Abstract
The PI3K-Akt pathway is activated in cancer by genetic or epigenetic events and efforts are under way to develop targeted therapies. PTEN tumor suppressor is the major brake of the pathway and a common target for inactivation in glioblastoma, one of the most aggressive and therapy-resistant cancers. To achieve potent inhibition of the PI3K-Akt pathway in glioblastoma, we need to understand its mechanism of activation by investigating the interplay between its regulators. We show here that PTEN modulates the PI3K-Akt pathway in glioblastoma within a tumor suppressor network that includes NHERF1 and PHLPP1. The NHERF1 adaptor, previously characterized by our group as a PTEN ligand and regulator, shows also PTEN-independent Akt-modulating effects that led us to identify the PHLPP1/PHLPP2 Akt phosphatases as NHERF1 ligands. NHERF1 interacts via its PDZ domains with PHLPP1/PHLPP2 and scaffolds heterotrimeric complexes with PTEN. Functionally, PHLPP1 requires NHERF1 for membrane localization and growth suppressive effects. PHLPP1 loss boosts Akt phosphorylation only in PTEN-negative cells and cooperates with PTEN loss for tumor growth. In a panel of low-grade and high-grade glioma patient samples, we show for the first time a significant disruption of all three members of the PTEN-NHERF1-PHLPP1 tumor suppressor network in high-grade tumors, correlating with Akt activation and patients’ abysmal survival. We thus propose a PTEN-NHERF1-PHLPP PI3K-Akt pathway inhibitory network that relies on molecular interactions and can undergo parallel synergistic hits in glioblastoma.
Keywords: glioblastoma, PTEN, NHERF1, PHLPP, Akt
INTRODUCTION
The phosphatidylinositol-3-OH kinase (PI3K)-Akt pathway is a major signaling cascade activated in a large variety of human cancers (Carnero et al., 2008). The oncogenic activation of the pathway can take place at different levels in tumor cells, either by mutation or amplification of the activators of the pathway or by inactivation of the inhibitors of the pathway (Georgescu, 2010). Once activated, the PI3K-Akt pathway promotes growth, survival, motility and invasion of the transformed cells. The enzymatic product of the PI3K, phosphatidylinositol 3,4,5-trisphosphate (PIP3), acts as second messenger and recruits the serine-threonine kinase Akt through binding to its pleckstrin-homology (PH)-domain. This binding releases the PH domain from masking the kinase domain (Franke, 2008), and allows the phosphorylation of Akt on Thr308 in the activation loop and on Ser473 in the carboxyl-terminal (CT) hydrophobic motif (Alessi et al., 1998; Sarbassov et al., 2005). The recently described PH domain leucine-rich repeat protein phosphatases (PHLPP) 1 and 2 have been shown to specifically dephosphorylate Akt on Ser473, appearing thus to be the second major brake of the pathway (Brognard et al., 2007; Gao et al., 2005). PHLPP phosphatases contain an amino-terminal (NT) Ras-association domain, a PH domain, a leucine-rich-repeat region, a PP2C phosphatase domain and a tail region ending in a PDZ (PSD-95/Disc-large/ZO-1)-binding motif. The PDZ-binding motif has been shown to be required for Akt dephosphorylation and the pro-apototic effects of PHLPP1 (Gao et al., 2005), but the mechanism underlying these processes has not been fully explored.
The most upstream inhibitor of the pathway, PTEN tumor suppressor, directly antagonizes the activity of PI3K by dephosphorylating PIP3 (Maehama and Dixon, 1998). PTEN mutation with loss of heterozygosity (LOH) is one of the major genetic alterations in glioblastoma (Li et al., 1997; Steck et al., 1997). PTEN is formed of an NT phosphatase domain, a phospholipid-binding C2 domain and a CT tail-region ending also in a PDZ-binding motif (Georgescu et al., 1999; Lee et al., 1999). Via the PDZ-binding motif, PTEN can associate to adaptor PDZ-domain containing proteins, including the Na+/H+ exchanger regulatory factor 1/ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 (NHERF1/EBP50) (Takahashi et al., 2006). The efficient inactivation of PI3K by PTEN was shown to be dependent on the presence of these adaptor proteins in normal cells, presumably by stabilizing PTEN in protein complexes at the plasma membrane. Consequently, loss or displacement of NHERF1 from the plasma membrane has been reported in a number of human cancers, including glioblastoma (Cardone et al., 2007; Hayashi et al., 2010; Molina et al., 2010b; Song et al., 2007), and has been associated with the activation of the PI3K pathway in connection with PTEN tumor suppressor (Molina et al., 2010b; Pan et al., 2008). In this study, we show that surprisingly NHERF1 loss has PTEN-independent effects on Akt activation. We demonstrate that NHERF1 interacts with PHLPP proteins and we further connect PTEN, NHERF1 and PHLPP1 in an inhibitory network with synergistic effects on the control of the PI3K-Akt pathway in glioblastoma.
RESULTS
Synergistic Akt activation by combined PTEN and NHERF1 silencing leads to the identification of PHLPP proteins as novel NHERF1 ligands
We have previously shown that NHERF1 stabilizes PTEN to the plasma membrane to suppress Akt activation (Molina et al., 2010b). Through this mechanism, we have implicated NHERF1 as a PTEN co-suppressor of the PI3K/Akt pathway (Georgescu et al., 2008; Molina et al., 2010b; Takahashi et al., 2006). In this study, we investigated the possibility of a co-potentiating effect of NHERF1 loss on Akt activation following loss of PTEN. Efficient depletion of either PTEN or NHERF1 in LN229 glioblastoma cells that express wild-type PTEN and membrane-distributed NHERF1 (Molina et al., 2010b), elevated moderately the basal Akt phosphorylation (Fig. 1A). Surprisingly, double PTEN and NHERF1 depletion induced a potent increase of Akt phosphorylation (Fig. 1A). We confirmed this synergistic Akt activation by double PTEN-NHERF1 depletion in DU145 prostate cancer cells in which single depletion of PTEN or NHERF1 had no significant effect on Akt phosphorylation (Fig. S1A). Additionally, we observed correlation between Akt activation and significantly increased proliferation in the PTEN-NHERF1 double-depleted cells (Fig. S1B). Overall, these results demonstrated synergistic activation of the PI3K/Akt pathway by double PTEN-NHERF1 depletion and suggested independent PI3K/Akt inhibitory activities for NHERF1. We thus postulated that NHERF1 might bind to additional unidentified PI3K/Akt suppressors.
