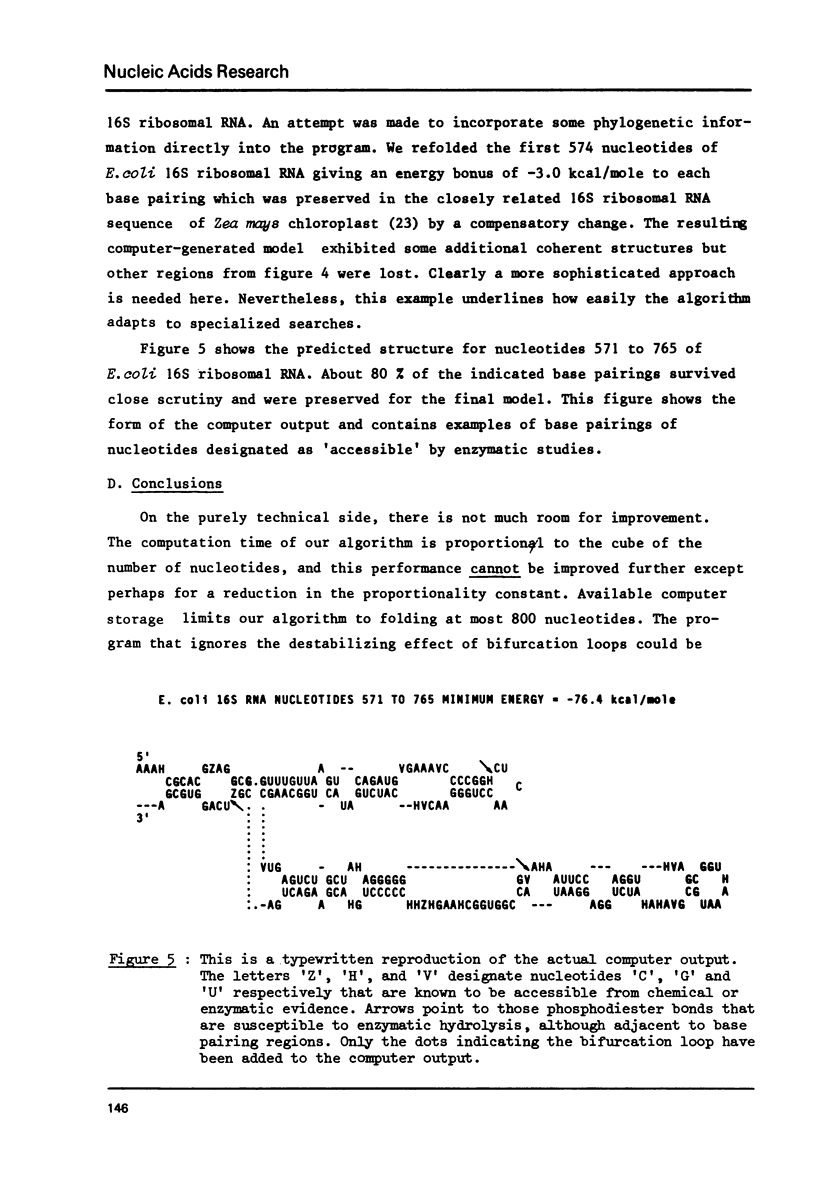
Abstract
This paper presents a new computer method for folding an RNA molecule that finds a conformation of minimum free energy using published values of stacking and destabilizing energies. It is based on a dynamic programming algorithm from applied mathematics, and is much more efficient, faster, and can fold larger molecules than procedures which have appeared up to now in the biological literature. Its power is demonstrated in the folding of a 459 nucleotide immunoglobulin gamma 1 heavy chain messenger RNA fragment. We go beyond the basic method to show how to incorporate additional information into the algorithm. This includes data on chemical reactivity and enzyme susceptibility. We illustrate this with the folding of two large fragments from the 16S ribosomal RNA of Escherichia coli.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyle J., Robillard G. T., Kim S. H. Sequential folding of transfer RNA. A nuclear magnetic resonance study of successively longer tRNA fragments with a common 5' end. J Mol Biol. 1980 Jun 5;139(4):601–625. doi: 10.1016/0022-2836(80)90051-0. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur J Biochem. 1979 Oct 15;100(2):399–410. doi: 10.1111/j.1432-1033.1979.tb04183.x. [DOI] [PubMed] [Google Scholar]
- Chapman N. M., Noller H. F. Protection of specific sites in 16 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J Mol Biol. 1977 Jan 5;109(1):131–149. doi: 10.1016/s0022-2836(77)80049-1. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Stiegler P., Carbon P., Ungewickell E., Garrett R. A. A topographical study of the 5'-region of 16 S rna of Escherichia coli in the presence and absence of protein S4. FEBS Lett. 1977 Sep 1;81(1):188–192. doi: 10.1016/0014-5793(77)80956-3. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Stiegler P., Carbon P., Ungewickell E., Garrett R. A. The topography of the 5' end of 16-S RNA in the presence and absence of ribosomal proteins S4 and S20. Eur J Biochem. 1980 Feb;103(3):439–446. doi: 10.1111/j.1432-1033.1980.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. 3. Small internal loops resulting from mismatches. J Mol Biol. 1973 Aug 5;78(2):301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Herr W., Chapman N. M., Noller H. F. Mechanism of ribosomal subunit association: discrimination of specific sites in 16 S RNA essential for association activity. J Mol Biol. 1979 Jun 5;130(4):433–449. doi: 10.1016/0022-2836(79)90433-9. [DOI] [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J., Yuki A., Brimacombe R. Studies on the environment of protein S7 within the 30-S subunit Escherichia coli ribosomes. Eur J Biochem. 1976 Apr 15;64(1):77–89. doi: 10.1111/j.1432-1033.1976.tb10276.x. [DOI] [PubMed] [Google Scholar]
- Rogers J., Clarke P., Salser W. Sequence analysis of cloned cDNA encoding part of an immunoglobulin heavy chain. Nucleic Acids Res. 1979 Jul 25;6(10):3305–3321. doi: 10.1093/nar/6.10.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Borer P. N., Dengler B., Tinoco I., Jr Stability of RNA hairpin loops: A 6 -C m -U 6 . J Mol Biol. 1973 Feb 5;73(4):483–496. doi: 10.1016/0022-2836(73)90095-8. [DOI] [PubMed] [Google Scholar]
- Ungewickell E., Ehresmann C., Stiegler P., Garrett R. Evidence for tertiary structural RNA-RNA interactions within the protein S4 binding site at the 5'-end of 16S ribosomal RNA of Escherichia coli.+. Nucleic Acids Res. 1975 Oct;2(10):1867–1888. doi: 10.1093/nar/2.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E., Garrett R. A. Further characterisation of the RNA structure in the binding region of protein S4 on 16 S ribosomal RNA of Escherichia coli. FEBS Lett. 1977 Sep 1;81(1):193–198. doi: 10.1016/0014-5793(77)80957-5. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]


