SUMMARY
Cardiogenesis within embryos or associated with heart repair requires stem cell differentiation into energetically competent, contracting cardiomyocytes. While it is widely accepted that the coordination of genetic circuits with developmental bioenergetics is critical to phenotype specification, the metabolic mechanisms that drive cardiac transformation are largely unknown. Here, we aim to define the energetic requirements for and the metabolic microenvironment needed to support the cardiac differentiation of embryonic stem cells. We demonstrate that anaerobic glycolytic metabolism, while sufficient for embryonic stem cell homeostasis, must be transformed into the more efficient mitochondrial oxidative metabolism to secure cardiac specification and excitation–contraction coupling. This energetic switch was programmed by rearrangement of the metabolic transcriptome that encodes components of glycolysis, fatty acid oxidation, the Krebs cycle, and the electron transport chain. Modifying the copy number of regulators of mitochondrial fusion and fission resulted in mitochondrial maturation and network expansion, which in turn provided an energetic continuum to supply nascent sarcomeres. Disrupting respiratory chain function prevented mitochondrial organization and compromised the energetic infrastructure, causing deficient sarcomerogenesis and contractile malfunction. Thus, establishment of the mitochondrial system and engagement of oxidative metabolism are prerequisites for the differentiation of stem cells into a functional cardiac phenotype. Mitochondria-dependent energetic circuits are thus critical regulators of de novo cardiogenesis and targets for heart regeneration.
Keywords: cardiogenesis, embryonic stem cells, heart regeneration, metabolic transcriptome, mitochondrial oxidative metabolism
INTRODUCTION
Embryonic stem cells differentiate into diverse cell types1 with distinct energetic requirements. A case in point is cardiomyogenic transformation.2–6 Commitment of embryonic stem cells to a cardiac lineage occurs early in embryogenesis, as the heart forms first to sustain nutrient and oxygen delivery.7,8
Transition from noncontractile stem cells requiring little energy to beating cardiomyocytes necessitates an energetic infrastructure capable of supporting the metabolic demands of increased electromechanical activity.9,10 Integrated transcriptional and metabolic circuits are critical for cell specification and differentiation, and nucleus–mitochondria intercommunication is required to translate signals that regulate cell fate.11–13 The generation of embryonic stem cell-derived functional and energetically competent cardiomyocytes is a promising step toward the development of regenerative therapy for heart disease.14,15 The metabolic and energetic requirements of cardiogenic programming are, however, only partly understood, limiting full exploitation of the regenerative potential of stem cell therapy.
This study aims to define the energetic requirements for and the metabolic microenvironment needed to support the cardiac differentiation of embryonic stem cells. We report that a switch from anaerobic to oxidative metabolism and a mitochondrial reorganization into a network intercalated with developing myofibrils are prerequisites for optimum differentiation into cardiomyocytes. Disruption of mitochondrial function hampered the formation of contracting progeny, precluding proper sarcomeric development. These studies thus provide novel insight into molecular mechanisms of metabolic remodeling integral to the execution of a cardiac differentiation program.
METHODS
Stem cell differentiation
Murine embryonic stem cells, maintained in Glasgow’s Minimum Essential Medium (BioWhittaker-Cambrex, Walkersville, MD) with sodium pyruvate, nonessential amino acids, 2-mercaptoethanol, 7.5% fetal bovine serum (Invitrogen Corporation, Carlsbad, CA), and leukemia inhibitory factor (ESGRO; Chemicon International, Inc, Temecula, CA), were differentiated in media containing 20% fetal bovine serum via an inverted hanging-drop method.2,3 Forming embryoid bodies were allowed to grow in suspension until plated. Cardiomyocytes, isolated from embryoid bodies dissociated with at least 96 U/ml collagenase type 4 (Worthington Biochemical Corporation, Lakewood, NJ) and 0.1 mg/ml pancreatin (Sigma-Aldrich, St Louis, MO), were separated by Percoll gradient and cultured on gelatin-coated polystyrene dishes or poly-L-lysine-coated glass slides.2,3
Lactate and protein assay
Cellular lactate production rate was measured using 164 mmol/l hydrazine, 64 mmol/l glycine, and 11 mmol/l NAD+ (pH 9.5), with 10 U/well lactate dehydrogenase16 (Roche Molecular Biochemicals, Mannheim, Germany). Optical density at 360 nm was read with use of the Multiskan Ascent® microplate photometer (Thermo Electron Oy Corporation, Vantaa, Finland), and protein concentration was determined with the Bio-Rad detergent-compatible protein assay kit (Bio-Rad, Hercules, CA).
