surface of Boyden chambers, incubated for 4 h, fixed, and examined by microscopy. The graph indicates the relative ratio of cells that migrated to the lower surface of the filter from five randomly selected fields in three independent experiments (means ± SD; *p < 0.01). (D) MDA-MB-231 cells that constitutively expressed GFP or GFP-tagged ARHGAP18 were established by infecting with recombinant retrovirus. Expression of ARHGAP18 was examined by immunoblot. The arrow indicates endogenous ARHGAP18 and the arrowhead GFP-ARHGAP18. Migration of these cells was examined using Boyden chambers. The graph indicates the relative ratio of cells that migrated to the lower surface of the filter from five randomly selected fields in three independent experiments (means ± SD; *p < 0.01).
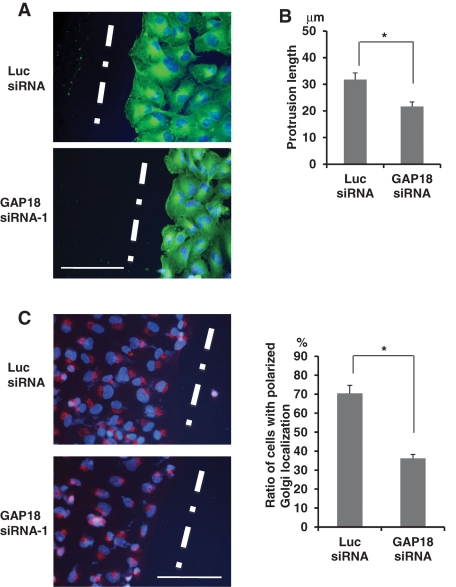
Cell polarization is an important prelude for cell migration; therefore we examined the role of ARHGAP18 on cell polarization during cell migration on the wound. We used KMST-6 human fibroblast cells because polarization upon wounding was more evident in this cell line. ARHGAP18 is essential for the regulation of migration and spreading of this cell line. Suppression of ARHGAP18 expression by siRNA transfection inhibited migration of KMST-6 cells (Supplemental Figure S2, A and B). Conversely, overexpression of ARHGAP18 enhanced cell migration (Supplemental Figure S2, C and D). In addition, ARHGAP18 knockdown suppressed spreading of this cell line (Supplemental Figure S2E). To examine polarization of KMST-6 cells in the absence of ARHGAP18, cells were transfected with either Luc or ARHGAP18 siRNA, and 72 h later a confluent monolayer of cells was scratched and then fixed 4 h later. More than 90% of Luc siRNA–transfected cells on the wound edge showed a polarized membrane protrusion toward the wound. In contrast, <20% of ARHGAP18-knockdown cells at the wound edge displayed polarized morphology (Figure 7A). Next we measured the protrusion length of cells on the wound edge. Luc siRNA–transfected cells exhibited protrusions with a length of 31.75 ± 0.36 μm (mean ± SD). In contrast, the protrusion length of ARHGAP18 knockdown cells was 21.6 5 ± 1.81 μm, ∼68% of that of the Luc siRNA–transfected cells (Figure 7B).
FIGURE 7:
ARHGAP18 controls cellular polarity for migration. (A) A confluent monolayer of KMST-6 cells transfected with each siRNA was scratched and incubated for 4 h. Cells were fixed and immunostained with anti–α-tubulin antibody and DAPI. White lines indicate wound direction. Scale bar, 100 μm. (B) The length of protrusions of cells on the wound edge treated as in A was measured. Fifty cells from randomly selected fields were measured in each of three independent experiments. Data are shown as average distances (mean ± SD) between the leading edge and the nucleus (*p < 0.01). (C) siRNA-transfected KMST-6 cells were wounded and incubated for 4 h. Cells were fixed and immunostained with anti-GM130 antibody and DAPI to evaluate the percentage of cells with Golgi located in the 120° arc facing the wound. One hundred cells on the wound edge were evaluated for Golgi localization in each of three independent experiments (mean ± SD; *p < 0.01). Left, representative images of immunostained cells 4 h after wounding. White lines indicate wound direction. Red, GM130; blue, DAPI. Scale bar, 100 μm.
