Abstract
T4 RNA ligases are commonly used to attach adapters to RNAs, but large differences in ligation efficiency make detection and quantitation problematic. We developed a ligation selection strategy using random RNAs in combination with high-throughput sequencing to gain insight into the differences in efficiency of ligating pre-adenylated DNA adapters to RNA 3′-ends. After analyzing biases in RNA sequence, secondary structure and RNA-adapter cofold structure, we conclude that T4 RNA ligases do not show significant primary sequence preference in RNA substrates, but are biased against structural features within RNAs and adapters. Specifically, RNAs with less than three unstructured nucleotides at the 3′-end and RNAs that are predicted to cofold with an adapter in unfavorable structures are likely to be poorly ligated. The effect of RNA-adapter cofold structures on ligation is supported by experiments where the ligation efficiency of specific miRNAs was changed by designing adapters to alter cofold structure. In addition, we show that using adapters with randomized regions results in higher ligation efficiency and reduced ligation bias. We propose that using randomized adapters may improve RNA representation in experiments that include a 3′-adapter ligation step.
INTRODUCTION
Bacteriophage T4 encodes two RNA end-joining enzymes, T4 RNA ligase 1 (Rnl1) (1) and T4 RNA ligase 2 (Rnl2) (2). Both enzymes catalyze the formation of a 3′- to 5′-phosphodiester bond between a 3′-hydroxyl group and a 5′-phosphoryl group in three nucleotidyl transfer steps (1,3,4). The function of Rnl1 is to counter a particular host defense mechanism induced after T4 phage infection. This host defense mechanism involves generating a break in the anticodon loop of tRNALys so that the viral genes of T4 cannot be translated. Rnl1, together with T4 polynucleotide kinase, repairs the cleaved anticodon loop of tRNALys in vivo (1,5,6). Although Rnl2 is phylogenetically related to DNA ligases, RNA-editing ligases and mRNA capping enzymes (7), the function of Rnl2 in vivo is not clear. The activity of both T4 RNA ligases has been exploited in vitro for use in applications such as RNA ligase-mediated rapid amplification of cDNA ends (8,9), ligation of oligonucleotide adapters to cDNA (10,11), various 5′-nt modifications of nucleic acids, RNA 3′-end modification (12) and small RNA sequencing library construction (13).
miRNAs are one class of small regulatory RNAs that mediate post-transcriptional gene regulation in higher eukaryotes (14). miRNAs base pair with a target mRNA when associated with the RNA-induced silencing complex (RISC) resulting in regulation of gene expression through mRNA degradation and translation repression (15,16). Studies of miRNAs in various organisms have revealed that the expression and regulatory functions of miRNAs are controlled at different developmental stages, in different cell types, tissues and species (14,17) and that misregulation of miRNA expression and function is a significant factor in many diseases (18). The emerging realization of miRNA functions in vivo makes the development of effective experimental methods to accurately detect and measure the expression of miRNAs important for future research.
High-throughput sequencing (HTS) has been an invaluable tool not only for the discovery of miRNAs but also for profiling their relative expression level (19–23). However, HTS-based miRNA profiling experiments are reported to be biased (24–26). The level of bias has been suggested to cause a miscalculation of miRNA abundance by as much as three or four orders of magnitude (24,25). Thus, relating the number of reads from HTS to the abundance of miRNA in the sample is problematic. Additional comparison studies showed that the bias is reproducible and independent of sequencing platforms and also that the bias is derived from the methods used for small RNA library preparation (24).
In general, small RNA library preparation methods for HTS start with the ligation of 5′- and 3′-adapters to add ‘handles’ that are used for priming during reverse transcription and PCR (24). Typically, adapters are attached to small RNAs by T4 RNA ligases using either a single-stranded adapter ligation approach, a splinted ligation approach or by poly adenylation of the RNA 3′-termini followed by 5′-end adapter ligation (13,24,25,27).
Two recent studies suggested that miRNA representation bias in HTS is primarily derived from the adapter ligation steps mediated by T4 RNA ligases and that the ligation might be biased in a sequence-dependent manner (25,26). However, these studies examined ligation bias using HTS, which means the results reflected the combined bias from both the 5′- and 3′-adapter ligation steps. Bias studies to date have, at most, been based on a pool of several hundred miRNAs or just a few known miRNAs (24,25). The limited sequence space of the reference pools in these recent reports is not sufficient to determine the exact nature of ligase bias.
In this work, we separated the two adapter ligation steps and focused on the ligation bias in 3′-adapter ligation reactions, which have been suggested to be more biased than 5′-adapter ligation reactions (26). We developed an in vitro selection strategy where the 3′-end of randomized RNA oligonucleotides were ligated to pre-adenylated DNA adapters using Rnl1 or four variants of a truncated form of Rnl2 (Rnl2tr) (28). We determined the sequences of ligated oligos using the Ion Torrent sequencing platform (29), and analyzed bias at the level of primary RNA sequence, RNA secondary structure and RNA-adapter cofold structures by comparing the ligated RNA sequences to the random input sequences.
Sequence analysis did not reveal appreciable RNA primary sequence preference in the 3′-adapter ligation reaction for any of the ligases we tested. Instead, ligation bias is primarily due to the cofold structure between a given RNA and the adapter. These findings are supported by results from in vitro ligation experiments on a representative set of miRNAs from miRBase (30). Furthermore, we demonstrate that in vitro ligation efficiency for specific miRNAs can be significantly affected by manipulating the adapter sequence to change the predicted RNA-adapter cofold structure and improve the ligation of otherwise poorly ligated miRNAs. Finally, we present an approach to improve ligation efficiency and reduce ligation bias of miRNA pools using a mixture of adapters with randomized 5′-regions.
MATERIALS AND METHODS
Ligated and random library preparation
Random RNA oligos were synthesized by Integrated DNA Technologies (Iowa, USA). To assess the sequence content of the random oligos, reactions adding a poly(A) tail to random RNA oligos G, C and U were performed using the protocol supplied by the manufacturer (New England Biolabs, Ipswich, MA, USA). Poly(C) tailing of random RNA oligo A was performed as previously described (31). All tailing reactions were incubated at 37°C for 2 h and the reactions were stopped by phenol/chloroform/isoamyl alcohol (IAA) extraction and precipitated by ethanol. After washing with 70% ethanol, the precipitated nucleic acid was resuspended in 20 µl H2O prior to undergoing preparation for Ion Torrent sequencing. For the ligase selected libraries, each ligation reaction contained 1.4 µM ligase, 5.5 µM of adenylated SR1 adapter, 12% PEG8000, 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT and 50 ng of each random RNA oligo in a total volume of 200 µl. Reactions were incubated at 25°C for 2 h and stopped as described above.