Cell respiration and adenine nucleotide content measurements
Respiratory capacity, with and without 2.5 μmol/l 2,4-dinitrophenol, was measured with the BD™ Oxygen Biosensor System (Becton-Dickinson Biosciences, San Jose, CA). Wells were covered with mineral oil to seal cells from atmospheric oxygen and were scanned with a Fluoroskan Ascent FL fluorometer (Thermo Electron). Nucleotide concentrations in perchloric acid extracts were determined by high-performance liquid chromatography (Hewlett-Packard Series 1100; Agilent Technologies, Palo Alto, CA) using a triethylamine bicarbonate elution buffer (pH 8.8).17
Confocal microscopy
For imaging of plasma and mitochondrial membrane potentials, cells were incubated with 1.3 μmol/l RH237, an imaging dye whose fluorescence decreases upon membrane depolarization (Invitrogen), 0.5 μmol/l JC-1, a mitochondrial membrane potential-sensitive dye (Invitrogen), or both, and washed with phosphate-buffered saline. Alternatively, cells were incubated with 3 μmol/l MitoTracker® Red CM-H2XRos (Cambrex Corporation, East Rutherford, NJ), which stains mitochondria in live cells, and fixed in 3% paraformaldehyde. Cells were stained with antibodies against α-actinin (Santa Cruz Biotechnology Inc., Santa Cruz, CA), transcription factor MEF2C (Cell Signaling Technology, Danvers, MA), or both, and with the nuclear probe 4′,6-diamidino-2-phenylindole-2HCl (Invitrogen). Fluorescent images were obtained with a Laser Scanning Microscope 510 (Carl Zeiss Microimaging, Thornwood, NY) and analyzed using Zeiss Laser Scanning Microscope Image Browser or MetaMorph® software (Universal Imaging Corporation, West Chester, PA).
Transmission electron microscopy
Embryonic stem cells and derived cardiomyocytes were postfixed in 1% osmium tetroxide, stained with 2% uranyl acetate, dehydrated in ethanol and propylene oxide, and embedded in epoxy resin. Thin sections were stained with lead citrate, and micrographs were taken with a JEOL 1200 EX II transmission electron microscope18 (JEOL USA, Peabody, MA).
Metabolic gene profiling
Total RNA isolated from embryonic stem cells or cardiomyocytes was screened using the GeneChip® Mouse Genome 430 2.0 array (Affymetrix Inc., Santa Clara, CA). Expression profiles were analyzed with the bioinformatics suite GeneSpring GX 7.3 (Silicon Genetics, Redwood City, CA). Gene lists were quality filtered to remove genes with expression levels below background and limited to report genes changing by 1.5-fold or greater during cardiac differentiation.
Statistics
Comparisons between groups were made with Student’s t-tests. Data are presented as mean (±SEM); n refers to sample size. Significance was predetermined at P < 0.05.