When cells are polarized for migration, the Golgi becomes reoriented between the nucleus and the direction of migration. We tested whether ARHGAP18 was required for the polarized localization of the Golgi during cell migration. To determine the localization of the Golgi, the cells were equally divided into three sections including the front sector between the nucleus and the leading edge. The Golgi in the front sector was considered to be in the polarized position. On wounding, around one third of the cells showed polarized localization of the Golgi, which would happen by chance because the cells were sectioned into three 120° arcs. Four hours after wounding, the cells were fixed and immunostained for the Golgi and nucleus to determine the localization. We found that ∼70% of Luc siRNA–transfected cells on the wound edge showed polarized localization of the Golgi. In contrast, the Golgi was randomly distributed in ARHGAP18 siRNA–transfected cells on the wound edge (Figure 7C). These results indicate that ARHGAP18 is necessary for the establishment of cell polarity and the production of membrane protrusions to induce migration of KMST-6 cells.
DISCUSSION
The present study demonstrates the essential role of ARHGAP18 in the regulation of the actin cytoskeleton and focal adhesion formation. Silencing of ARHGAP18 expression by two different siRNAs induced stress fiber formation. Conversely, overexpression of wild-type ARHGAP18, but not GAP-defective ARHGAP18, resulted in the disruption of stress fiber and focal adhesion formation. We found that enhanced formation of actin stress fibers by ARHGAP18 knockdown was abolished by either addition of Rho kinase inhibitor or expression of dominant-negative RhoA. These results indicate that ARHGAP18 regulates RhoA activity to control actin cytoskeleton organization. Although previous research using immunohistochemical analysis demonstrated that ARHGAP18 is specifically expressed in the epididymis (Li et al., 2008), we found that ARHGAP18 was expressed in many cell lines as well as various tissues. This finding suggests that ARHGAP18 is ubiquitously required to maintain the integrity of the actin cytoskeleton.
To confirm that ARHGAP18 stimulates the hydrolytic activity of RhoA, we performed an in vitro assay to measure the GTP hydrolysis of RhoA, Rac1, and Cdc42 using a recombinant ARHGAP18 protein. However, we did not observe GAP activity toward any of these GTPases (Supplemental Figure S3). We speculated that posttranslational modifications or interactions with other proteins are necessary for ARHGAP18 to activate GTP hydrolysis on RhoA. In this context, it was reported that MgcRacGap, which is essential for the completion of cell division and exhibits GAP activity toward Rac1 and Cdc42 in vitro, is converted to a RhoA GAP by phosphorylation of a specific serine residue during cytokinesis (Minoshima et al., 2003). Although we failed to detect GAP activity of ARHGAP18 on RhoA in vitro, we observed clear reduction in RhoA activity, but not Rac1 or Cdc42, in cells with overexpression of ARHGAP18. In addition, we found that ARHGAP18 associates with active RhoA. Combined with the phenotypic changes induced by suppression or overexpression of ARHGAP18, these results clearly show that ARHGAP18 stimulates the intrinsic hydrolytic activity of RhoA in cells.