Ion Torrent sequencing library preparation
Ligated products and tailed random oligos were reverse transcribed into cDNA using the ProtoScript™ M-MuLV Taq Reverse transcription (RT–PCR) Kit (New England Biolabs) following the protocol supplied with the kit. The primers for reverse transcription contained the Ion Torrent (Life Technologies, Carlsbad, CA, USA) ‘trP1’ sequence (CCTCTCTATGGGCAGTCGGTGAT) and sequence complimentary to the adapter sequence for ligated libraries or complimentary to the poly A or C tailed region for the random oligo libraries (Supplementary Table S1, Ligated RT, Random A,C,G or U RT). The cDNA products were amplified by 10 cycles of PCR using LongAmp® Taq master mix (New England Biolabs) with primers ‘IT Forward’, which added the Ion Torrent ‘A’ sequence, and ‘IT Reverse’ (Supplementary Table S1). PCR products were gel purified using either E-Gel® SizeSelect 2% agarose gels (Life Technologies) or 6% acrylamide gels. The purity and concentration of purified PCR products were analyzed using an Agilent 2100 Bioanalyzer (Agilent Biotechnologies, Santa Clara, CA, USA). Each purified library was diluted to a concentration of 44 pM and was prepared for Ion Torrent sequencing by using the Ion Xpress™ Template Kit (Life Technologies) and following the protocol supplied by the manufacturer. Libraries were sequenced on an Ion PGM™ using Ion 314™ or Ion 316™ chips deposited at full density.
Adenylation of DNA oligos
The DNA adapters were synthesized by Integrated DNA Technologies (Iowa, USA) with a phosphorylated 5′-end and a blocking amino group at the 3′-end. The adapters were adenylated using a 5′-DNA Adenylation Kit (New England Biolabs) as described previously (32). The adenylation reactions were stopped by adding 1 µg of proteinase K (New England Biolabs) per µl of adenylation reaction and incubated at 37°C for 30 min. DNA was further purified by two extractions with phenol/chloroform/IAA followed by ethanol precipitation. The adenylated oligos were separated from unadenylated ones in 20% Tris-borate-EDTA (TBE)–urea acrylamide gels. Bands corresponding to adenylated oligos were isolated, crushed and soaked in 1 ml water overnight at room temperature with constant rotation. After soaking, DNA was extracted from soaking solution using phenol/chloroform/IAA and precipitated by ethanol.
Ligation reactions
In vitro ligation reactions containing defined RNA substrates were carried out in 10 µl reactions containing 0.5 µM miRNA, 1 µM adenylated DNA adapter, 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 40 U of Murine RNase Inhibitor (New England Biolabs), 12.5% PEG8000 and 0.1 µM (Figures 1, 6 and 7) or 1.3 µM ligase (Figure 8). Reactions were incubated at 25°C for 2 h and stopped by adding same volume of 2×RNA loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, bromophenol blue, xylene cyanol). The ligation reactions were then loaded on a 15% TBE–urea gel to resolve ligated product, unligated RNA and unligated DNA adapter. The nucleic acid in the gel was stained with SYBR® Gold (Life Technologies) and scanned on a Typhoon™ 9400 Variable Mode Imager (GE Healthcare, NJ, USA). The intensity of each band was quantified using Quantity One software (BIO-RAD, Hercules, CA, USA) in order to determine the ligation efficiency. The amount of miRNA in the ligated product (Iligated miRNA) was normalized using the following equation. Iligated miRNA = Iligated × lengthmiRNA/(lengthmiRNA + lengthadapter). Ligation efficiency was calculated using the equation, ligation efficiency = Iligated miRNA/ImiRNA.
Figure 1.
3′-adapter ligation efficiencies of miRNAs. (A) Each miRNA was incubated in a ligation reaction containing Rnl2tr with or without SR1 adapter. The ligation products were separated on 15% TBE–urea gels and visualized with SYBR Gold. Ligated products correspond to high molecular weight bands, which only appear in reactions with SR1 adapter. Unligated miRNAs and SR1 adapters remain as lower molecular weight bands. (B) The ligation efficiency of each miRNA was determined and plotted. The data are represented as the average ± standard deviation from two experimental replicates.
Figure 6.
Comparison of miRNA ligation efficiencies using SR1 versus SR1-S adapter. (A) Sequences and predicted secondary structures of SR1 and SR1-S adapters. The underlined sequence is shared by both adapters. The secondary structures of SR1 and SR1-S are presented in bracket and dot form where brackets represent base paired nucleotides and dots represent unpaired nucleotides. (B) Ligation reactions of miRNAs with the SR1 or SR1-S adapter were performed using Rnl2tr. Ligation products were resolved in 15% TBE–urea gels and stained with SYBR Gold. (C) Ligation efficiency was calculated and plotted. The data points plotted represent average ligation efficiency ± standard deviation from two independent experiments.
Figure 7.
Improving miRNA ligation efficiency using redesigned adapters. (A) Ligation of miRNAs with SR1 adapter or a new adapter specifically designed for each miRNA. Ligation reactions were performed using Rnl2tr. Ligation products were resolved in 15% TBE–urea gels, stained with SYBR Gold to visualize the nucleic acids. (B) Ligation efficiency was determined and plotted. The data points represent average ligation efficiency ± standard deviation from two independent experiments.
Figure 8.
Improvement of miRNA ligation efficiencies using a randomized adapter, SR1-R. (A) Ligation reactions were performed with Rnl2tr and the SR1 or SR1-R adapter. Ligation products were resolved in 15% TBE–urea gels and stained with SYBR Gold to visualize the nucleic acids. (B) Ligation efficiencies of 24 miRNAs with the SR1 or SR1-R adapter were determined and plotted. The data are represented as the average ± standard deviation from two experimental replicates.
Ligation reactions in the presence of small RNA mixtures contained 40 U of RNase Inhibitor (New England Biolabs), 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 12.5% PEG8000, 375 fmol of sRNA extracted from mouse ES cells (ES-E14TG2a, ATCC, Manassas, VA, USA), 0.75 fmol of 5′-32P radio-labeled miRNA, 50 pmol of SR1 or SR1-R adapter, and 6.5 pmol of Rnl2tr in 10 µl reaction volume. Reactions were incubated at 25°C for 2 h and stopped as described above. The ligated and unligated radio-labeled miRNAs were separated in 15% TBE–urea gels. Gels were exposed to a storage phosphor screen (GE Healthcare, NJ, USA) and the intensities of bands were quantified using Quantity One software (BIO-RAD, Hercules, CA, USA). The ligation efficiencies of miRNAs were calculated from the equation, ligation efficiency = Iligated/(Iligated + Iunligated).