RESULTS
Switch from anaerobic glycolytic to mitochondrial oxidative metabolism
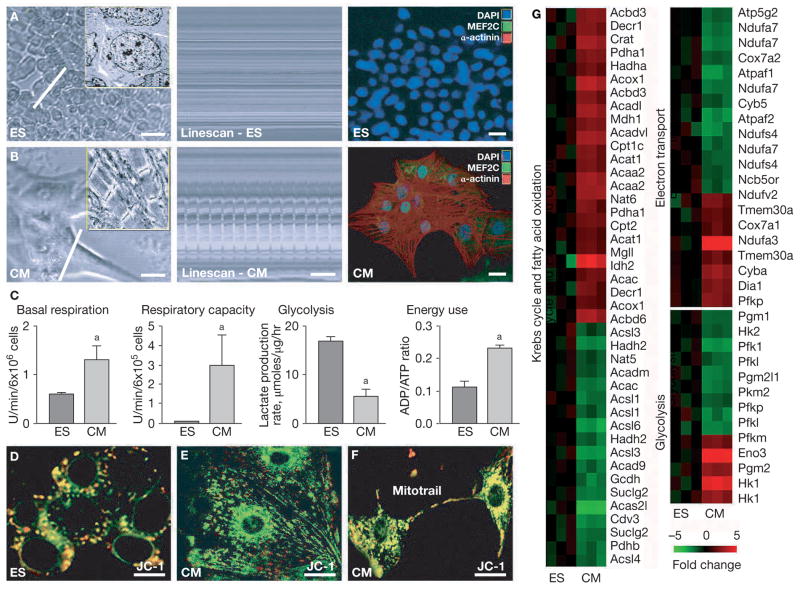
As might be predicted by the elementary, low-energy nature of undifferentiated cell types lacking contractile machinery, microscopy showed that embryonic stem cells had high nucleus-to-cytosol ratios and scarce mitochondria (Figure 1A). With differentiation into embryoid bodies, stem cells undergo transformation into specialized cell types, including beating cardiac progeny.2 Upon isolation, stem cell-derived cardiomyocytes displayed sarcomeric-mitochondria aligned structures and underwent rhythmic contractions, which imposed high-energy demands (Figure 1B). Specific to cardiomyocytes, a switch in energy metabolism was associated with increased mitochondrial oxygen consumption and reduced glycolysis. The basal oxygen consumption rate was 1.29 ± 0.29 fluorescence units/min per 6 × 106 cells in cardiomyocytes, compared with 0.61 ± 0.02 fluorescence units/min per 6 × 106 cells in embryonic stem cells (P <0.05; Figure 1C). The maximal respiratory capacity of cardiomyocytes was approximately 40-fold higher than that of embryonic stem cells, 2.99 ± 1.55 versus 0.08 ± 0.01 fluorescence units/min per 6 × 105 cells (P <0.05; Figure 1C). The rate of lactate production, an index of anaerobic glycolytic metabolism, was reduced in cardiomyocytes compared with that in their embryonic source (5.54 ± 1.36 versus 17.08 ± 0.83 μmol/μg/h, P <0.05; Figure 1C). These changes translated into higher overall ATP and ADP levels (38.8 ± 2.6 and 9.2 ± 0.4 nmol/mg protein, respectively, for cardiomyocytes and 30.6 ± 2.5 and 3.2 ± 0.3 nmol/mg protein, respectively, for embryonic stem cells). Also, the increased energy turnover during cardiac differentiation was reflected in a higher ADP/ATP ratio in cardiomyocytes than in stem cells (Figure 1C). Probing mitochondrial membrane potential, an indicator of respiratory chain maturation, revealed abundant active mitochondria with zones of increased energy utilization in cardiomyocytes, particularly in the perinuclear and interfibrillar spaces (Figure 1E), in contrast to embryonic stem cells (Figure 1D). Cardiomyocytes also demonstrated intercellular nanotubular connections facilitating mitochondrial cell-to-cell exchange (Figure 1F) visualized by time-lapse microscopy (not illustrated). Thus, although anaerobic metabolism is sufficient to sustain the primitive energetics of embryonic stem cells, contracting cardiomyocyte progeny require energy-efficient oxidative metabolism in order to sustain their higher energetic needs.
Figure 1.