Cell spreading is regulated by dynamic remodeling of the actin cytoskeleton. On cell adhesion to the ECM, integrin-mediated signals induce production of active membrane protrusion. After this initial cell spreading, cellular contraction induces retraction of membrane protrusions to form a stable morphology (Ito et al., 2010). Rho family GTPases are critical factors for rearrangement of the actin cytoskeleton during cell spreading. For example, Rac1 is activated upon cell adhesion to the ECM, and the activation is required for the production of membrane protrusion in the early phase of spreading (Arthur et al., 2002). Conversely, RhoA is inactivated after cell–ECM interaction to relieve cellular contraction, allowing cells to spread. Arthur and Burridge (2001) reported that expression of dominant-negative p190RhoGAP in Rat1 fibroblast cells suppressed a transient decrease in RhoA activity after cell attachment and inhibited membrane extension. Similar to these results, ARHGAP18-knockdown cells were defective in production of membrane protrusions and showed sustained activation of RhoA after cell–ECM interaction. Consistent with this activation of RhoA, premature formation of stress fibers was observed in the early phase of spreading, when control cells were still devoid of the formation. Addition of Rho kinase inhibitor rescued the spreading defect by ARHGAP18 knockdown. These results strongly indicate that ARHGAP18-mediated inactivation of RhoA plays a pivotal role in the promotion of cell spreading. Tyrosine phosphorylation of p190RhoGAP by c-Src is essential for the activation of p190RhoGAP to reduce RhoA activity, promoting membrane extension for spreading (Arthur et al., 2000). We examined tyrosine phosphorylation of ARHGAP18 upon cell attachment; however, we did not observe any phosphorylation (Supplemental Figure S4). We also used anti-MPM2 antibody, which detects phosphorylated serine or threonine followed by proline. Although phosphorylation of ARHGAP18 was detected by anti-MPM2 antibody (Supplemental Figure S4), there was no difference in phosphorylation between cells in suspension and spread. Whether phosphorylation regulates the activity of ARHGAP18 requires further investigation; however, interactions with other molecules may regulate the activity or localization of ARHGAP18 to control RhoA activity during cell spreading.
Cell migration is a complex process that involves membrane extension and the formation of focal adhesions on the leading edge, as well as retraction and detachment at the rear of the cell (Ridley et al., 2003; Raftopoulou and Hall, 2004). We found that overexpression or suppression of ARHGAP18 expression resulted in the promotion or inhibition of cell migration, respectively. In addition, ARHGAP18 was localized to the leading edge, and ARHGAP18 silencing suppressed membrane extension in the direction of migration. Recent studies using biosensors revealed spatiotemporal activation of Rho GTPases at the leading edge during cell migration (Kraynov et al., 2000; Nalbant et al., 2004; Pertz et al., 2006). RhoA is activated at the very front of the leading edge, whereas activation of Cdc42 and Rac1 was observed around 2 μm behind the edge with a 40-s delay relative to the RhoA activation (Machacek et al., 2009). These spatiotemporal activations of Rho family GTPases at the leading edge are essential for efficient cell migration; however, it remains to be determined which GEFs and GAPs are crucial for activation. Our results indicate that ARHGAP18 may be one of the essential factors that coordinates RhoA activity at the leading edge to promote membrane protrusion for migration. We also found that ARHGAP18 was required for the polarized localization of the Golgi during cell migration. It is well documented that Cdc42 is a central player in the polarization of cells (Etienne-Manneville, 2004). Cdc42 activates the Par6/aPKC complex, followed by the inactivation of GSK3β and accumulation of adenomatous polyposis coli at the plus end of microtubules, to induce polarization of migrating cells (Etienne-Manneville and Hall, 2003). We examined the activity of Cdc42 in Luc and ARHGAP18 siRNA–transfected cells during cell migration, but we did not detect any difference in activity (Supplemental Figure S5). ARHGAP18 may regulate signal pathways either downstream or independent of Cdc42 to control the polarization of migrating cells. In addition to Cdc42, the RhoA/mDia pathway has been reported to regulate cellular polarity by mediating stabilization of microtubules (Palazzo et al., 2001; Gundersen et al., 2005). Therefore activation of the RhoA/mDia pathway by knocking down ARHGAP18 may induce aberrant stabilization of microtubules to inhibit polarization.
In summary, we demonstrated that ARHGAP18 is a novel RhoA GAP required for remodeling the actin cytoskeleton in response to integrin engagement. ARHGAP18 is localized on the leading edge of cells during spreading and migration; therefore it is likely that ARHGAP18 inactivates RhoA on the membrane edge of cells to control cellular shape changes. In future work, it will be interesting to explore how the activity of ARHGAP18 is regulated to modulate RhoA activity in time and space to control cell migration and spreading.