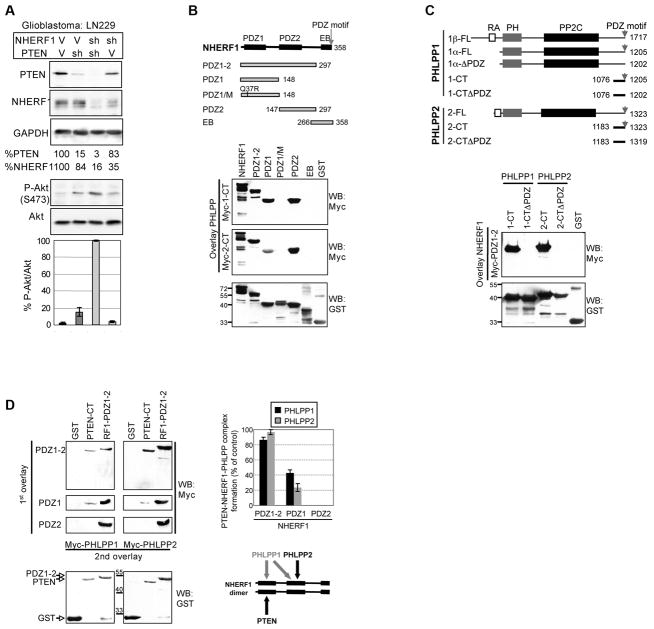
Figure 1.
Synergistic Akt activation by combined PTEN and NHERF1 knockdown points to NHERF1 as a scaffolding platform for PTEN and PHLPP proteins. A. Western blot of cell lysates from LN229 glioblastoma cells with knockdown of PTEN and NHERF1 by shRNA (sh). V, vector controls. The % of actin-normalized PTEN and NHERF1 levels relatively to vector control levels are shown. The graph shows the Akt activation as phosphorylated Akt S473 levels normalized to total Akt levels (P-Akt/Akt) from two independent infections. B-C. NHERF1 associates with PHLPP proteins. Schematic organization of NHERF1 (B) and PHLPP (C) with amino acid boundaries. EB, ERM-binding region; FL, full length; ΔPDZ, PDZ-binding-motif-deleted; RA, Ras-associating domain; PH domain; PP2C, protein phosphatase 2C domain. Reciprocal overlays with either My-tagged PHLPP1-CT or PHLPP2-CT on filter-immobilized NHERF1 GST-fusion proteins (B) or with Myc-tagged NHERF1-PDZ1-2 on filter-immobilized PHLPP1 and PHLPP2 GST-fusion proteins (C) were probed with Myc-antibody to detect interactions. Re-probing of the membranes with anti-GST antibody showed equal amounts of immobilized GST-fusion proteins. D. Bridging two-step overlay assay in which the indicated filter-immobilized proteins were overlaid in a first step (1st overlay) with the NHERF1 PDZ domains and, in a second step (2nd overlay), with Myc-PHLPP1-CT or Myc-PHLPP2-CT. Probing with Myc antibody revealed the bound Myc-tagged PHLPP1/2. The filters stripped and reprobed with GST antibody show the input amounts of immobilized proteins. The graph shows the PTEN-band intensities normalized to the corresponding input and expressed as % from the normalized values of the positive control. The cartoon is a simplified view of complex assembly by NHERF1.
NHERF1 is composed of two NT PDZ domains and a CT ERM-binding region ending in a PDZ-binding motif (Morales et al., 2007; Reczek et al., 1997; Weinman et al., 1998) (Fig. 1B). To identify inhibitors of the PI3K-Akt pathway that bind NHERF1, we searched for PI3K-Akt suppressor molecules with PDZ-binding motifs. Interestingly, the PHLPP proteins known to dephosphorylate Akt on Ser473 (Brognard et al., 2007; Gao et al., 2005), have PDZ-motifs that conform to the consensus motif for binding NHERF1 PDZ domains (Table 1) (Hall et al., 1998). We therefore expressed recombinant CT regions of PHLPP1 and PHLPP2 and tested their direct interaction to a battery of NHERF1 individual domains (Fig. 1B). PHLPP1 interacted with both NHERF1 PDZ domains while PHLPP2 interacted mainly with NHERF1 PDZ2 domain (Table 1 and Fig. 1B). We also confirmed this differential interaction by precipitation of endogenous PHLPP1/2 proteins with GST-fusion NHERF1 domains (Fig. S2). Conversely, we determined that the PHLPP PDZ-binding motifs are the major interaction sites for NHERF1 PDZ1-2 domains (Fig. 1C).
Table 1.
PHLPP1/2 contain consensus PDZ-motifs for binding NHERF1 PDZ domains
| NHERF1 ligand | PDZ-binding motif | Interacting NHERF1 PDZ domain | Reference |
|---|---|---|---|
| PHLPP1 | …YDTPL | PDZ1, PDZ2 | This study |
| PDGFR | …EDSFL | PDZ1 | (Maudsley et al., 2000) |
| PHLPP2 | …FDTAL | PDZ2 | This study |
| β-catenin | …FDTDL | PDZ2 | (Shibata et al., 2003) |
| PTEN | …QITKV | PDZ1 | (Takahashi et al., 2006) |
| NHERF1 | …LFSNL | PDZ2 | (Morales et al., 2007) |
Bold residues indicate the NHERF1 PDZ-binding consensus motif DT/SXL.