Embryonic stem cell cardiac differentiation is coordinated with metabolic transcriptome reprogramming and mitochondrial oxidative metabolism. (A) ES have no contractile activity on linescans and MEF2C, a cardiac transcription factor, and cardiac α-actinin, a tissue-specific protein of the contractile apparatus, are absent. (B) Cardiomyocytes derived from ES have distinct structures on light microscopy (upper panel) and electron microscopy (inset), contractile activity on linescans, and MEF2C and cardiac α-actinin are abundant in the nucleus and sarcomeres, respectively, on immunofluorescent microscopy. (Bars 10 μm [A,B left], 2 μm [A,B insets], 2 s [B middle], and 5 μm [A,B right]). (C) The basal respiratory rate and maximum respiratory capacity of cardiomyocytes were markedly higher, while the lactate production from anaerobic glycolysis was lower than in ES, underscoring distinct metabolic identities of the progeny compared with the embryonic source. A higher ADP:ATP ratio reflected an increased rate of energy use in cardiomyocytes than ES. (D) High-membrane-potential mitochondria (red) in ES on confocal microscopy. (E) Mitochondria have lower membrane potential (green) in cardiomyocytes, associated with increased energy use. (F) Intercellular cardiomyocyte connections with mitochondrial traffic (mitotrail) indicate cell–cell metabolic cross-talk in cardiogenesis. (D–F, bars 20 μm, sample >3.) (G) Microarray analysis of total mRNA in ES and cardiomyocytes revealed specific changes in genetic programming of the cellular energetic system. Genes were hierarchically clustered as mRNA copy numbers of cardiomyocytes versus ES transcripts (n = 3 in each group). Redundant probe sets were included to illustrate the individual dynamics of expression profiles. aP<0.05; n=3–12. Abbreviations: CM, cardiomyocytes; ES, embryonic stem cells; mRNA, messenger RNA.
Metabolic transcriptome reconfiguration with cardiogenic specification
At the transcriptional level, cardiomyocytes displayed a messenger RNA fingerprint distinct from that of embryonic stem cells, including divergent expression patterns of genes involved in the Krebs cycle, the electron transport chain, fatty acid oxidation, and glycolysis (Figure 1G). Metabolic energetic remodeling was apparent with increased copy numbers for mitochondrial respiratory chain-associated genes, including the cytochrome oxidase subunit Cox6a2, diaphorase (NADH:cytochrome b-5 reductase, Dia1), and NADH dehydrogenase (Ndufa3), with concomitant downregulation of alternative subunits of NADH dehydrogenase (Ndufa7, Ndufs4) and mitochondrial ATP synthase (Atpaf1, Atpaf2; Figure 1G). Cardiomyocytes demonstrated upregulation of genes involved in lipolysis and fatty acid oxidation, including monoglyceride lipase (MgII ), acyl-coenzyme A oxidase (Acox1), acyl-coenzyme A dehydrogenase (Acadl, Acadvl ), and carnitine acetyltransferase (Crat; Figure 1G). Compared to stem cells, cardiomyocytes displayed downregulation in genes involved in lipid biosynthesis, such as acetyl-coenzyme A synthetase (Acas2l ) and fatty acid-coenzyme A ligase (Acsl6; Figure 1G). Among Krebs cycle components, cardiomyocytes showed upregulation of pyruvate dehydrogenase (Pdha1) and malate dehydrogenase (Mdh1; Figure 1G). Modulation of oxidative metabolism was associated with restructuring of the glycolytic transcriptome, including downregulation of nonmuscle glycolytic enzymes, such as liver and platelet phosphofructokinase (Pfkl and Pfkp), and upregulation of cardiac muscle-specific enolase 3 and hexokinase 1 isoforms (Eno3, Hk1; Figure 1G). Thus, remodeling of the metabolic transcriptome provides a molecular basis for the increased contribution of oxidative metabolism, further supported by the relative prevalence of glycolytic genes coding for enzymes coupled with mitochondrial energetics.
Development and maturation of the mitochondrial network during cardiogenesis
Genetic reprogramming associated with cardiac differentiation of stem cells was reflected in the modulation of genes involved in mitochondrial structural organization and network formation (Figure 2A). Genes with roles in mitochondrial fission or inhibition of fusion were downregulated, including Dynamin-1-like protein (Dnm1l), by approximately 40% (P <0.003), mitochondrial protein 18 kDa (Mtp18) by approximately 66% (P <0.004), optic atrophy type 1 (Opa1) by approximately 34% (P <0.009), and mitochondrial fragmentation-causing death-associated protein 3 (DAP3) by approximately 48% (P <0.003; Figure 2A). Mitofusin2 (Mfn2), linked to mitochondrial fusion, was upregulated by approximately 272% (P <0.005), while the signal for the cristae maturing mitofilin (IMMT) did not reach statistical significance (Figure 2A). Ultrastructurally, mitochondria were spherical and cristae-poor in embryonic stem cells but became elongated with abundant and organized cristae in cardiomyocytes (Figure 2B), indicating a higher energetic potential per mitochondrial volume with cardiac differentiation. Live imaging of embryonic stem cells revealed discrete mitochondria, whereas mitochondrial networks filled the expanded cytoplasm of derived cardiomyocytes (Figure 2C). Mitochondria progressed from a random to a peri-nuclear and, ultimately, transcellular arrangement paralleling development of the contractile apparatus during cardiogenic maturation (Figure 2D). Synchronized development was reflected in mitochondrial intercalation with emerging sarcomeres (Figure 2E) and integration of the cardiac excitation-contraction machinery with the energetic system (Figure 2F).
