Abstract
Summary
Although there have been major advances in elucidating the functional biology of the human brain, relatively little is known of its cellular and molecular organization. Here we report a large-scale characterization of the expression of ~1,000 genes important for neural functions, by in situ hybridization with cellular resolution in visual and temporal cortices of adult human brains. These data reveal diverse gene expression patterns and remarkable conservation of each individual gene’s expression among individuals (95%), cortical areas (84%), and between human and mouse (79%). A small but substantial number of genes (21%) exhibited species-differential expression. Distinct molecular signatures, comprised of genes both common between species and unique to each, were identified for each major cortical cell type. The data suggest that gene expression profile changes may contribute to differential cortical function across species, in particular, a shift from corticosubcortical to more predominant corticocortical communications in the human brain.
Introduction
The brain is perhaps the most critical contributor to the uniqueness of human beings (Varki et al., 2008). Individual human brains exhibit variations in their structural and functional organization (Mazziotta et al., 2001; Mori et al., 2005), and there is significant genomic variation across human populations (Durbin et al., 2010; Sudmant et al., 2010). The genetic diversity within the human population, combined with variation in environmental influences, sets the stage for functional diversity across individual human brains.
During primate evolution the brain has expanded greatly both in size and complexity, with the greatest expansion occurring in the human cerebral cortex (Carroll, 2003; Herculano-Houzel, 2009; Rakic, 2009). The cerebral cortex is overall a 6-layered structure whose regional structure is characterized by the differential prominence of these layers and by relative abundance and distribution of many different types of neurons. The expansion of the human cortex is associated with the prominent appearance and expansion of prefrontal, parietal, and temporal association regions that are considered to support the greater cognitive abilities of humans. A large body of evidence has shown that excitatory and inhibitory cortical neurons, across and within cortical layers, can be divided into many different types based on their diverse molecular, morphological, physiological and connectional properties, and together form the complex neural circuits of the cerebral cortex (Ascoli et al., 2008; Douglas and Martin, 2004; Molyneaux et al., 2007; Somogyi and Klausberger, 2005).
Gene expression change has been considered a major driver of species differentiation and evolution (King and Wilson, 1975). However, comparative microarray studies have shown a high degree of conservation across species in overall gene expression, although there is considerable upregulation of brain gene expression in humans compared to non-human primates (Khaitovich et al., 2006; Preuss et al., 2004; Vallender et al., 2008). Microarray studies have also revealed distinct gene expression profiles and coexpression networks associated with different human brain regions, major cell types, and cellular processes (Johnson et al., 2009; Miller et al., 2010; Oldham et al., 2006; Oldham et al., 2008). To date, such gene expression profiling studies have typically been performed on homogenized tissues, unable to discern cellular heterogeneity as well as spatial localization within the brain. From studies in mouse, it is clear that analysis of gene expression with high spatial resolution can identify cell type and other differences in gene expression among brain regions and among mouse strains with different genetic backgrounds (Lein et al., 2007; Morris et al., 2010). It is thus critical to examine gene expression with cellular resolution in the human brain to elucidate its contribution to cell types, neural circuits, and brain function.
To this end, we profiled the expression of approximately 1,000 genes in the human visual cortex and mid temporal cortex at cellular resolution using a standardized in situ hybridization (ISH) platform, and created a public online database (http://human.brain-map.org/ish) for this extensive set of high-resolution human gene expression data. Through detailed analysis of individual genes, we compared gene expression among individuals and cortical areas, as well as between human and mouse using the Allen Mouse Brain Atlas dataset. We found very little qualitative gene expression variation between cortical regions and among individual human subjects. A substantially higher degree of gene expression variation was observed when comparing putatively homologous human and mouse cortical areas. From these, differential gene expression signatures for different cortical cell types were derived. This large dataset provides a rich public resource for the study of specific gene functions and their relation to cortical cell type diversity.