To test whether NHERF1 could engage complexes with both PTEN and PHLPP, we used a bridging two-step-overlay assay (Fig. S3) (Morales et al., 2007; Takahashi et al., 2006). GST-PTEN-CT containing PTEN’s PDZ-binding motif that specifically interacts with NHERF1 PDZ1 domain (Table 1) was immobilized on filter together with GST and GST-NHERF1-PDZ1-2, as negative and positive controls for PHLPP binding, respectively (Fig. 1D). Immobilized proteins were first overlaid with NHERF1 PDZ1-2, PDZ1 or PDZ2, washed, and subsequently overlaid with Myc-tagged PHLPP1-CT or PHLPP2-CT. NHERF1 PDZ1-2 domains efficiently assembled complexes having both PTEN and the PHLPP proteins (Fig. 1D). The single PDZ1 domain could bridge some complexes mainly with PHLPP1 that associates with both NHERF1 PDZ domains. The single PDZ2 domain that does not bind PTEN did not assemble complexes. The amount of PHLPP proteins engaged in NHERF1-PTEN complexes (Fig. 1D graph) correlated to the overall ability of NHERF1 PDZ domains to dimerize that is increased when both tandem PDZ domains are present (Fouassier et al., 2000; Shenolikar et al., 2001; Takahashi et al., 2006). However, the possibility of monomeric NHERF1 bridging complexes with PHLPP2 via PDZ2, and PTEN via PDZ1, cannot be excluded (Fig. 1D scheme).
NHERF1 interacts with PHLPP1/2 in vivo and regulates their plasma membrane recruitment and growth suppressive effect
We further tested the interaction between NHERF1 and PHLPP1/PHLPP2 in vivo. PHLPP1 has 2 splice isoforms, a shorter α isoform and a long β isoform that contains an extended NT region (Fig. 1C and S4) (Brognard and Newton, 2008). We expressed FLAG-tagged NHERF1 with either PHLPP1α-FL (full-length) or PHLPP1α-ΔPDZ in 293T cells. NHERF1 co-immunoprecipitated with PHLPP1α-FL and significantly less with the PHLPP1α-ΔPDZ form that lacks the PDZ-binding motif (Fig. 2A). A NHERF1 PDZ-domain double mutant also failed to co-immunoprecipitate with PHLPP1α-FL (not shown). We next found that NHERF1 interacts with both the long PHLPP1β isoform (Fig. 2B, upper panels) and PHLPP2 (Fig. 2B, lower panels). We have previously reported a relatively reduced NHERF1 association with PDZ-domain ligands, including PTEN, due to a closed conformation of NHERF1, in which NHERF1’s PDZ-binding motif masks its PDZ domains (Morales et al., 2007) (Table 1). By co-immunoprecipitating endogenous proteins from LN229 cells, we found that NHERF1 immunoprecipitated with two different antibodies associates with PHLPP1 (Fig. 2C). The quantification of the co-immunoprecipitated PHLPP1 relative to the input indicated that this endogenous fraction represents approximately 27.2%±7.3% of the ectopic PHLPP1 fraction co-immunoprecipitated with overexpressed FLAG-tagged NHERF1. These experiments suggested that a small but consistent fraction of endogenous NHERF1 is present in open conformation in complex with PHLPP1 in cells.
Figure 2.
NHERF1 interacts with PHLPP1/2 in vivo. A. Reciprocal co-immunoprecipitation (IP) between PHLPP1α-FL and 1α–ΔPDZ forms and FLAG-tagged NHERF1 overexpressed in 293T cells. TL, total lysate; HC, heavy chain. B. Co-immunoprecipitation of FLAG-tagged NHERF1 with PHLPP1β (upper panels) and PHLPP2 (lower panels). C. Co-immunoprecipitation of endogenous PHLPP1 and NHERF1 from LN229 total cell lysate. Two rabbit NHERF1 antibodies–I (Abcam) and II (Affinity BioReagents)–and rabbit IgG control were used. D. Immunofluorescence analysis (x400 with oil immersion) with Myc antibody (red), NHERF1 antibody (green) and DAPI (blue) of LN18 cells overexpressing the proteins indicated on top. Note recruitment of PHLPP1α but not PHLPP1α-ΔPDZ to the plasma membrane by Myr-NHERF1 (arrows).
NHERF1 is expressed at the plasma membrane in physiological conditions and is frequently displaced to the cytoplasm in cancer cells (Georgescu et al., 2008). Since PHLPP1/2 associate with NHERF1, we examined their subcellular localization in glioblastoma cells with different NHERF1 localizations. LN229 cells express predominantly membrane-localized NHERF1, whereas LN18 and LN428 cells express NHERF1 mainly in the cytoplasm (Fig. S5A). The distribution of PHLPP1β and PHLPP2 paralleled the compartmentalization of endogenous NHERF1 in these cells (Fig. S5A, graph). Importantly, the expression of a membrane-targeted Myr-NHERF1 form in LN18 and LN428 cells redistributed both endogenous PHLPP proteins from the cytoplasm to the membrane (Fig. S5B). Co-expression of Myr-NHERF1 with either PHLPP1α-FL or PHLPP1α-ΔPDZ in LN18 cells resulted in recruitment to the plasma membrane of PHLPP1α-FL but not PHLPP1α-ΔPDZ by Myr-NHERF1 (Fig. 2D), suggesting direct interaction between PHLPP1α and NHERF1 at the plasma membrane.