Figure 2.
Development and maturation of mitochondrial network in stem cell cardiac differentiation. (A) Gene array analysis of selected genes related to mitochondrial fission, fusion, and/or cristae maturation. Compared with the embryonic stem cell source, genes involved in mitochondrial fission and membrane structure remodeling in cardiomyocytes (Dnm1l, Mtp18, Opa1, and DAP3) were downregulated, whereas those involved in mitochondrial fusion and cristae maturation were typically upregulated (Mfn2 and IMMT). (B) Transmission electron microscopy revealed spherical mitochondria with underdeveloped cristae in embryonic stem cells versus elongated, cristae-rich mitochondria in cardiomyocytes. (C) Live-cell imaging showed discrete organelles with no apparent pattern of mitochondrial arrangement in ES versus an expanded network of aligned mitochondria in cardiomyoctyes. (D) Tracking cardiomyocyte development revealed an organization of the mitochondrial network (upper panels) ranging from random (left), to perinuclear (center), to transcellular (right), along with concomitant maturation of myofibrillar structure (lower panels). Mitochondria were visualized with MitoTracker Red, myofibrils with α-actinin staining (green), and nuclei with DAPI (blue). (E) Development of mechanoenergetic coupling through intercalation of mitochondria with myofibrils in cardiomyocytes observed by confocal microscopy (upper panel). Profile of fluorescence intensity (lower panel corresponds to arrow in upper panel) indicates an alternating distribution of mitochondria (red) and myofibrils (green). (F) Integration of mitochondrial (green) and electrical (red) activities in a beating area of an embryoid body (EB). Cardiac beating area delineated by the tight correlation of electrical activity staining with RH237, a probe for plasma membrane potential, and mitochondrial imaging with JC-1 (upper panel). Profiles of overlapping fluorescence intensity for both signals within the cardiac beating area are depicted in the lower panel (lower panel corresponds to line in upper panel). aP <0.05. Abbreviations: CM, cardiomyocytes; ES, embryonic stem cells.
Disruption of mitochondrial function compromises cardiac differentiation
Beginning on the fourth day of the hanging-drop method, continuous treatment of attached differentiating embryoid bodies with mitochondrial poisons resulted in abnormal formation of beating areas and a disrupted mitochondrial network, ultimately precipitating defective sarcomerogenesis with low yields of functional cardiomyocytes (Figure 3). Compared with untreated controls, disruption of the electron transport chain between cytochromes b and c by the antibiotic antimycin (50 nmol/l) produced an approximately 20% reduction in the number of beating embryoid bodies, halved the content of cardiac-specific α-actinin, and abrogated sarcomere formation (Figure 3A). Furthermore, prevention of NADH oxidation by rotenone (250 nmol/l) inhibition of the electron transfer in Complex I reduced the number of beating embryoid bodies by approximately 80%, reduced cardiac α-actinin intensity to 11 ± 4 AU from 19 ± 4 AU in controls, and depleted sarcomere content by approximately 60% (Figure 3A). Imaging of embryoid bodies with mitochondrial and membrane potential-sensitive dyes revealed that antimycin or rotenone treatment caused mitochondrial fragmentation—preventing network organization and compromising the assembly of cohesive beating zones in the developing mesoderm (Figure 3B). Exposure to respiratory chain poisons generated aberrant progeny characterized by reduced volume and abnormal distribution of mitochondria (Figure 3C), associated with impaired myofibrillogenesis (Figure 3D). Consequently, in contrast to vigorous contractions of normally developed beating areas in control embryoid bodies, antimycin or rotenone treatment of embryoid bodies stunted contractility, reducing the peak amplitude of contractions by approximately 90% and 80%, respectively (Figure 3E).
Figure 3.