Results
High resolution imaging of gene expression in the human cortex
Expression analysis of 995 genes was done by ISH in multiple postmortem adult brain specimens from two functionally distinct cortical regions: visual cortex (containing Brodmann’s areas 17 and 18), and mid temporal cortex (containing mostly Brodmann’s area 21, with parts of area 22 and sometimes 20), from individuals without any known neuropathology or history of neuropsychiatric illnesses. The visual cortex was chosen as one of the most studied and well conserved cortical regions, allowing comparison of human brain gene expression with analogous data in mouse (Lein et al., 2007). The mid temporal cortex was selected as a second, functionally distinct association region to assess regional difference in gene expression. Each gene was assayed in 2 to 6 brains, with most assayed in 4 brains, split approximately equally between men and women. A total of 46 donor cases (27 men, 19 women) were used (see Table S1 for case details). About 92 genes were profiled per case. The resulting online resource includes approximately 31,000 ISH images from this study.
Four categories of genes of broad scientific and clinical interest were selected: markers of cortical cell types (237 genes), gene families important to neural function, genes related to diseases of the central nervous system (382 genes), and genes identified from comparative genomics (111 genes) (Table S2). There was considerable overlap of genes across these categories. The cortical cell-type marker genes comprised genes identified in the Allen Mouse Brain Atlas (Lein et al., 2007) as having specific or enriched expression in particular cortical layers, as well as known or putative interneuron, glial, and vascular markers. The neural function genes included ~80% of all ion channel genes, genes for nearly all neurotransmitter receptors, and genes for G-protein-coupled receptors (GPCRs), transporters, synaptic proteins, secreted peptide or protein ligands and their receptors, among others. The disease-related genes were identified based on published literature as being relevant to schizophrenia, depression, epilepsy, neurodegenerative diseases, autism spectrum disorders, mental retardation, microcephaly, and other diseases. They include those genes linked to a brain disease, known to be involved in physiological pathways implicated in a disease, or encoding known drug targets. Lastly, the comparative genomics category comprised genes identified in the literature as showing accelerated evolution, being under positive selection, or showing microarray-based gene expression difference either between rodents and primates or between non-human and human primates (Abrahams et al., 2007; Burki and Kaessmann, 2004; Caceres et al., 2003; International HapMap Consortium, 2005; Dorus et al., 2004; Gibbs et al., 2007; Gilbert et al., 2005; Khaitovich et al., 2005; Pollard et al., 2006; Redon et al., 2006). Genes with human lineage specific amplifications compared to non-human primates were also included (Popesco et al., 2006).
Tissue quality validation and cross-sample variation
To control for the inherent tissue quality variation across donors and specimens, several control genes were assayed in every tissue block to benchmark tissue and ISH quality (Figure S1A). Control sections from all tissue blocks were hybridized to probes for GAP43 and CTNND2, a strongly and a moderately expressed gene, respectively. In addition, control sections of visual cortex were hybridized with PCP4 to delineate the boundary of area 17 (primary visual cortex) and area 18 (secondary visual cortex), and control sections of temporal cortex were hybridized with CARTPT based on prior literature (Hurd and Fagergren, 2000).
To establish a metric for evaluating tissue quality for ISH analysis, a modified intensity score (Imod) was calculated for each control gene and each tissue sample (Supplemental Information). The average Imod score for all control genes for each tissue block was used as an index of tissue quality. An extremely low average Imod score for a tissue block almost always correlated with lower expression intensities of many other genes profiled in that block compared to other blocks. There was no direct correlation between postmortem interval (PMI) or RNA integrity number (RIN) and the Imod score (data not shown). Therefore, tissue blocks for which Imod was 2.0 standard deviations (SD) beyond the median Imod value [Imod < 2 (i.e., below -2 SD) or Imod > 4 (i.e., above +2 SD)] were excluded from the study (Figure S1B).
Analysis of the control genes across all tissue blocks revealed differing degrees of expression consistency across different blocks (Figure S1C). GAP43 and PCP4 showed a higher degree of consistency in expression pattern and level, with deviations in only 7% and 21% of tissue samples, respectively. CTNND2 and CARTPT showed less consistency, with deviations in 31% and 41% of samples, respectively. Variation in expression levels of CTNND2 often correlated with Imod of the tissue block, indicating that as a moderate to low expressing gene CTNND2 may be more sensitive to tissue quality variation. Expression of CARTPT appeared to be independent of tissue quality variation.