In LN229 cells, depletion of the endogenous membrane-localized NHERF1 significantly decreased PHLPP1β from the membrane fraction (Fig. 3A), suggesting membrane stabilization of PHLPP1 by NHERF1 in glioblastoma cells. To confirm these findings in normal cells, we fractionated NHERF1(+/+) and (−/−) MEFs. The membrane levels of both PHLPP1 and PTEN were reduced in NHERF1(−/−) MEFs (Fig. 3B), confirming the stabilization of these proteins at the membrane by NHERF1.
Figure 3.
NHERF1 stabilizes PHLPP1 at the membrane and regulates its growth suppression. A-B. Subcellular fractionation of LN229 glioblastoma cells transduced with vector (V) or NHERF1 shRNA (shRF1) (A), and of NHERF1 wild-type (WT) and knockout (KO) primary MEFs (B) in cytosolic (C) and membrane (M) fractions. Erk2 and N-cadherin were used as cytosolic and membrane fractionation markers, respectively. The membrane and cytoplasmic PHLPP1β or PTEN levels were normalized to the levels of N-cadherin and Erk2, respectively. C. Stable PHLPP1α (α) expression in A172 glioblastoma cells with and without prior NHERF1 knockdown by shRNA (sh). V, corresponding vector control. D. Proliferation assay with the 4 sets of cells from (A) showing significant proliferation suppression by PHLPP1α only in the presence of NHERF1.
PHLPP1 overexpression has been shown to suppress growth or promote apoptosis in a variety of cancer cells (Gao et al., 2005; Liu et al., 2009; Qiao et al., 2010). To examine whether NHERF1 regulates the growth suppressive effects of PHLPP1, we overexpressed PHLPP1α in cells with or without depletion of endogenous NHERF1 (Fig. 3C). We used the PTEN-negative A172 glioblastoma cells that have high endogeonous NHERF1 expression at the plasma membrane (Molina et al., 2010b). Virtually complete NHERF1 depletion increased cell proliferation, consistent with a tumor suppressor role of NHERF1 at the plasma membrane (Molina et al., 2010b), in this case independent of PTEN (Fig. 3D). PHLPP1α overexpression significantly suppressed cell proliferation but only in the presence of NHERF1 (Fig. 3D), indicating that NHERF1 is required for PHLPP1 growth suppressive effects.
PHLPP1 loss synergizes with PTEN loss for Akt activation and aggressive tumor growth
To show that the PHLPP proteins are the suppressors recruited by NHERF1 to synergize with PTEN for PI3K-Akt pathway control, we depleted PTEN and PHLPP1 in LN229 cells in which we have previously demonstrated synergistic Akt activation following PTEN-NHERF1 depletion (Fig. 1A). Several PHLPP1 shRNAs were tested and shRNA#2 that showed the highest depletion efficiency was used in further experiments (Fig. S6A). By itself, single PHLPP1 depletion was not sufficient to elevate Akt phosphorylation (Fig. 4A and S6B), suggesting that PTEN effectively suppresses PI3K-Akt signaling. Remarkably, knockdown of PHLPP1 in the LN229 PTEN-depleted cells robustly activated Akt by increasing phosphorylation on both Thr308 and Ser473 activation sites (Fig. 4A). Similarly, PTEN-PHLPP1 double depletion in DU145 cells elevated Akt phosphorylation levels similarly to the double PTEN-NHERF1 depletion (Figs. S1A and S7).
Figure 4.
Synergistic Akt activation and tumor growth by concomitant PTEN and PHLPP1 silencing. A. PTEN followed by PHLPP1 shRNA (sh) knockdown in LN229 cells. V, corresponding vector control. The PTEN and PHLPP1 GAPDH-normalized levels, and % of P-Akt-S473/Akt activation are shown (from two independent infections). B. Soft agar colony formation images (100x) scoring the colonies as small, medium and large, 18 days after seeding the indicated shRNA-depleted LN229 cells. Single cells were not counted. C. Kaplan-Meier survival of mice (n=3) inoculated with the indicated shRNA-depleted LN229 cells. Significant difference and median survival are indicated. D. Immunohistochemistry (x200) of serial sections from tumors isolated from the mice in (C) with GFP antibody (recognizing the GFP moiety of the pGIPZ PTEN shRNA- cloning vector) and with P-Akt S473 antibody. Arrowheads indicate invasive cells.
We further examined whether the synergistic Akt activation by coupled PTEN and PHLPP1 loss is relevant for cancer cell growth. Loss of both PTEN and PHLPP1 substantially increased the anchorage-independent growth of LN229 cells, whereas depletion of PTEN alone had no effect (Fig. 4B). Interestingly, single depletion of PHLPP1 had an intermediate effect on colony formation, suggesting that PHLPP1 has PI3K-Akt pathway-independent growth suppressive effects. LN229 cells form tumors upon injection in the brain of immunosuppressed mice and therefore the 4 sets of cells, control and PTEN/PHLPP1 single –and double–depleted cells, were injected intracranially in SCID mice to assess their in vivo tumorigenicity. The Kaplan-Meier survival curves of the 4 cohorts of mice showed the shortest median survival in mice inoculated with the PTEN-PHLPP1 double-depleted cells correlating perfectly with the results of the colony formation assay (Fig. 4C). These results were confirmed in mouse survival experiments using LN229 cells single-infected with vector, PTEN, PHLPP1 or PHLPP2 shRNAs (not shown). Dissection of the brain tumors developing in these mice showed strong Akt activation by labeling with P-Akt(S473) antibody (Fig. 4D) or P-Akt(T308) antibody (Fig. S8) only in the tumors derived from the PTEN-PHLPP1 double-depleted cells. These tumors grew much faster than the tumors developing in the other 3 cohorts of mice, indicating that the activation of the PI3K-Akt pathway strongly enhances the aggressiveness of tumor cells. Examination of the tumors for cell dispersal, the main cause of glioblastoma relapse in patients, revealed that both PTEN single-depleted and PTEN-PHLPP1 double-depleted cells markedly infiltrated the meninges and the brain parenchyma either as isolated cells or surrounding the vessels, as compared to control or PHLPP1 single-depleted cells (Fig.4D, arrowheads and Figs. S8–S9). This indicated that within the PTEN-PHLPP1 network, PTEN is the main brake of glioblastoma cell dispersal, most likely through suppressing the PI3K-Akt pathway (Molina et al., 2010a).