Bioinformatics
The 3′-adapter, or homopolymer tail sequences and 5′-constant regions were trimmed off using Galaxy (33–35). Only trimmed reads that were 21 nt in length were considered in subsequent analyses. RNA CONTRAfold (http://contra.stanford.edu/contrafold/index.html) was used for RNA secondary structure prediction. Default settings were used for the prediction. To predict RNA and adapter cofold structures, the Vienna RNAcofold (http://rna.tbi.univie.ac.at/cgi-bin/RNAcofold.cgi) was used with the default setting of minimum free energy algorithms and folding temperature at 25°C (36,37). The algorithm of Vienna RNAcofold prediction is based on the minimum free energy model (38). In our analysis, the 1999 Turner Model was used as the energy parameter during prediction (38).
Mouse ES cell small RNAs preparation
Total RNA was extracted from mouse embryonic stem cells (ES-E14TG2a, ATCC) using TRI Reagent (Sigma-Aldrich, MO, USA). The total RNA was subjected to Dnase I (New England Biolabs) digestion at 37°C for 30 min. RNA was further purified by acid-phenol:choloform (Life Technologies) extraction and ethanol precipitation. Pellets were resuspended in H2O and the integrity of total RNA was assessed by checking the ribosomal RNAs in 1% agarose gels. Small RNAs <40 nt were isolated by flashPAGE™ fractionation (Life Technologies) and precipitated by 1.5 volume of isoproponal, 1/10 volume of 3 M sodium acetate and 25 µg of linear acrylamide (Amresco, Solon, OH, USA). After precipitation and washing, the small RNAs were resuspended in water and the RNA concentration was determined by Qubit Fluorometer (Life Technologies). Each pmol of small RNAs were further treated with 1 U of alkaline phosphatase, Calf Intestinal (CIP, New England Biolabs) at 37°C for 1 h. The reaction was stopped by acid–phenol:choloform extraction and small RNAs were collected by ethanol precipitation. After resuspension in H2O, the concentration of purified small RNAs was determined by Quibit Fluorometer (Life Technologies).
RESULTS
The efficiency of 3′-adapter ligation varies between different miRNAs
To illustrate the variation in 3′-adapter ligation efficiency for different miRNAs, we selected 25 miRNAs from miRBase (30) and performed ligations using Rnl2tr (Figure 1A). As shown in Figure 1B, the ligation efficiency of a given miRNA to a pre-adenylated DNA adapter is highly variable, ranging from ligation of as little as 0% of the input to as much as 100%. These results confirm that there is significant bias in miRNA 3′-adapter ligation reactions that cannot be easily explained by miRNA primary sequence or predicted secondary structure (Supplementary Table S2).
Ligation and HTS of oligonucleotide pools to study ligation bias
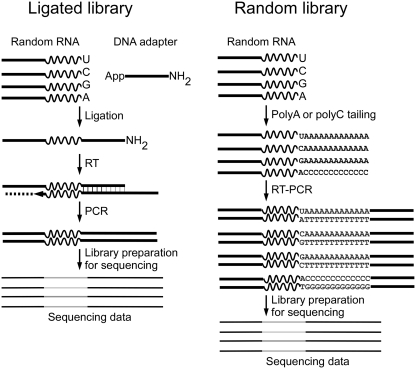
To study the bias of T4 RNA ligases, we designed an in vitro ligation selection assay that uses a pool of random RNA oligos to which a 3′-adapter is ligated (Figure 2). Following reverse transcription and amplification, the sequence of the ligated products was determined using the Ion Torrent sequencing platform (29). The random RNA oligo pool contained equimolar amounts of four random RNA oligos, which consisted of the same constant 21 nt region at the 5′-end, followed by a 20 nt random region, and a U, C, G or A at the 3′-end (Supplementary Table S1). A fixed nucleotide at the 3′-end of oligo is required for oligo synthesis. Therefore, it was necessary to hand mix an equimolar amount of four random oligos to generate an oligo pool containing 21 random positions.
Figure 2.
Scheme of in vitro ligation selection and sequencing library preparation. For each ligase selected library, an equal amount of 4 random RNA oligos containing a constant region (solid line), a randomized region (wavy line) and a known 3′-nt were combined to make a random oligo pool and used as substrates in a ligation reaction with pre-adenylated SR1 DNA adapter using a specific T4 RNA ligase. The ligated products were reverse transcribed and amplified to introduce the required primer regions for Ion Torrent sequencing. To determine the sequence content of the random RNA oligo pool, each of the four RNA oligos was sequenced independently. First, the oligos were poly A tailed for the random RNA oligo U, C and G or poly C tailed for the random RNA oligo A using poly(A) polymerase. The tailed RNA oligos were then reverse transcribed using primers complementary to the polymer tails (Supplementary Table S1). The cDNA libraries were amplified and processed in the same manner as the ligase selected libraries described above.
To assess the frequency of nucleotides at each randomized position in the oligo pool, the random RNA oligos were subjected to Ion Torrent sequencing without undergoing adapter ligation (Figure 2). Each oligo was tailed with poly nucleotides using poly(A) polymerase. The random RNA oligo ending with U, C or G was tailed with ribonucleotide A, while the random RNA oligo ending with A was tailed with ribonucleotide C in order to distinguish the last A from the tailed region. We performed poly-adenylation reactions under conditions that minimized 3′-end nucleotide bias (39). These conditions resulted in essentially complete tailing of the input oligos (data not shown). After tailing, the RNAs were reverse transcribed and the cDNAs were used for Ion Torrent sequencing library preparation. After sequencing, 29 737 sequences from each of the four random oligos were pooled to generate a random input library with 118 948 reads in total (Supplementary Table S3).
Our ligation selection reactions used 9.0 × 1013 molecules of random oligos in order to cover all the 4.4 × 1012 possible sequence combinations from 21 random positions. For each library, a ligation reaction was performed using an excess of RNA ligase and 5′-pre-adenylated DNA adapter (SR1) over random RNA oligos. The SR1 adapter was blocked at its 3′-end by an amino group to prevent it from participation intra- and inter-molecular ligation with other SR1 molecules (25,28,39). Ligated products were reverse transcribed into cDNA and sequenced on the Ion PGM™. Rnl1 and four variants of Rnl2tr were tested in the selection assay. After quality control and adapter trimming, we obtained 105–106 sequences for each library. The summary of sequence reads and quality control of libraries are shown in Supplementary Table S3.