Disturbed execution of the cardiac differentiation program with inhibition of the mitochondrial respiratory chain. (A) After antimycin (50 nmol/l) or rotenone (250 nmol/l) treatment the percentage of beating EBs (n>80), α-actinin expression (n=6–10) and sarcomeric content (n = 7–10) were reduced compared with those in untreated controls. (B) Imaging of embryoid bodies with mitochondrial and plasma-membrane-potential probes (JC-1 and RH237, respectively) revealed that respiratory chain inhibition caused mitochondrial fragmentation and defective assembly of cardiac beating areas. (C) Cardiomyocytes isolated from EBs treated with antimycin or rotenone and cultured in the presence of the corresponding inhibitors had aberrant mitochondrial content, localization, and network architecture as detected by JC-1. (D) Sarcomere formation was compromised with mitochondrial inhibition. The intensity of α-actinin staining was decreased and fewer cardiomyocytes had detectable sarcomeric structures. (Bars 20 μm.) (E) Effects of respiratory chain inhibition on contractions in cardiac beating areas. Contractions were measured as changes in cell edge position using confocal microscopy, and analyzed with the region of interest function in the software. Abbreviations: CM, cardiomyocytes; EBs, embryoid bodies.
DISCUSSION
This study demonstrates that while anaerobic metabolism is sufficient to sustain embryonic stem cell energetics, it must be transformed into mitochondria-mediated oxidative metabolism to secure cardiac specification. Induction of cardiogenic pathways was linked to metabolic transcriptome restructuring and organization of the mitochondrial network. Integration of mitochondria with the nascent electromechanical apparatus conferred the functional competence required to develop a cardiac phenotype. Disruption of respiratory chain function and prevention of mitochondrial network organization compromised the developing energetic infrastructure, causing deficient sarcomere formation, improper beating area assembly, and defective contractility. Establishing the prerequisites for the generation of energetically competent cardiomyocytes provides novel insight into molecular processes underlying the transition of stem cells into cardiac progeny.
In accord with previous reports,3,19,20 here we demonstrated that embryonic stem cells have a rudimentary energetic system characterized by few mitochondria, low oxygen consumption, and greater reliance on glycolysis. The apparent independence from oxidative metabolism observed in our study positions embryonic stem cells as ideal candidates for survival and differentiation in an energetically challenging environment, such as the infarcted myocardium, providing a basis for their use in cardiac repair.2,4,9,10,21
Compared to the embryonic source, derived cardiomyocytes displayed a divergent expression pattern of genes, with nonmuscle and cardiac-muscle-specific metabolic enzyme isoforms, respectively, down- and upregulated. Besides regulating cardiac differentiation, the increased expression of cardiac transcription factors promotes mitochondrial function and cellular organization.12,22,23 In stem cell-derived cardiomyocytes, downregulation of genes with roles in mitochondrial fission or inhibition of fusion led to intracellular and intercellular networks facilitating expansion of the cardiogenic phenotype. These cell-to-cell connections enabling the transport of mitochondria and leading to the acquisition of a cardiomyogenic phenotype by progenitor cells had been observed previously.24 Genes that influence mitochondrial morphology, thereby promoting cardiogenesis, included Dnm1l (a dynamin-related GTPase) and Mtp18 (a transcriptional downstream target of PI3-kinase), which contribute to fission.25,26 Also, the dynamin-related Opa1, which maintains the mitochondrial inner membrane structure,27 and DAP3, which is involved in mitochondrial fragmentation during apoptosis,28 were down-regulated. Conversely, mitofusin 2, an essential protein for mitochondrial fusion, protection, and maintenance of morphology,29 was upregulated, while mitofilin, a mitochondrial protein abundant in the heart mitochondrial proteome,30 exhibited less-pronounced change. The significance of coordinated mitochondrial expansion is underscored by the critical role of nucleus-mitochondria interaction in promoting muscle-specific gene expression, thereby securing myogenic differentiation.31 The hypothesis that cardiac development is dependent on mitochondrial function was further validated using the respiratory chain inhibitors antimycin or rotenone, which disrupted proper cardiogenesis but still gave rise to viable embryoid bodies. Furthermore, the state of mitochondrial activation influences the intracellular redox potential.32 Promotion of an intracellular oxidized state, such as by extrinsic signaling molecules and reactive oxygen species generated by NADPH oxidases, has been shown to be conducive to cardiac differentiation.32–35 Thus, the state of mitochondrial physiology is an important determinant of developmental competence influencing the outcome of the differentiation process.36
CONCLUSIONS
In summary, embryonic stem cell-based cardiac differentiation was used as a paradigm to ascertain the metabolic requirements for tissue specification. We demonstrated that mitochondrial network maturation integrated with energy-consuming excitation-contraction coupling is mandatory for securing functional progeny of a cardiac lineage. These findings contribute novel insight into the vital role of energetic circuits in genomic reprogramming during cardiac differentiation, establishing metabolic targets for directed cardiogenesis and regeneration.