Differential expression across genes
Analysis of the entire gene set reveals a broad range of expression patterns, consistent with the diverse functions of these genes. To compare expression effectively, and because of the intrinsic variation of postmortem human tissues and limited sample sizes, a rigorous manual scoring system was used to characterize the level (Figure S1D) and spatial pattern (Figure S1E) of expression for each gene (see Table S2 for detailed scoring of each gene). Overall, 79% of the genes were expressed above background. There was little correlation between expression characteristics and gene category. Similar distributions of expression patterns and levels were represented within each gene category (Figure 1A), indicating that related genes could contribute differentially to the functions of different cell populations.
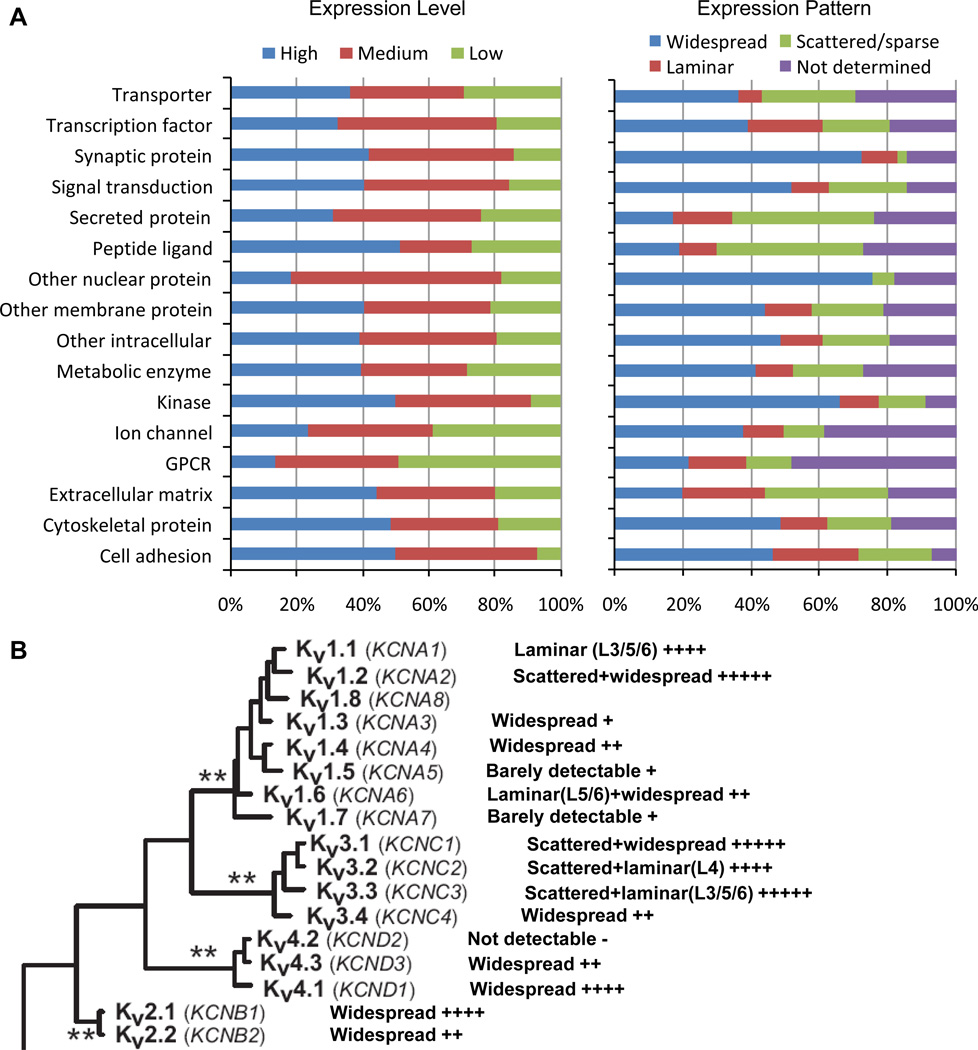
Figure 1.