MATERIALS AND METHODS
Cells, antibodies, and chemicals
HeLa, MDA-MB-231, KMST-6, and HEK293T cells were propagated in DMEM supplemented with 10% fetal bovine serum. Saos-2 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. Saos-2 and KMST-6 cells were kindly provided by the Cell Resource Center for Biomedical Research, Tohoku University, Sendai, Japan. To generate an anti-ARHGAP18 antibody, the N-terminus of ARHGAP18 (amino acids 70–116) fused with GST was produced in bacteria, and recombinant protein was purified using glutathione agarose beads (Sigma-Aldrich, St. Louis, MO). The protein was mixed with Freund's adjuvant (Sigma-Aldrich) and injected into a rabbit four times every 2 wk. To purify the anti-GAP18 and anti-GST antibodies, we used HiTrap NHS-activated HP columns (GE Healthcare BioScience, Uppsala, Sweden) coupled with recombinant GST-GAP18 or GST. Other antibodies were purchased from the following manufacturers: anti-vinculin, anti–α-tubulin, and anti–β-actin antibodies, Sigma-Aldrich; anti-GFP antibody, Nacalai Tesque (Tokyo, Japan); and anti-GM130, anti-Rac1, anti-Cdc42, and anti-RhoA antibodies, BD Transduction Laboratories (San Jose, CA). Rhodamine-conjugated phalloidin was obtained from Invitrogen (Carlsbad, CA). Rho kinase inhibitor Y27632 was kindly provided by K. Kaibuchi (Graduate School of Medicine, Nagoya University, Nagoya, Japan).
siRNA screening
A library of siRNAs targeting GAPs and GEFs was purchased from Invitrogen. HeLa cells were cultured in 24-well plates and transfected with 20 nM of each siRNA with Lipofectamine RNAiMAX (Invitrogen). Seventy-two hours later, cells were fixed with 4% paraformaldehyde and stained with fluorescein isothiocyanate (FITC)–labeled paclitaxel (Invitrogen) to visualize cells. Pictures were taken using an Olympus (Tokyo, Japan) IX71 fluorescence microscope .
DNA constructs
Human ARHGAP18 was amplified by PCR from a HeLa cDNA library. Full-length ARHGAP18 was cloned into a pQCXIP retrovirus vector with a GFP tag on the N-terminus (Clontech, Mountain View, CA). To produce GFP-tagged ΔGAP, the region of 1–322 amino acids was amplified by PCR and ligated into the pQCXIP vector. R365A was produced by PCR-based site-directed mutagenesis. Plasmids that encoded for GST-tagged active RhoA (Q63L) and GFP-tagged inactive RhoA (T19N) were kindly provided by K. Kaibuchi.
Generation of stable cell lines
Saos-2 cells that constitutively expressed each protein were established by retrovirus infection. pQCXIP vectors that encoded each cDNA were transfected to 293T cells in combination with the pVPack-GP and pVPack-Ampho vectors (Stratagene, Tokyo, Japan) using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the supernatants were added to Saos-2 cells with 2 μg/ml polybrene (Sigma-Aldrich), and infected cells were selected with 1 μg/ml puromycin for 3 d. MDA-MB-231 and HeLa cells that constitutively expressed either GFP or GFP-tagged, full-length ARHGAP18 were also established by the same protocol. To produce HeLa/shLuc and HeLa/shGAP18 cells, oligonucleotides encoding shRNA specific for human ARHGAP18 (5′-GGGTTATAAAGTCAAAGCCATTGTA-3′) and luciferase (5′-CTTACGCTGAGTACTTCGA-3′) were cloned into the pSIREN-RetroQ retroviral vector (Clontech). Recombinant retrovirus was produced, and infected HeLa cells were selected with 1 μg/ml puromycin for 3 d.
siRNA transfection
The sequences of the siRNAs used to suppress ARHGAP18 expression were 5′-GGGUUAUAAAGUCAAAGCCAUUGUA-3′ (siRNA-1), 5′-CAUUGACAGCGCUAUUAGAAC-3′ (siRNA-2), and 5′-GCAAAUGUCAUGCACUUAUTT-3′ (siRNA for mouse ARHGAP18). The sequence of the control siRNA targeting luciferase was 5′-CUUACGCUGAGUACUUCGATT-3′. Cells were transfected with 20 nM siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. siRNA-1 for ARHGAP18 (stealth siRNA) was obtained from Invitrogen, and the other siRNAs were from Sigma-Aldrich.