KEY POINTS.
While anaerobic metabolism is sufficient to sustain embryonic stem cell energetics, it must be transformed into mitochondria-mediated oxidative metabolism to secure cardiac specification
Induction of cardiogenic pathways is linked to restructuring of the metabolic transcriptome and organization of the mitochondrial network
Maturation and integration of mitochondria with the nascent electromechanical apparatus confer the functional competence required to develop a cardiac phenotype
Disruption of respiratory chain function and prevention of mitochondrial network organization compromise the developing energetic infrastructure, causing deficient sarcomere formation, improper beating area assembly, and defective contractility
As mandatory components of genomic reprogramming during cardiac differentiation, energetic circuits are metabolic regulators of cardiogenesis and targets for heart regeneration
Acknowledgments
This work was supported by the National Institutes of Health (NIH), American Heart Association, Marriott Heart Disease Research Program, Marriott Foundation, Ted Nash Long Life Foundation, Ralph Wilson Medical Research Foundation, Asper Foundation, and the Mayo Clinic Clinician-Investigator Program.
Footnotes
Competing interests
The authors declared they have no competing interests.
References
- 1.Donovan PJ, Gearhart J. The end of the beginning for pluripotent stem cells. Nature. 2001;414:92–97. doi: 10.1038/35102154. [DOI] [PubMed] [Google Scholar]
- 2.Behfar A, et al. Stem cell differentiation requires a paracrine pathway in the heart. FASEB J. 2002;16:1558–1566. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Terzic C, et al. Structural adaptation of the nuclear pore complex in stem cell-derived cardiomyocytes. Circ Res. 2003;92:444–452. doi: 10.1161/01.RES.0000059415.25070.54. [DOI] [PubMed] [Google Scholar]
- 4.Hodgson DM, et al. Stable benefit of embryonic stem cell therapy in myocardial infarction. Am J Physiol. 2004;287:H471–H479. doi: 10.1152/ajpheart.01247.2003. [DOI] [PubMed] [Google Scholar]
- 5.Van Laake LW, et al. Cardiomyocytes derived from stem cells. Ann Med. 2005;37:499–512. doi: 10.1080/07853890500327843. [DOI] [PubMed] [Google Scholar]
- 6.Lev S, et al. Differentiation pathways in human embryonic stem cell-derived cardiomyocytes. Ann NY Acad Sci. 2005;1047:50–65. doi: 10.1196/annals.1341.005. [DOI] [PubMed] [Google Scholar]
- 7.Kirby ML. Molecular embryogenesis of the heart. Pediatr Dev Pathol. 2002;5:516–543. doi: 10.1007/s10024-002-0004-2. [DOI] [PubMed] [Google Scholar]
- 8.Foley A, Mercola M. Heart induction: embryology to cardiomyocyte regeneration. Trends Cardiovasc Med. 2004;14:121–125. doi: 10.1016/j.tcm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Dzeja PP, Terzic A. Phosphotransfer networks and cellular energetics. J Exp Biol. 2003;206:2039–2047. doi: 10.1242/jeb.00426. [DOI] [PubMed] [Google Scholar]
- 10.Naya FJ, et al. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 11.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 12.Spitkovsky D, et al. Activity of complex III of the mitochondrial electron transport chain is essential for early heart muscle cell differentiation. FASEB J. 2004;18:1300–1302. doi: 10.1096/fj.03-0520fje. [DOI] [PubMed] [Google Scholar]
- 13.St John JC, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–153. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- 14.Behfar A, et al. Administration of allogenic stem cells dosed to secure cardiogenesis and sustained infarct repair. Ann NY Acad Sci. 2005;1049:189–198. doi: 10.1196/annals.1334.018. [DOI] [PubMed] [Google Scholar]