T4 RNA ligases do not show appreciable bias in RNA substrates at the primary sequence level
We first looked for evidence that the ligases have any primary sequence preference within the 21 nt random region of the RNA substrates. To do so, we calculated the frequency of each nucleotide at each position from 105 to 106 sequences in each library (Supplementary Table S4). In Figure 3A and Supplementary Figure 1, the raw nucleotide frequency each position in random region was plotted in enoLOGO format (40). The frequency of a particular nucleotide at each position is proportional to the size of the letter representing that base. The distribution of nucleotide frequencies in ligated libraries is very similar to that of the random input library. For the random input library, the nucleotide frequencies from positions 1 to 19 shares a similar distribution with A and U being close to 25%, but slightly less C (19–20%) and higher G (30–34%) (Supplementary Table S4). The frequencies at position 20 showed a slightly different distribution pattern compared to positions 1–19 with 22% A, 22% C, 29% G and 26% U. The nucleotide percentage of position 21 was 25% for all 4 nt reflecting equal numbers of sequences that were combined from four random oligo libraries.
Figure 3.
Nucleotide frequencies at each position in the randomized region of random and ligase selected libraries. (A) The nucleotide frequencies calculated from Ion Torrent sequencing runs of the random and ligase selected libraries were plotted in enoLOGOS format (40). The y-axis represents the frequency of each nucleotide proportional to the height of their representative letters, A, U, G and C. (B) The nucleotide frequencies of the ligase selected libraries were corrected to the frequencies in the randomized input library. The value of enrichment plotted on the y-axis is the normalized nucleotide frequency (RNnp) subtracting 0.25, ‘RNnp −0.25’. If (RNnp −0.25) of a nucleotide n at position p is equal to 0, it indicates the ligase doesn’t have preference for nucleotide n at position p. If RNnp −0.25) is greater or less than 0, it means that the nucleotide is preferred or not preferred at position p, respectively. The x-axis in A and B represents the position of nucleotides in the random region from 5′ to 3′.
We then normalized the nucleotide frequency in the ligated libraries to that of the random input library and determined the enrichment of nucleotides at every position using a previously described method (41). Briefly, the relative ratio of each nucleotide n at position p in each ligase selected library was determined from the nucleotide frequency in the ligated RNA pool  versus the frequency in the random input pool
versus the frequency in the random input pool  by using the equation:
by using the equation:
The RNnp ratio at each position was normalized to 1 by equation:
The value of (RNnp −0.25) was plotted according to the nucleotide positions for each ligase (Figure 3B). If (RNnp −0.25) of a nucleotide n at position p is equal to 0, it indicates the ligase does not have any preference for nucleotide n at position p. If (RNnp −0.25) is greater or less than 0, it means that nucleotide n is preferred or not preferred at position p, respectively. As shown in Figure 3B, Rnl1 and the four variants of Rnl2tr show minimal preference for any particular nucleotide at any particular position in our RNA substrates. We interpret these observations to mean that the primary sequence of RNA substrate has minimal impact on the ligation efficiency.
Ligation efficiency is affected by the secondary structure within an RNA substrate
Given the striking differences we observed in miRNA 3′-adapter ligation efficiency and having failed to observe significant sequence bias in our sequencing experiments, we next asked whether T4 RNA ligases prefer certain secondary structures within RNA substrates as suggested previously (25). Considering the function of T4 Rnl1 is to repair a break in the anticodon loop of tRNALys, it is possible that ligases such as Rnl1 prefer RNA substrates similar in structure to its biological substrate. To explore the possible secondary structure preferences of ligases, sequences from each library were analyzed by CONTRAfold to predict their secondary structures (42). After folding, structural predictions of the 42 nt sequences were sorted into groups based on the number of unpaired nucleotides at their 3′-end (Figure 4). If the 3′-end nucleotide is predicted to be paired, there are ‘0’ unpaired nucleotides and if no pairing was predicted within the RNA, there are ‘42’ unpaired nucleotides. The percentage of each group in a specific library was calculated and the enrichment of that structure group was determined by comparing its percentage to that in the random input library using the following equation, ‘(Observed–Expected)/Expected’. ‘Observed’ is the percentage of a specific group in a ligated library and ‘Expected’ is its corresponding percentage in the random input library. If the calculated value is positive or negative, it means the structure is over- or under-represented in the ligated library, respectively. As shown in Figure 4, it is clear that all tested ligases share a similar preference. Specifically, RNAs with fewer than three unpaired nucleotides at the 3′-end are under-represented in the ligated libraries while RNAs with three or more unpaired nucleotides at the 3′-end appear at their expected frequency or are over-represented. Thus, RNA ligases generally prefer RNA with a relatively accessible 3′-end and one source of bias in ligation is secondary structure at the 3′-end of an RNA substrate.
Figure 4.
Enrichment of RNA 3′-end predicted secondary structures in ligated libraries. Each sequence from the ligated libraries and random library was subjected to RNA CONTRAfold analysis. RNA structural predictions were classified based on the number of unpaired nucleotides at their 3′-end as labeled in the x-axis. A value of ‘42’ on the x-axis represents RNAs that lack any secondary structure according to CONTRAfold prediction. The value of enrichment was determined by the equation ‘(Observed–Expected)/Expected’, where ‘Observed’ is the percentage of a specific category in a ligated library and ‘Expected’ is the percentage of the same category in the random input library.
Analysis of RNA and adapter cofolding
When comparing predicted structure at the 3′-end to the ligation efficiency of an RNA (Figure 1 and Supplementary Table S2), it is clear that the preference of ligases for RNAs lacking 3′-end secondary structure cannot completely explain the ligation bias we observed. We hypothesized that another possible source of bias could result from the interaction between the RNA substrate and the adapter. Thus, we predicted the cofold structures of our sequenced library members with the SR1 adapter to examine the correlation between cofold structures and ligation efficiencies.