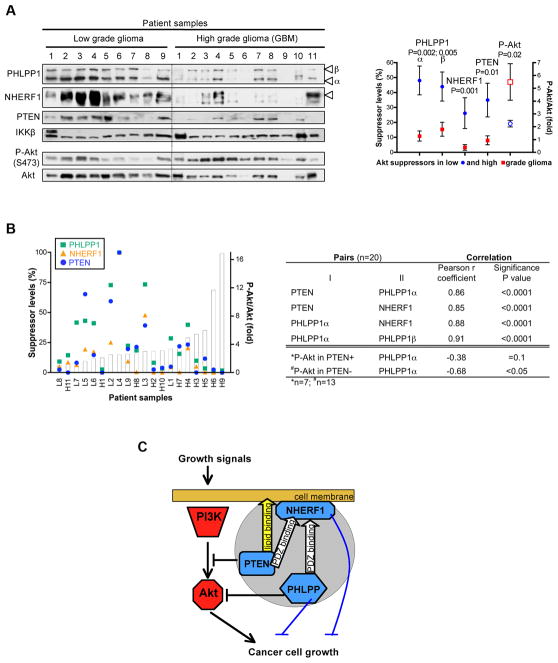
The PTEN-NHERF1-PHLPP1 network is altered in glioblastoma
To determine the relevance of the PTEN-NHERF1-PHLPP1 network in tumors, we examined the levels of these tumor suppressors and the activation of Akt in patient samples, segregated as low-grade glioma and high-grade glioma or glioblastoma (Table 2 and Fig. 5A). Strikingly, a significant decline of all three suppressors and rise of activated Akt was noted in glioblastoma as compared to low-grade glioma samples (Fig. 5A, graph). In individual samples, an overall inverse tendency was observed between the levels of the suppressors and the activation of Akt (Fig. 5B, graph). Correlation analysis found high correlation between the levels of the suppressors, two by two, in all the samples, implying a coordinated regulation of these suppressors (Fig. 5B, table). Importantly, whereas none of the suppressor levels taken separately correlated with phospho-Akt levels (not shown), the levels of PHLPP1α correlated with the activation of Akt in the PTEN-negative samples, implying synergy in tumors between PTEN and PHLPP1 for Akt activation (Fig. 5B, table).
Table 2.
Patient clinicopathological correlates.
| Tumor grade | Patients* (n=20)
|
|
|---|---|---|
| Low (9) | High (11) | |
| Age (yrs) | 51.3±4.8 | 64.0±6.2 |
| Survival (mos) | >70.9±10 | 6.5±5.8 |
values are means±SD
Figure 5.
The PTEN-NHERF1-PHLPP inhibitory network is disabled in glioblastoma. A. Western blot analysis with indicated antibodies of protein extracts (8-10μg) from 9 low-grade glioma and 11 glioblastoma (GBM) samples. The average PHLPP1, NHERF1 and PTEN suppressor levels normalized to IKKβ and P-Akt/Akt levels show statistical significant differences in low-grade versus high-grade tumors. B. The correlation between PHLPP1, NHERF1 and PTEN suppressor levels and P-Akt/Akt levels for individual tumor samples, as sorted by increasing P-Akt/Akt levels (graph), is summarized in the table. Note significant inverse correlation between PHLPP1α levels and Akt phosphorylation in PTEN-negative tumors. C. Model depicting the molecular and functional interrelationships between the triple inhibitory network (gray circle) of PI3K-Akt. The white arrows indicate recruitment of PTEN and PHLPP to the membrane by NHERF1 via PDZ-motif/PDZ-domain interactions and the yellow arrow denotes an alternative lipid membrane association mechanism for PTEN. The Akt-independent suppression of tumor growth is shown with blue blocking lines.
To confirm these results, we examined the PHLPP levels in 9 glioblastoma cell lines (Fig. S10). In PTEN-positive cells, there was no correlation between PHLPP levels and Akt activation. In contrast, a significant inverse correlation was found in PTEN-negative cells between PHLPP1β, the major PHLPP1 isoform in glioblastoma cell lines, or PHLPP2 levels and Akt phosporylation. Overall, these results suggested the presence of an Akt suppressor network formed by PTEN, NHERF1 and PHLPP1 that is targeted for elimination in glioblastoma.
To explore whether the decreased protein levels of PHLPP, NHERF1 and PTEN correlate with decreased mRNA abundance in glioblastoma, we performed gene expression analysis of normal, low-grade (grade III) astrocytoma and glioblastoma samples of the GSE4290 dataset (Sun et al., 2006) from the publically available GEO database (Barrett et al., 2007) (Fig. S11). An important decrease of transcript levels was apparent for PHLPP1 and NHERF1 in glioblastoma as compared to normal or low-grade glioma samples, strongly suggesting that the observed decline in protein expression in glioblastoma is a consequence of mRNA reduction. A small decrease in PTEN gene expression was also observed, and likely underestimated, due to cross-hybridization of PTEN probes with PTEN pseudogene mRNA. PHLPP2 showed also a marked transcript decrease, present from earlier stages of low-grade glioma compared to control samples. Importantly, NHERF2, the homolog of NHERF1, showed no variation of gene expression in these tumors.