Range of gene expression profiles. (A) Percentages of genes with different expression levels (high, medium or low) or patterns (widespread, laminar, scattered/sparse, or not determined due to low or no expression) among different gene families. See also Figure S1, Table S1 and S2. (B) Diverse expression levels and patterns of the voltage-gated potassium channel subfamily. The phylogenetic tree (including the asterisks) is adopted from Yu and Catterall (Yu and Catterall, 2004) with permission.
For example, the voltage-gated potassium channels, a 17-gene subfamily of the ion channel gene family, comprises four groups with distinct physiological properties: Kv1, Kv2, Kv3 and Kv4 (Yu and Catterall, 2004) (Figure 1B). Genes within each group exhibited differential expression, suggesting that different cell types have different compositions of potassium channels which could result in different excitability properties. The Kv3-type genes showed predominantly scattered patterns, consistent with preferential expression in interneurons. The Kv1-type genes included those showing interneuron preference (e.g. KCNA2), laminar enrichment in excitatory neurons (e.g. KCNA1 and KCNA6), as well as low or no expression.
Comparing expression across cortical regions
Many genes exhibit differential expression across cortical layers and thus can delineate anatomical boundaries and fine cytoarchitectural differences. In this study, the most prominent boundary is between primary and secondary visual cortices. Area 17 is anatomically characterized in anthropoid primates by a uniquely expanded layer 4 that is further divided into sublayers 4A, 4B, 4Cα and 4Cβ. Over 15% of the genes showed a clear expression pattern change at this boundary (Figure 2A and Table S2), often due to higher or lower expression in layer 4 compared to other layers. For example, DKK3 exhibited a fairly uniform expression across layers 2 through 6, except layer 4, in which markedly lower expression was seen in area 17. PCP4 expression is generally enriched in layers 5 and 6, but is also prominently expressed in layer 4c in area 17. SYT6 has much stronger expression in layer 6 of area 17 than that of area 18. CTNNB1 has prominent expression in layer 4 of area 18 instead of area 17.
Figure 2.
Cortical regional variation of gene expression. (A) Examples of genes defining the regional boundary between areas 17 and 18. For each gene, a Nissl-stained section (upper left panel) is shown alongside an ISH section (lower left panel) with enlarged views of areas 17 and 18 (middle and right panels). Arrowheads point to the visible borders between areas 17 and 18. (B) Examples of genes with differential expression in areas 17 and 18. TLE4 has denser expression in layer 6 of area 17 than 18. Expression of SST is largely absent in layer 4 of area 17 but not 18. (C) Examples of genes with differential expression in areas 18 and 21. GRIN3A has more enriched expression in layer 5 of area 21 but not 18. Expression of SCN3B is lower in layer 4 of area 18 but not 21. (D) Examples of genes with differential expression among areas 17, 18 and 21. Cortical layers are labeled on the right side of the figure.
In the regional comparison among areas 17, 18, and 21, 84% of the genes exhibited consistent expression patterns across the three areas. For the 16% of genes showing an expression pattern difference between any two of the three areas (Table S2), most differences could be attributed to the unique cytoarchitecture of area 17 (Figure 2A, 2B). Gene expression patterns were similar between areas 18 and 21 for the vast majority of the genes, with only subtle cytoarchitectural difference. Only 4% of genes exhibited expression pattern difference between areas 18 and 21 (Table S2), for example GRIN3A and SCN3B (Figure 2C), with a very small subset showing differential expression in all three regions, such as PENK, KCNC3 and SYT2 (Figure 2D).
Comparing expression across individuals
Individual donor comparison to assess expression difference for each gene is confounded by tissue quality variation, especially for expression level. To assess whether an expression level difference reflected tissue integrity or a donor difference, a Pearson rank correlation was performed on genes showing significant difference in intensity level (Supplemental Information). Only genes with level variations showing no significant correlation with tissue integrity were considered as having donor difference.