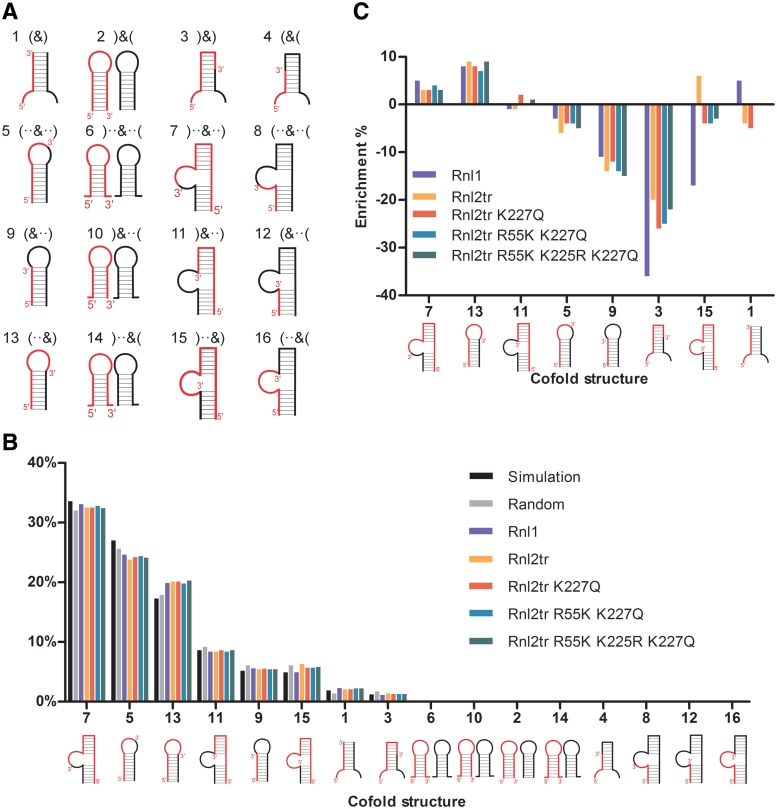
Using the Vienna RNAcofold algorithm (36,37), we cofolded all sequences from all sequenced libraries with the SR1 adapter and classified the cofold structures according to the regional secondary structure at the ligation junction. These structures are summarized and presented in dot-bracket notation and schematic representation (Figure 5A). Brackets indicate paired nucleotides and dots represent unpaired nucleotides. An ampersand symbol, ‘&’, denotes the ligation junction. Structure class 2, 6, 10 and 14 are distinct from the rest of the classes because the RNA and adapter do not form heterodimers.
Figure 5.
RNA-adapter cofold structures. Each sequence from random and ligated libraries was cofolded with the SR1 adapter using the Vienna RNAcofold. Based on the structural differences of predicted secondary structures at the ligation junction, 16 possible cofold structure classes are listed in (A). Each cofold structure class is numbered and presented in bracket and dot notion, in which brackets represent base pair(s) and dots represent unpaired nucleotide(s). The ‘&’ symbol represents the ligation junction between the RNA 3′-end and the adapter 5′-end. Multiple dots and brackets represent two or more unpaired or paired nucleotides in a row and the directionality of the brackets (open or closed) indicates the pairing orientation. Generalized schematic diagrams of corresponding cofolding structures are shown under the bracket and dot notation, in which RNA is in red and the DNA adapter is in black. The base pairings are shown as thin black lines. (B) Distribution of RNA and adapter cofold structures in simulated and sequenced libraries showing the percentage of library members assigned to each structural class. This distribution was used to calculate enrichment. (C) Enrichment of cofold structures in ligated libraries. The enrichment of each cofold structures was calculated using the equation, ‘(Observed–Expected)/Expected’, where ‘Observed’ is the percentage of a cofold structure in the ligated library and ‘Expected’ is the percentage of the corresponding structure in the random input library. Numbers on the x-axis correspond to the cofold structure classes in A and their schematic illustrations are shown under the numbers.
Only 8 out of the 16 structure classes were present in all sequenced libraries as shown in the distribution plot in Figure 5B. Structure classes 5, 7 and 13 are the three most abundant and they make up 19.2–33.1% of the total number of classes in each library. Structure classes 1, 3, 9, 11 and 15 were less abundant and ranged from 1.1% to 8.6% in each library. We did not observe structure classes 2, 4, 6, 8, 10, 12, 14 and 16 in any of our sequenced libraries.
To assess the cause of the absent structure classes in our sequenced libraries, and exclude that the absence was caused by a lack of sequence coverage, we generated random libraries in silico. Ten simulated random libraries, each consisting of 300 000 random sequences, were generated by a computer containing the same 5′-constant sequence and 21-nt random region. Each sequence was then cofolded with the SR1 adapter and the average distribution of cofold structures from the 10 libraries was compared to our sequenced random input library (Figure 5B). Distribution of the simulated random libraries is similar to that of our sequenced random library confirming that our sequenced random library indeed represents a random pool. The absence of observed cofold structure classes is therefore not due to the sequence coverage nor bias introduced by poly(A) polymerase. In addition, the absent cofold structure classes do not result from the presence of 5′-constant region in the RNA (Supplementary Figure S2) since the presence or absence of the 5′-constant region did not affect their presence in our cofold predictions. The absent cofold structure classes are structures where the RNA and adapter are predicted to not form heterodimers or where adapters are internally structured. Since the SR1 adapter does not form internal secondary structure or homodimers according to secondary structure prediction (Figure 6A), it makes sense that these structure classes would not be represented when RNAs are cofolded with the SR1 adapter. These observations lead us to conclude that the distribution of cofold structures is largely due to the SR1 adapter and limitations in what structures that it can form.
Enrichment of RNA-adapter cofold structures in the ligated libraries
To determine whether the ligases prefer or discriminate against a specific cofold structure, we first compared the percentage of each cofold structure class in the ligated libraries to that in the random library. We further calculated enrichment for each cofold structure class using ‘(Observed–Expected)/Expected’, in which ‘Observed’ is the percentage of a specific cofold structure class from a ligated library and ‘Expected’ is the percentage of the corresponding class from the random library. When the value is greater or less than 0, it means the structure class is over- or under-represented, respectively. If the value is equal to 0, it means there is no preference for that structure class. As shown in Figure 5C, it is evident that two structure classes, 7 and 13, were slightly over-represented in the libraries of all tested ligases and none of the ligases showed much preference toward structure class 11. Three structure classes, 3, 5 and 9, were found to be under-represented in the ligated libraries, though structure 5 was only slightly so. The natural substrate of T4 Rnl1, cleaved tRNALys, is predicted to cofold into structure class 5 (5). Interestingly, the ligases did not differ in their preference for structure class 5 as reflected by the representation of this class in our sequenced libraries. However, the ligases showed different preferences toward structure classes 1 and 15. For instance, class 15 was under-represented in the library using T4 Rnl1 but was over-represented when using Rnl2tr, but all of the mutant variants of T4 Rnl2 showed similar preferences to structure class 15 as Rnl1.