- 15.Menard C, et al. Transplantation of cardiac-committed mouse embryonic stem cells to infarcted sheep myocardium: a preclinical study. Lancet. 2005;366:1005–1012. doi: 10.1016/S0140-6736(05)67380-1. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Balcarcel RR. 96-well plate assay for sublethal metabolic activity. Assay Drug Dev Technol. 2004;2:353–361. doi: 10.1089/adt.2004.2.353. [DOI] [PubMed] [Google Scholar]
- 17.Dzeja PP, et al. Energetic communication between mitochondria and nucleus directed by catalyzed phosphotransfer. Proc Natl Acad Sci USA. 2002;99:10156–10161. doi: 10.1073/pnas.152259999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Terzic C, et al. Directed inhibition of nuclear import in cellular hypertrophy. J Biol Chem. 2001;276:20566–20571. doi: 10.1074/jbc.M101950200. [DOI] [PubMed] [Google Scholar]
- 19.Sathananthan H, et al. The fine structure of human embryonic stem cells. Reprod Biomed Online. 2002;4:56–61. doi: 10.1016/s1472-6483(10)61916-5. [DOI] [PubMed] [Google Scholar]
- 20.Sturmey RG, Leese HJ. Energy metabolism in pig oocytes and early embryos. Reproduction. 2003;126:197–204. doi: 10.1530/rep.0.1260197. [DOI] [PubMed] [Google Scholar]
- 21.Singla DK, et al. Transplantation of embryonic stem cells into the infarcted mouse heart: formation of multiple cell types. J Mol Cell Cardiol. 2006;40:195–200. doi: 10.1016/j.yjmcc.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15:148–159. doi: 10.1093/humrep/15.suppl_2.148. [DOI] [PubMed] [Google Scholar]
- 23.Goffart S, et al. Regulation of mitochondrial proliferation in the heart: power-plant failure contributes to cardiac failure in hypertrophy. Cardiovasc Res. 2004;64:198–207. doi: 10.1016/j.cardiores.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 24.Koyanagi M, et al. Cell-to-cell connection of endothelial progenitor cells with cardiac myocytes by nanotubes: a novel mechanism for cell fate changes? Circ Res. 2005;96:1039–1041. doi: 10.1161/01.RES.0000168650.23479.0c. [DOI] [PubMed] [Google Scholar]
- 25.Smirnova E, et al. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Cell Biol. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tondera D, et al. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci. 2005;118:3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- 27.Misaka T, et al. The dynamin-related mouse mitochondrial GTPase OPA1 alters the structure of the mitochondrial inner membrane when exogenously introduced into COS-7 cells. Neurosci Res. 2006;55:123–133. doi: 10.1016/j.neures.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Mukamel Z, Kimchi A. Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. J Biol Chem. 2004;279:36732–36738. doi: 10.1074/jbc.M400041200. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14:R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- 30.John GB, et al. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seyer P, et al. Mitochondrial activity regulates myoblast differentiation by control of c-Myc expression. J Cell Physiol. 2006;207:75–86. doi: 10.1002/jcp.20539. [DOI] [PubMed] [Google Scholar]
- 32.Smith J, et al. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc Natl Acad Sci USA. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauer H, et al. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 2000;476:218–223. doi: 10.1016/s0014-5793(00)01747-6. [DOI] [PubMed] [Google Scholar]
- 34.Noble M, et al. Redox regulation of precursor cell function: insights and paradoxes. Antioxid Redox Signal. 2005;7:1456–1467. doi: 10.1089/ars.2005.7.1456. [DOI] [PubMed] [Google Scholar]
- 35.Li J, et al. The NADPH oxidase NOX4 drives cardiac differentiation: Role in regulating cardiac transcription factors and MAP kinase activation. Mol Biol Cell. 2006;17:3978–3988. doi: 10.1091/mbc.E05-06-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonergan T, et al. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol. 2006;208:149–153. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]