Immunofluorescence analysis
Cells were grown on glass coverslips coated with fibronectin, fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.5% Triton X-100 for 3 min, and blocked with phosphate-buffered saline (PBS) containing 7% fetal bovine serum for 30 min. Cells were incubated with primary antibody in PBS for 1 h, washed three times with PBS, incubated with FITC- or Alexa Fluor 594–labeled secondary antibody in PBS for 1 h, and then analyzed using a fluorescence microscope (BX60; Olympus). To visualize the actin cytoskeleton, fixed cells were incubated with rhodamine-conjugated phalloidin for 30 min and washed.
Rho GTPase activity assay
Cells were lysed with pull-down lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1% NP-40, 5 mM MgCl2, protease inhibitor cocktail [Roche, Basel, Switzerland], and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and incubated with GST-PAK-PBD or GST-Rhotekin-RBD fusion protein bound to glutathione-agarose beads for 1 h at 4°C. The beads were washed with pull-down buffer four times and then subjected to Western blot analysis with each antibody to detect active Rho GTPases. Total protein was detected by immunoblotting of whole cell lysates.
Pull-down assay for the ARHGAP18 and active RhoA interaction
HeLa cells that constitutively expressed GFP-tagged ARHGAP18 were lysed with lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, protease inhibitor cocktail, 1 mM PMSF) and incubated with GST or GST-active RhoA (Q63L) fusion protein bound to glutathione-agarose beads for 1 h at 4°C. Beads were washed with lysis buffer four times and then subjected to Western blot with anti-GFP antibody to detect the interaction between ARHGAP18 and RhoA.
Time-lapse analysis
HeLa cells were transfected with siRNAs; 3 d later, the suspended cells were seeded onto fibronectin-coated glass–based dishes (Iwaki, Tokyo, Japan). Spreading was monitored using a time-lapse microscope system (IX81-ZDC, Olympus) with a noncooled camera (Retiga Exi Fast, Q-Imaging, Surrey, Canada). Images were acquired and analyzed using the MetaMorph Imaging System (Molecular Devices, Sunnyvale, CA).
Cell spreading assay
Cells were seeded onto a 48-well pate coated with fibronectin at a density of 2.0 × 104 cells per well and fixed 1 h later. Spread and nonspread cells were counted in five randomly selected fields. Nonspread cells were defined as small, round cells with few or no membrane protrusions, whereas spread cells were defined as large cells with extensive visible lamellipodia. Y27632 (Rho kinase inhibitor) or dimethyl sulfoxide (DMSO) was added 15 min after seeding cells onto the plate. The data are presented as the average of the results from three independent experiments.
Cell attachment assay
Cells were seeded onto a 24-well plate coated with fibronectin at a density of 1.0 × 105 cells per well. After 20 min, unattached cells were removed by tapping the plate and rinsing the wells with PBS twice. Attached cells were counted in five representative high-power fields, and three independent experiments were performed.
Cell migration assays
Wound-healing assays were performed by scratching confluent monolayers of cells with a pipette tip and were incubated at 37°C with 5% CO2. Eighteen hours later, wound widths were measured in three randomly selected points from three independent experiments. To measure cell migration using Boyden chambers, 5.0 × 104 cells were seeded onto the upper surface of the chamber. The lower surface of the filter was coated with fibronectin. Four hours after seeding, cells were fixed with 70% methanol and stained with 0.5% of crystal violet. Cells that migrated to the lower surface of the filters were counted in five randomly selected fields from three independent experiments.
Golgi reorientation measurements
Measurement of Golgi reorientation was performed as described previously (Funasaka et al., 2010). In brief, a confluent monolayer of cells that had been cultured on fibronectin-coated glass slides were scratched with a pipette tip and incubated for 4 h. Cells were fixed and stained for GM130 to visualize the Golgi. Cells on the wound edge were divided equally into three sectors, including the front sector between the nucleus and the leading edge. The Golgi in the front sector was determined to be in the polarized position. One hundred cells in 20 randomly selected fields were evaluated for Golgi localization in each experiment, and three independent experiments were performed.