DISCUSSION
The PI3K-Akt pathway is one of the most important pathways controlling cell growth, and a wealth of evidence has shown that it is also one of the most frequently activated pathways in cancers (Carnero et al., 2008). Because of its constitutive activation in a wide variety of cancers, it recently became an important target for anti-cancer therapy (Courtney et al., 2010). In this study, we attempted to characterize the multimodal mechanism of PI3K-Akt activation in cancer cells by the inactivation of the upstream tumor suppressors of the pathway. PTEN is the pathway’s most frequently altered tumor suppressor in cancers, in many instances by mutation with LOH, leading to complete absence of the protein (Li et al., 1997; Steck et al., 1997). Surprisingly, depletion of PTEN by shRNA in cancer cells with wild-type PTEN did not generate a massive activation of the PI3K-Akt pathway, indicating extra layers of inhibition. We identified these additional negative regulators that synergize with PTEN depletion to boost Akt activation, as the adaptor protein NHERF1 that binds to PTEN (Takahashi et al., 2006), and the PHLPP Akt phosphatases that, in turn, bind to NHERF1.
NHERF1 directly binds to PTEN and PHLPP1/2 via PDZ domain-PDZ-binding motif interactions, and possibly scaffolds ternary complexes at the membrane to suppress the activation of the PI3K-Akt pathway. The mechanisms of PTEN and PHLPP membrane recruitment by NHERF1 appear to be different. For PTEN, NHERF1 might stabilize the direct interactions of PTEN’s membrane-binding domains with the plasma membrane following a conformational change triggered by the dephosphorylation of PTEN (Lee et al., 1999; Molina et al., 2010b; Rahdar et al., 2009; Takahashi et al., 2006; Walker et al., 2004) (Fig. 5C). For PHLPP, our fractionation studies support constitutive membrane localization modulated by NHERF1. PHLPP1/2 co-fractionate with endogenous NHERF1, re-localize to the membrane with membrane-targeted Myr-NHERF1 and shift to the cytoplasm when membrane-localized NHERF1 is depleted. Importantly, complex formation with NHERF1 appeared essential for regulating the growth suppressive effects of PHLPP1, as the absence of NHERF1 prevented PHLPP1 from inhibiting cell growth.
Previous reports have described unchanged Akt phosphorylation following PHLPP silencing or overexpression in a multitude of cancer cells (Liu et al., 2009; Qiao et al., 2010). In our hands, PHLPP1 silencing in a series of PTEN-positive cells of different origins had no effect on Akt phosphorylation, suggesting that PHLPP loss cannot efficiently activate Akt in the presence of PTEN. Notably, PHLPP1 silencing following PTEN silencing significantly and massively activated Akt by phosphorylation on both Ser473 and Thr308. Similarly, synergistic Akt activation occurred when PTEN and NHERF1 were depleted. In both circumstances, the synergistic PI3K-Akt pathway activation promoted synergistic tumor cell growth. Intriguingly, the effects of single molecule depletion on cell growth did not correlate with Akt activation. Thus, NHERF1 or PTEN depletion had similar Akt activation effects but NHERF1 significantly increased cell proliferation in both LN229 cells ((Molina et al., 2010b) and data not shown) and DU145 cells, as compared to PTEN depletion. Even more apparent, PHLPP1 depletion that had no detectable effect on Akt activation in these cells significantly increased their growth and tumorigenesis. It results that single NHERF1 or PHLPP1 losses activate PI3K-Akt-independent signaling pathways that lead to cancer cell growth. Recently, PHLPP1/2 were shown to interact with the tumor suppressor Mst1 and exert pro-apoptotic effects via activation of p38 and JNK (Qiao et al., 2010). Conceivably, NHERF1 loss might influence cell growth by deregulation of PHLPP proteins. In addition, due to its interactions with other molecules involved in cancer progression, including β-catenin (Kreimann et al., 2007; Shibata et al., 2003), growth factor receptors (Lazar et al., 2004; Maudsley et al., 2000) and NF2 (Murthy et al., 1998), involvement of other pathways could account for the PI3K-Akt-independent effects of NHERF1 on tumor growth (Georgescu et al., 2008; Kreimann et al., 2007). These observations suggest that the loss of PHLPP1 and NHERF1 in glioblastoma that we report here for the first time, most likely induce both PI3K-Akt-dependent effects on cell growth in cooperation with PTEN loss, and PI3K-Akt-independent effects, explaining thus the pronounced aggressiveness of these tumors and the dismal patient survival (Table 2).
In conclusion, our results show that silencing of PTEN separately does not potently activate Akt. Remarkably, simultaneous knockdown of PTEN and NHERF1, or of PTEN and PHLPP1 that is recruited by NHERF1 at the plasma membrane, significantly activates Akt and increases cell tumorigenicity. Coupled to our results from glioblastoma patient samples, we thus demonstrate for the first time an upstream inhibitory network formed by the suppressors of the PI3K-Akt pathway PTEN, NHERF1 and PHLPP1, and a requirement for their concurrent loss to cause potent activation of the pathway. In cancers, the interplay between these suppressors result from modulating the levels of PHLPP proteins in PTEN-negative cells or from loss of membrane-localized NHERF1. We propose thus a model in which PTEN appears the key suppressor of the PI3K-Akt pathway whereas the levels of the PHLPP proteins and the levels and/or subcellular localization of NHERF1, which regulates both PTEN and PHLPP, add a second layer of regulation (Fig. 5C). In glioblastoma, both this synergistic PI3K-Akt inhibitory network and the previously unrecognized Akt-independent contributions of NHERF1 and PHLPP1 need to be taken into consideration for the design of a PI3K-Akt-targeted therapy and the management of patients.