RNA-adapter cofold structure influences ligation efficiency
Our observations that certain cofold structure classes were over- or under-represented in our sequenced libraries prompted us to examine the influence of RNA-adapter cofolding on ligation efficiency. We designed a new adapter, SR1-S, which shares the same sequence as SR1 at the 5′-end from positions 1 to 12, but with a modified 3′-sequence so that, in contrast to SR1, SR1-S is predicted to have secondary structure. We presume that internal secondary structure in the adapter would reduce its ability to productively cofold with RNAs (Figure 6A and Supplementary Table S5). In Figure 6B, SR1-S migrated faster than the SR1 adapter, which is the same length, likely because of incomplete denaturation or renaturation. SR1-S stained more strongly than an equivalent amount of SR1 in gels, consistent with the 2× increased staining of dsDNA versus ssDNA with SYBR Gold (43). We then compared the ligation efficiencies of 18 miRNAs with both the SR1 and SR1-S adapter. As shown in Figure 6B and C, we observed large overall decreases in ligation efficiency using SR1-S. The average ligation efficiency of the miRNAs decreased from 27% with SR1 to 8% with SR1-S. The decrease in ligation was most dramatic for miRNAs which ligated efficiently with SR1, such as miR-31, let-7, miR-139-3p, miR-122, miR-253, miR-1614 and miR-2419. The fold decrease in ligation efficiency for these RNAs ranged from 3- to 22-fold. miRNAs that had low ligation efficiencies with SR1 also ligated poorly with SR1-S. The finding that we can modulate ligation efficiency by changing secondary structure within the adapter without changing primary sequence at the 5′-end is at odds with a previous report (26). This report concluded that RNA ligase bias is due to primary sequence-specific preferences at the ligation junction, specifically the first two nucleotides of the 5′-end of adapter sequence and the last two nucleotides of the 3′-end of the RNA (26). Our results, especially when considered with the nucleotide frequency results from our sequencing experiments (Figure 3), contradict this conclusion. Together, our results demonstrate that structures within and between the RNA acceptor and the adapter are more important than primary sequence in influencing ligation efficiency. These results support a model where favorable heterodimeric RNA and adapter cofold structures promote efficient ligation.
To further test our model, we tried to improve the ligation efficiency of seven miRNAs that ligate poorly with SR1 by designing a new adapter for each miRNA. In each case, the adapter was designed so that the predicted RNA-adapter cofold structure was changed from an under-represented class to an over-represented class (Table 1). The predicted cofold structures of six miRNAs with SR1 belonged to one of two under-represented cofold structure classes, either class 1 or 5. The adapters we designed for each of these miRNAs changed their predicted RNA-adapter cofold structures to the over-represented structure class 13. The seventh miRNA, miRNA-5183, was already predicted to cofold with SR1 in structure class 13, despite the fact that it ligates poorly with SR1. When we examined predicted loop size of miRNAs within structure class 13, and correlated this with ligation efficiency, we noted a trend that some loop sizes appear to be unfavorable for ligation (data not shown). The adapter we designed for miR-5183 adjusted the size of the loop in the cofold structure class 13.
Table 1.
Predicted cofold structures of miRNA with SR1 or redesigned adapter
| miRNA/adapter | Cofold structure | Structure No. |
|---|---|---|
| miR-103b/SR1 | (((((....(.((..((((((((&))))).))))))..))))).. | 1 |
| miR-103b/new adapter | (((((....(.(((.((..(((.&)))))...))))..))))).. | 13 |
| miR-653/SR1 | ..((.(((((...........&.........)).))).))... | 5 |
| miR-653/new adapter | .......(((...(((.(((.&))))))..))).......... | 13 |
| miR-567/SR1 | ............((((.(((((.&............))))))))) | 5 |
| miR-567/new adapter | .(.(((.....((...((((...&)))).))...))))... | 13 |
| miR-4803/SR1 | ..((((....))))(((((((&))).)).))............ | 1 |
| miR-4803/new adapter | .......(((...(((.(((.&))))))..))).......... | 13 |
| miR-5183/SR1 | .....((((..((.(((....&))).))...))))........ | 13 |
| miR-5183/new adapter | .....((((.....((((((.&))))))..))))......... | 13 |
| miR-495/SR1 | ...((((.(((((..((.....&..))..))))).)).)).... | 5 |
| miR-495/new adapter | ......(((((((((.......&))))).......))))..... | 13 |
| miR-712/SR1 | ...........((((((((..&.....))))))))........ | 5 |
| miR-712/new adapter | ...........(((.(((((.&)))))..)))........... | 13 |
Cofold structure prediction of an miRNA with the SR1 adapter or a specifically designed new adapter are shown in bracket and dot notation, where brackets represent base paired nucleotides and dots represent unpaired nucleotides. The ‘&’ represents the ligation junction between the RNA 3′-end and the adapter 5′-end. The corresponding cofold structure category number is listed as defined in Figure 5A.
Overall, the average ligation efficiency of these miRNAs increased from 2.7% with SR1 to 18.7% with the newly designed adapters (Figure 7 and Supplementary Table S1). The increase in ligation efficiency for miRNAs ranged from 2.2- to 17.1-fold. We interpret these results to indicate that a RNA-adapter pair that is predicted to cofold in an under-represented class is unfavorable for ligation. These data confirm the important role of RNA-adapter cofolding during ligation. We suggest that the method demonstrated here is useful for improving the ligation of any known RNA sequence and could be applied to situations where accurate quantification of a set of known RNAs is important.
Improved miRNA ligation efficiency using 5′-end randomized adapters
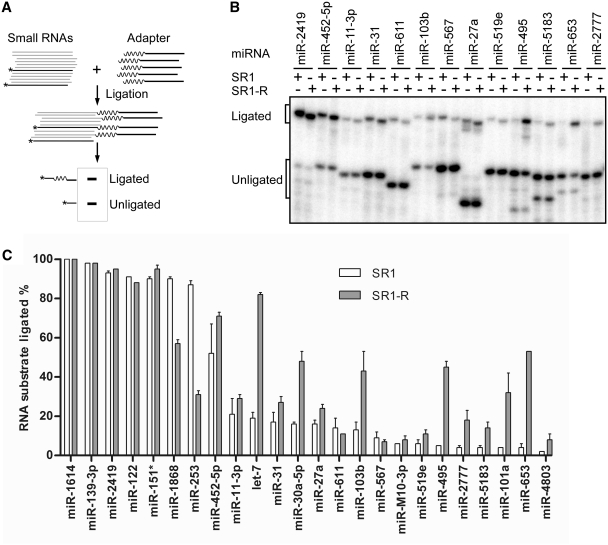
Our results support the hypothesis that cofold structures are a major factor contributing to T4 RNA ligase bias. We therefore attempted to minimize the bias using a pool of adapters with randomized 5′-ends in order to decrease the likelihood that a particular miRNA will be incompetent for ligation with a single sequence adapter. In other words, we attempt to minimize bias by supplying many adapters to increase the likelihood of more favorable cofold structures between adapters and all RNAs in the sample. We designed a pre-adenylated adapter, SR1-R, which contains the same sequence as SR1 except the first six nucleotides at 5′-end are randomized (Supplementary Table S1).