Measurement of protrusion length
siRNA-transfected cells were scratched and fixed 4 h later. Cells were immunostained with anti–α-tubulin antibody and 4′,6-diamidino-2-phenylindole (DAPI). To quantify the length of protrusion, the distance of the leading edge from the nuclei of cells on the wound edges was measured. Fifty cells in 10 randomly selected fields were evaluated for the protrusion length in each experiment, and three independent experiments were performed.
Supplementary Material
Acknowledgments
We thank the members of the Division of Cancer Biology for technical assistance and helpful discussions and K. Kaibuchi for kindly providing the plasmids. This research was partly funded by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used:
- GAP
GTPase activating protein
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- GTP
guanosine triphosphate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0364) on August 24, 2011.
REFERENCES
- Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 2010;67:545–554. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell. 2001;12:2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Noren NK, Burridge K. Regulation of Rho family GTPases by cell-cell and cell-matrix adhesion. Biol Res. 2002;35:239–246. doi: 10.4067/s0716-97602002000200016. [DOI] [PubMed] [Google Scholar]
- Arthur WT, Petch LA, Burridge K. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr Biol. 2000;10:719–722. doi: 10.1016/s0960-9822(00)00537-6. [DOI] [PubMed] [Google Scholar]
- Barrett T, Xiao B, Dodson EJ, Dodson G, Ludbrook SB, Nurmahomed K, Gamblin SJ, Musacchio A, Smerdon SJ, Eccleston JF. The structure of the GTPase-activating domain from p50rhoGAP. Nature. 1997;385:458–461. doi: 10.1038/385458a0. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;12:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Buchsbaum RJ. Rho activation at a glance. J Cell Sci. 2007;120:1149–1152. doi: 10.1242/jcs.03428. [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedhar S, Hannigan GE. Integrin cytoplasmic interactions and bidirectional transmembrane signalling. Curr Opin Cell Biol. 1996;8:657–669. doi: 10.1016/s0955-0674(96)80107-4. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42—the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Funasaka K, Ito S, Hasegawa H, Goldberg GS, Hirooka Y, Goto H, Hamaguchi M, Senga T. Cas utilizes Nck2 to activate Cdc42 and regulate cell polarization during cell migration in response to wound healing. FEBS J. 2010;277:3502–3513. doi: 10.1111/j.1742-4658.2010.07752.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, Burridge K. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 2006;406:425–437. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Gomes ER. Regulation of microtubules by Rho GTPases in migrating cells. Novartis Found Symp. 2005;269:106–116. discussion 116–126, 223–130. [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Ito S, Takahara Y, Hyodo T, Hasegawa H, Asano E, Hamaguchi M, Senga T. The roles of two distinct regions of PINCH-1 in the regulation of cell attachment and spreading. Mol Biol Cell. 2010;21:4120–4129. doi: 10.1091/mbc.E10-05-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- Kimura K, et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- Li X, Liu Q, Liu S, Zhang J, Zhang Y. New member of the guanosine triphosphatase activating protein family in the human epididymis. Acta Biochim Biophys Sin. 2008;40:855–863. [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 1996;15:2208–16. [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y, et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- Narumiya S, Ishizaki T, Watanabe N. Rho effectors and reorganization of actin cytoskeleton. FEBS Lett. 1997;410:68–72. doi: 10.1016/s0014-5793(97)00317-7. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- Peck J, Douglas GT, Wu CH, Burbelo PD. Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships. FEBS Lett. 2002;528:27–34. doi: 10.1016/s0014-5793(02)03331-8. [DOI] [PubMed] [Google Scholar]
- Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Schaller MD, Ginsberg MH. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Sepulveda JL, Wu C. The parvins. Cell Mol Life Sci. 2006;63:25–35. doi: 10.1007/s00018-005-5355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K, Der CJ. Rho-family GTPases: it's not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.