MATERIALS/SUBJECTS AND METHODS
Cells, plasmids, retroviral infections and shRNA silencing
The glioblastoma cells lines LN18, LN229, LN308, LN428, U87-MG, U251-MG and A172 were previously described (Molina et al., 2010b). These and LN382T (Dr. Erwin Van Meir, Emory University, Atlanta, GA; 2004) and SNB19 (ATCC; 1998), as well as the prostate cancer cell line DU145 (Dr. David Nanus, Weiss Medical College, New York, NY; 2000), were authenticated in October 2009, as described (Molina et al., 2010b). All cells were grown in DMEM supplemented with 10% FBS. Primary mouse embryonic fibroblasts (MEFs) were isolated from 14 day-old embryos as previously described (Takahashi et al., 2006). The plasmids for Myr-NHERF1 in pCXp (puromycin selection) retroviral vector and shRNAs for PTEN and NHERF1 were previously described (Molina et al., 2010b). All NHERF1 and PTEN constructs for recombinant protein expression were also described (Takahashi et al., 2006). Human PHLPP1α and PHLPP2 cDNAs were purchased from Open Biosystems and rat PHLPP1β was obtained from Dr. Kimiko Shimizu (Shimizu et al., 1999). Full-length PHLPP1α and ΔPDZ mutant lacking the last 3 residues that form the PDZ-binding motif were inserted in pcDNA3 and pCXb (blasticidin resistance) mammalian expression vectors. Myc-tagged PHLPP2 full length was inserted in pcDNA3 vector. CT fragments encoding 130 residues for PHLPP1 and 141 residues for PHLPP2, and their corresponding ΔPDZ mutants, were cloned in pGEX-6P-1 vector with or without an NT Myc tag for recombinant protein expression. The two distinct PTEN and NHERF1 shRNAs in pGIPZ lentiviral vector (neomycin resistance) (Open Biosystems) and pSIREN-RetroQ retroviral vector (puromycin resistance) (Clontech), respectively, were previously described (Molina et al., 2010b). PHLPP1 shRNA#2 (CCGAGCTGTTTAACAAATAAA), #3 and #4 (CGCTGTCCTTTGTCA TATCAA) in the pLKO lentiviral vector (puromycin resistance) were purchased from Open Biosystems. Transfections and retroviral/lentiviral infections were described (Georgescu et al., 1999; Molina et al., 2010b).
Proliferation and soft agar colony assays
The MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) proliferation assay was previously described (Molina et al., 2010b). Soft agar colony assays were performed as described (Georgescu et al., 1999) in triplicate plates. Colonies were counted from 8 fields/plate (total 24 fields/cell line) taken at 50× magnification and classified into small (<2 mm), medium (2–5 mm), and large (>5 mm). Images were acquired with a Zeiss Axiovert 200 microscope using the Cool SNAP ES Photometrics Camera (Ropert Scientific) and Meta Imaging Corporation Series software (Universal Imaging).
Protein analysis
The protocols for cell lysis, Western blotting, overlay assays, GST-pull down and cellular fractionation were previously described (Takahashi et al., 2006). For immunoprecipitation, cells were homogenized in phosphate lysis buffer (50mM Na2HPO4, 1mM Sodium Pyrophosphate, 20mM NaF, 2mM EDTA, 2mM EGTA, 1% Triton X-100) containing protease and phosphatase inhibitors (1mM phenylmethylsulfonyl fluoride, 21 μ/ml aprotinin, 1mM sodium orthovanadate, and 0.1 mM sodium molybdate). Clarified cell lysates were incubated for 2 h at 4°C with indicated antibodies and ultraLink protein AG beads (Pierce). Beads were washed three times with the phosphate lysis buffer and fresh 2.5× Laemmli buffer was added for SDS PAGE analysis. Antibodies were obtained as follows: PTEN (A2B1), Erk1 (C-16), Erk2 (C-14), GAPDH (sc-47724) (Santa Cruz Biotechnology), phospho-Akt (Ser473 and Thr308), Akt/PKB (Cell Signaling), PHLPP1, PHLPP2 (Bethyl Laboratories), N-cadherin (Zymed), NHERF1 (Abcam ab3452, Affinity BioReagents or BD Biosciences), M2 anti-FLAG (Sigma), Myc (Invitrogen), and actin (Chemicon).
Immunofluorescence analysis
The immunofluorescence analysis of cells was performed as described (Takahashi et al., 2006) with some modifications. Formaldehyde-fixed cells permeabilized with 0.1% Triton X-100 in PBS for 5 min were treated with Image-iT FX signal enhancer (Invitrogen) for 30 min and blocked with 10% goat serum in PBS-gel (0.2% gelatin in PBS) for 30 min. The primary antibodies c-Myc (9E10) (Santa Cruz Biotechnology) and NHERF1 (Affinity BioReagents) and secondary antibodies Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 555 goat anti-mouse IgG (Molecular Probes) were used. Image stacks were acquired with a Zeiss Axiovert 200M inverted microscope and deconvolved with the AxioVision Rel 4.5 SP1 software.
Orthotopic tumorigenicity assay in SCID mice
2×106 LN229 cells in 10 μl PBS were stereotactically injected in the right frontal lobe of 7-week-old SCID mice. Mice were euthanized once neurological signs of tumor burden were observed (lethargy, severe paralysis), and the brains were collected.
Immunohistochemistry
Mouse brains were embedded in paraffin and 4μm sections were deparaffinated in xylene and then hydrated through a series of ethanol solutions (100%, 95%, 70%) and water. Endogenous peroxidase activity was blocked in 3% H2O2 in methanol for 10 min. The samples were also incubated (steamed) in antigen retrieval solution (Dako Target Retrieval) for 20 min, then rinsed in water and transferred to PBS. The Histomouse Max Broad Spectrum DAB kit (Zymed) was used according to the manufaturer specifications. Primary antibodies were GFP (Chemicon) and phospho-Akt Ser473 and Thr308 (Cell Signaling).
Patient samples
Frozen brain tumor samples were washed twice in cold PBS and then homogenized in sample lysis buffer (50 mM Tris-HCl pH 7.5, 7M urea, 2M thiourea, 1% CHAPS) containing protease and phosphatase inhibitors. The samples were labeled as low-grade glioma, which comprise grade II oligodendroglioma and grade III anaplastic astrocytoma or mixed oligoastrocytoma, and high-grade glioma, which are all grade IV glioblastoma. All patients were recorded in MD Anderson Cancer Center between 1991 and 2011 and their mean age and survival per group are shown (Table 2).