We performed ligation reactions using Rnl2tr to compare the ligation efficiency of each miRNA with SR1 and SR1-R (Figure 8A). miRNAs that ligated efficiently with SR1 were also efficiently ligated with SR1-R, while miRNAs that ligated poorly with SR1 generally showed improved ligation efficiency with SR1-R (Figure 8B). Reflecting this observation, the average ligation efficiency increased from 67% with SR1 to 78% with SR1-R. Furthermore, the index of dispersion, defined as the ratio of the variance to the mean, decreased from 0.13 with SR1 to 0.034 with SR1-R. The decreased index of dispersion indicates less divergence in ligation efficiencies among miRNAs and suggests that using a randomized SR1-R adapter reduces the bias of Rnl2tr in ligation.
While the ligations of pure miRNA substrates demonstrated the potential benefits of using randomized adapters, we further examined whether the same benefits were retained for a particular miRNA when a pool of small RNA substrates is present. In order to test this, we radio-labeled an miRNA of interest at the 5′-end with 32P so that we could use small amounts (0.75 fmol) of the miRNA in a mixture containing small RNAs extracted from mouse embryonic stem (ES) cells (Figure 9A). Ligation reactions were performed using 500-fold excess of mouse ES cell small RNAs and excess amount of adapter compared to radio-labeled miRNA. The ligated and unligated radio-labeled miRNA were separated in 15% TBE–urea gels (Figure 9B). The average ligation efficiency increased from 36% with SR1 to 46% with SR1-R. The ligation efficiency was increased for nine miRNAs, unchanged for 13 miRNAs and somewhat decreased for two miRNAs (Figure 9C). All miRNAs with <40% ligation efficiency with SR1 exhibited improved or unchanged ligation efficiencies when ligated to SR1-R. In agreement with the results of the experiments with pure miRNAs, the ligation bias was reduced as measured by the index of dispersion, which decreased from 0.42 with SR1 to 0.23 with SR1-R. In summary, the use of randomized adapters when ligating adapters to the 3′-end of miRNAs with Rnl2tr results in generally improved ligation efficiency and decreased ligation bias.
Figure 9.
Improvement of miRNA ligation efficiency using a randomized adapter in the presence of mouse ES cell small RNAs. (A) Scheme of ligation reactions in the presence of mouse ES cell small RNAs. Each reaction contained 0.75 fmol of a 5′-32P labeled miRNA mixed with a 500-fold excess of mouse ES cell small RNAs and either the SR1 or the SR1-R adapter. Gray lines represent the ES cell small RNAs and the black line with an asterisk represents the radio-labeled miRNA. The SR1-R adapter is shown in black with a wavy line representing the random region at the 5′-end. Ligation products were resolved on 15% TBE–urea acrylamide gels, exposed to phosphor storage screens, and scanned. The ligated radio-labeled miRNA appears as a higher molecular weight band than unligated miRNA. (B) Representative results of ligation gels as described in A. (C) Comparison of ligation efficiency of miRNAs with the SR1 and SR1-R adapters. The intensity of ligated and unligated bands in each lane was quantified and ligation efficiencies were determined by calculating the percentage of ligated miRNA from the total miRNA. The data are represented as the average ± standard deviation ligation efficiency from two independent experimental replicates.
DISCUSSION
HTS technology has revolutionized miRNA discovery and expression analysis. Compared to traditional gene expression profiling methods such as hybridization based methods, microarrays and quantitative PCR, HTS offers the advantages of high sensitivity, the ability to identify novel miRNAs and provides information about miRNA editing and 3′-end modification simultaneously (21). Despite these advantages, recent studies have revealed the existence of bias in HTS when quantifying the level of miRNA expression directly from sequence reads (24,25).
A recent study using a pool of synthetic miRNAs concluded that inconsistencies in miRNA quantitation in HTS experiments are primarily due to biases in the adapter ligation steps and not due to downstream steps such as reverse transcription, PCR, or the sequencing reaction itself (25). Previous studies examined the bias after complex sample preparation protocols, which reflect a combined bias from two ligation steps using two ligases (24–26). In this study, we used a random mixture of RNA substrates to examine the bias of T4 RNA ligases during 3′-adapter ligation in isolation. Our selection strategy enabled us to include 9 × 1013 randomized RNA sequences in one ligation reaction, which provides complete sequence coverage for all possible RNAs 21 nt in length in contrast to previous studies (24–26).
To study bias in 3′-adapter ligations, it was critical to accurately determine the content of the random input sequences in our ligation reaction. To do so we employed a homopolymer tailing approach. Alternatively, we attempted to assess the nucleotide content of the random pools using a direct reverse transcription method with two different 3′-overhang degenerate nucleotide stem–loop RT primers (44). Hairpin RT primers with either 6 or 10, 3′-overhanging degenerate nucleotides, were designed to hybridize to the 3′-ends of unknown RNAs, and serve as reverse transcription primers. Libraries prepared with the degenerate stem–loop RT primers showed bias for G and C nucleotides in the degenerate priming region (Supplementary Figure S1). We interpreted this to reflect a bias that results from primer annealing, where more stable G·C base pairs were favored over A·T pairs. In addition to being a poor option for assessing the content of a random oligo pool, using a stem–loop primer with a randomized 3′-end for annealing will introduce additional bias when used to quantify miRNAs. In contrast, the poly(A) polymerase tailing method showed no detectable bias (Supplementary Figure S1). For this reason, we used the random library sequences obtained by the poly(A) polymerase tailing method for all subsequent analysis.
Strikingly, we found that T4 RNA ligases show no significant preference for RNA primary sequence, contradicting a previous report (26). Instead, we provided experimental evidence for the important role of RNA and adapter cofold structures that were suggested to be influential in an article published while this manuscript was in preparation (25). What distinguishes our work from these recent studies is that we separated the 3′-ligation step so that we could study its inherent bias in the absence of potentially confounding effects from 5′-ligation that was used by Hafner (25) and Jayaprakash (26). In addition, our expanded analysis of a larger group of possible ligation substrates allowed us to accurately assess primary sequence preference. We were then able to predict, test, and prove that particular cofold structural classes are disfavored for ligation with T4 RNA ligases, while others are neutral or slightly favored. Together, these factors explain why we arrived at different conclusions than Jayaprakash et al. (26). We performed folding analysis based on their results, but are unable to comment on whether their results can be explained based on our finding of structural bias because of what we believe to be confounding effects of 5′-ligation and different ligation conditions. Future definition of the bias in ligation of adapters to the 5′-ends of RNAs, and interpretation of how that bias may have affected the conclusions of both Hafner et al. (25) and Jayaprakash et al. (26) will necessitate further study. The cumulative results of our experiments demonstrate that, for T4 RNA ligases, the adapter and its ability to interact with an RNA substrate has a major influence on ligation efficiency. The concept of redesigning adapters can be practically applied to improve the ligation efficiency of a specific miRNA with known sequence.