Statistical analysis
Data are representative of at least two or three independent experiments with essentially similar results. The shRNA silencing data were derived from independent sets of lentiviral/retroviral infections with two different shRNAs. Data were examined for normality of distribution and expressed as mean±SEM, unless mentioned otherwise. Parametric or non-parametric methods, including t-test with or without Welch’s correction for variances significantly different and Mann-Whitney test, respectively, were used to analyze the differences between groups. Data correlations were assessed by Pearson correlation coefficient. Statistical significance was considered for P< 0.05. Confidence intervals for all tests were 95%. Animal survival was analyzed by the Gehan-Breslow-Wilcoxon test and plotted with the GraphPad Prism program. ImageJ program was used for the densitometric analysis.
Supplementary Material
Acknowledgments
Financial support: NCI-CA107201 and the corresponding ARRA supplement (M-M. Georgescu) and NCI-CA16672 that partially supported the DNA sequencing and animal breeding.
Footnotes
CONFLICT OF INTEREST
The authors have no competing financial interests in relation to this work.
References
- Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007;35:D760–5. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008;19:223–30. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Cardone RA, Bellizzi A, Busco G, Weinman EJ, Dell'Aquila ME, Casavola V, et al. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol Biol Cell. 2007;18:1768–1780. doi: 10.1091/mbc.E06-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–98. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouassier L, Yun CC, Fitz JG, Doctor RB. Evidence for ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) self-association through PDZ-PDZ interactions. J Biol Chem. 2000;275:25039–25045. doi: 10.1074/jbc.C000092200. [DOI] [PubMed] [Google Scholar]
- Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473–88. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Georgescu MM. PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes & Cancer. 2010;1:1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu MM, Morales FC, Molina JR, Hayashi Y. Roles of NHERF1/EBP50 in cancer. Curr Mol Med. 2008;8:459–468. doi: 10.2174/156652408785748031. [DOI] [PubMed] [Google Scholar]
- Hall RA, Ostedgaard LS, Premont RT, Blitzer JT, Rahman N, Welsh MJ, et al. A C-terminal motif found in the beta2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc Natl Acad Sci USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Molina JR, Hamilton SR, Georgescu MM. NHERF1/EBP50 Is a New Marker in Colorectal Cancer. Neoplasia. 2010;12:1013–22. doi: 10.1593/neo.10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimann EL, Morales FC, de Orbeta-Cruz J, Takahashi Y, Adams H, Liu TJ, et al. Cortical stabilization of beta-catenin contributes to NHERF1/EBP50 tumor suppressor function. Oncogene. 2007;26:5290–5299. doi: 10.1038/sj.onc.1210336. [DOI] [PubMed] [Google Scholar]
- Lazar CS, Cresson CM, Lauffenburger DA, Gill GN. The Na+/H+ exchanger regulatory factor stabilizes epidermal growth factor receptors at the cell surface. Mol Biol Cell. 2004;15:5470–5480. doi: 10.1091/mbc.E04-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Maudsley S, Zamah AM, Rahman N, Blitzer JT, Luttrell LM, Lefkowitz RJ, et al. Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol Cell Biol. 2000;20:8352–8363. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, Hayashi Y, Stephens C, Georgescu MM. Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia. 2010a;12:453–463. doi: 10.1593/neo.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, Morales FC, Hayashi Y, Aldape KD, Georgescu MM. Loss of PTEN binding adapter protein NHERF1 from plasma membrane in glioblastoma contributes to PTEN inactivation. Cancer Res. 2010b;70:6697–703. doi: 10.1158/0008-5472.CAN-10-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales FC, Takahashi Y, Momin S, Adams H, Chen X, Georgescu MM. NHERF1/EBP50 Head-to-Tail Intramolecular Interaction Masks Association with PDZ Domain Ligands. Mol Cell Biol. 2007;27:2527–37. doi: 10.1128/MCB.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, et al. NHE-RF, a regulatory cofactor for Na(+)-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- Pan Y, Weinman EJ, Dai J. NHERF1 (Na+/H+ exchanger regulatory factor 1) inhibits platelet-derived growth factor signaling in breast cancer cells. Breast Cancer Res. 2008;10:R5. doi: 10.1186/bcr1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Wang Y, Xu X, Lu J, Dong Y, Tao W, et al. Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol Cell. 2010;38:512–23. doi: 10.1016/j.molcel.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci U S A. 2009;106:480–5. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Shenolikar S, Minkoff CM, Steplock DA, Evangelista C, Liu M, Weinman EJ. N-terminal PDZ domain is required for NHERF dimerization. FEBS Let. 2001;489:233–236. doi: 10.1016/s0014-5793(01)02109-3. [DOI] [PubMed] [Google Scholar]
- Shibata T, Chuma M, Kokubu A, Sakamoto M, Hirohashi S. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 2003;38:178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 1999;458:363–9. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- Song J, Bai J, Yang W, Gabrielson EW, Chan DW, Zhang Z. Expression and clinicopathological significance of oestrogen-responsive ezrin-radixin-moesin-binding phosphoprotein 50 in breast cancer. Histopathology. 2007;51:40–53. doi: 10.1111/j.1365-2559.2007.02730.x. [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Morales FC, Kreimann EL, Georgescu MM. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006;25:910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SM, Leslie NR, Perera NM, Batty IH, Downes CP. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem J. 2004;379:301–7. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman EJ, Steplock D, Tate K, Hall RA, Spurney RF, Shenolikar S. Structure-function of recombinant Na/H exchanger regulatory factor (NHE-RF) J Clin Invest. 1998;101:2199–2206. doi: 10.1172/JCI204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.