In many experimental situations, the starting material to be ligated is a pool of unknown RNAs, or a mixture of RNAs that is so complex that designing an adapter for each RNA is impractical. Our 5′-randomized adapter approach increases the chance that an appropriate adapter for ligation is present for each miRNA. While largely effective, the ligation of some individual miRNAs to 5′-randomized adapters was not improved in the context of excess small RNA as we predicted. A possible explanation is that there may be interference from other small RNAs in the pool that interact with the miRNA and inhibit productive cofolding with adapters. Overall, however, we observed that ligation bias was reduced with randomized adapters.
Recently, methods that include barcoding when preparing samples for HTS have been shown to be efficient and affordable for sequencing multiple samples simultaneously (45). A very recent report showed that barcodes introduced at the ligation step resulted in significant bias on miRNA expression profiles in high-throughput multiplex sequencing (46). The effect of cofold structures on ligation explains the observation of bias. For that reason, introducing barcodes in the 3′-adapter for HTS warrants careful consideration, especially when one tries to compare the relative miRNA expression level from different samples prepared with different barcoded adapters. We therefore suggest introducing barcodes in the reverse transcription or PCR step to avoid introducing ligation bias among samples.
The procedures described here for studying T4 RNA ligase bias should be applicable to other ligases and other ligation conditions, for instance ligation of adapters to unknown RNA 5′-ends. These procedures represent important early steps toward resolving the issue of ligation bias by seeking alternative or modified ligases.
In summary, our findings show that the bias introduced by T4 RNA ligases in HTS experiments is due to structural properties within and between RNA substrates and the adapters used in ligation. Our model of what constitutes a compatible RNA-adapter pair was successfully used to design adapters to improve the ligation of RNAs with a known sequence. The randomized adapter that we designed demonstrated promise toward improving ligation efficiency and reducing bias when ligating a pool of RNAs. This approach may be extended by producing minimized sets of adapters for the study of specific pools of RNAs. For instance, a set of adapters could be designed so that the each member of the miRNA repertoire of an organism would have a corresponding high efficiency adapter included in the mixture. Our approaches should also be applicable to RNAs other than miRNA, including mRNAs fragmented for strand-specific RNA sequencing library preparation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1 and 2.
FUNDING
Funding for open access charge: New England Biolabs Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joanna Bybee, Eileen Dimalanta and Laurie Mazzola for assistance with Ion Torrent sequencing; Brenda Baker for help in DNA adapter adenylation; Alexander Zhelkovsky for discussion of DNA adapter adenylation using Mth ligase; Bo Wu for regents; Richard Roberts, Bill Jack, Sriharsa Pradhan and Larry McReynolds for critical reading of the article.
REFERENCES
- 1.Silber R, Malathi VG, Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl Acad. Sci. USA. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho CK, Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cranston JW, Silber R, Malathi VG, Hurwitz J. Studies on ribonucleic acid ligase. Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J. Biol. Chem. 1974;249:7447–7456. [PubMed] [Google Scholar]
- 4.Sugino A, Snoper TJ, Cozzarelli NR. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 1977;252:1732–1738. [PubMed] [Google Scholar]
- 5.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Gorovsky MA. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE) Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XH, Chiang VL. Single-stranded DNA ligation by T4 RNA ligase for PCR cloning of 5′-noncoding fragments and coding sequence of a specific gene. Nucleic Acids Res. 1996;24:990–991. doi: 10.1093/nar/24.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tessier DC, Brousseau R, Vernet T. Ligation of single-stranded oligodeoxyribonucleotides by T4 RNA ligase. Anal. Biochem. 1986;158:171–178. doi: 10.1016/0003-2697(86)90606-8. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita Y, Nishigaki K, Husimi Y. Fluorescence-, isotope- or biotin-labeling of the 5′-end of single-stranded DNA/RNA using T4 RNA ligase. Nucleic Acids Res. 1997;25:3747–3748. doi: 10.1093/nar/25.18.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 14.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 15.van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim. Biophys. Acta. 2008;1779:668–677. doi: 10.1016/j.bbagrm.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matranga C, Zamore PD. Small silencing RNAs. Curr. Biol. 2007;17:R789–793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 19.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 20.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 21.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoeckius M, Maaskola J, Colombo T, Rahn HP, Friedlander MR, Li N, Chen W, Piano F, Rajewsky N. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat. Methods. 2009;6:745–751. doi: 10.1038/nmeth.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 25.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr693. September 21 (doi:10.1093/nar/gkr693; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 28.Viollet S, Fuchs RT, Munafo DB, Zhuang F, Robb GB. T4 RNA ligase 2 truncated active site mutants: improved tools for RNA analysis. BMC Biotechnol. 2011;11:72. doi: 10.1186/1472-6750-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bindereif A, Schon A, Westhof E. Handbook of RNA Biochemistry: Stduent Edition. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 2009. [Google Scholar]
- 32.Zhelkovsky AM, McReynolds LA. Simple and efficient synthesis of 5' pre-adenylated DNA using thermostable RNA ligase. Nucleic Acids Res. 2011;39:e117. doi: 10.1093/nar/gkr544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19 10 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster Fast folding and comparison of RNA secondary sStructures. Monatshefte f. Chemie. 1994;125:22. [Google Scholar]
- 37.Bernhart SH, Tafer H, Muckstein U, Flamm C, Stadler PF, Hofacker IL. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol. Biol. 2006; 1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 39.Munafo DB, Robb GB. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010;16:2537–2552. doi: 10.1261/rna.2242610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhuang F, Karberg M, Perutka J, Lambowitz AM. EcI5, a group IIB intron with high retrohoming frequency: DNA target site recognition and use in gene targeting. RNA. 2009;15:432–449. doi: 10.1261/rna.1378909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–e98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 43.Tuma RS, Beaudet MP, Jin X, Jones LJ, Cheung CY, Yue S, Singer VL. Characterization of SYBR Gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators. Anal. Biochem. 1999;268:278–288. doi: 10.1006/abio.1998.3067. [DOI] [PubMed] [Google Scholar]
- 44.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33: e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AM, Heisler LE, St Onge RP, Farias-Hesson E, Wallace IM, Bodeau J, Harris AN, Perry KM, Giaever G, Pourmand N, et al. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010;38:e142. doi: 10.1093/nar/gkq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alon S, Vigneault F, Eminaga S, Christodoulou DC, Seidman JG, Church GM, Eisenberg E. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011;21:1506–1511. doi: 10.1101/gr.121715.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.