Overall, 46 genes (5%) were determined to show donor variation in expression pattern or level (Table S2 and Supplemental Information). For example, RELN, which shows layer 1-specific expression, was also detected in layer 4C cells in some cases; DISC1 exhibited robust expression in scattered layers 1 and 2 cells in a subset of cases; and VAMP1 had variable expression levels across different cases (Figure 3). The donor difference for RELN was validated by RT-qPCR on laser capture microdissected layer 4C tissues from the 3 donors shown in Figure 3 (ΔCp values between RELN and GAPDH control are 8.3 ± 0.3 for Donor 1, 9.2 ± 0.2 for Donor 2, and 9.1 ± 0.2 for Donor 3, means ± SD; p < 0.01 for both Donors 1–2 and 1–3 comparisons, Student’s t-test).
Figure 3.
Individual variation of gene expression. Three gene examples are shown, each with sections from 3 different donor specimens. Numbers shown next to each donor are Imod values for each tissue block. For VAMP1 both areas 17 and 18 are shown, whereas for RELN and DISC1 only area 17 is shown. Arrow points to the expression of RELN in layer 4C of just one donor. Cortical layers are labeled on the edges of the figure.
Comparing expression between species
To explore the degree of conservation between human and a widely used animal model system, the mouse, a systematic comparison of gene expression between human and mouse visual cortex was performed on 941 genes with orthologs and expression data in both species. Due to the inevitable tissue quality difference between the two species, and the demonstrated impact of tissue quality on expression intensity, only particularly striking expression level differences (e.g., + versus +++ or more) were called out. The comparison included both primary and secondary cortical areas (i.e., both areas 17 and 18 in human), and any significant difference in either area was recorded.
Overall, gene expression showed a high degree of concordance between mouse and human visual cortices, with only 21% of the genes (199 genes) showing moderate or marked difference in either expression patterns or levels (Table S2). A wide range of pattern differences was observed, often restricted to particular anatomical domains or cell populations (Figure 4A). Examples include increased expression in human in specific layers (e.g., ANXA1, NNAT) or overall (e.g., BCL6); decreased expression density (e.g., NPY); decreased expression in specific layers (e.g., CALB1, GRIK1, RELN); shifted localization from one layer to another (e.g., CRYM, SYT2) or from laminar to scattered (e.g., HCN1, PDYN), etc. As an extreme example, CARTPT expression was variable in all 3 aspects – among regions (human areas 17/18 versus 21, or mouse visual versus somatosensory cortices), individuals, and species (Figure 4B).
Figure 4.
Gene expression difference between human and mouse. (A) Examples of genes with a variety of differential expression patterns between human and mouse visual cortices. Human area 17 is shown for ANXA1, BCL6, GRIK1, HCN1, NNAT, PDYN, and RELN. Human area 18 is shown for CALB1, CRYM, and NPY. Mouse areas are corresponding to each gene’s human area shown. Corresponding layers of human and mouse are marked on left. (B) The CARTPT gene has different expression patterns both across species and among different cortical regions within a species. M S1, mouse primary somatosensory cortex. M V1, mouse primary visual cortex. M TEa, mouse temporal association cortex. H 17, H 18, H 21, human cortical areas 17, 18, and 21. Cortical layers are labeled on the left for mouse and right for human. (C) Percent of genes in each gene category exhibiting human-mouse difference in expression (level or pattern combined) in visual cortex. The number of genes showing expression difference and the total number of genes in that category are shown in parentheses above each bar. See also Figure S2. (D) Cross-species comparison in both visual (vis) and temporal (te) cortices. The pie chart shows the number and percentage of genes (out of 611 total) with human-mouse expression difference (level or pattern combined) in either visual or temporal comparisons.
The proportion of genes with similar or variable cross-species expression was generally consistent across different gene categories (Figure 4C). For example, the disease genes (354) and comparative genomics genes (95) showed similarly low cross-species variability, with only 13% and 23% exhibiting expression difference, respectively. In marked contrast, the cell-type marker gene category exhibited significantly more cross-species difference. Approximately 50% of the 237 genes identified as cortical markers in the Allen Mouse Brain Atlas expressed differently in human. Among these, 59% of the laminar markers and 41% of the putative interneuron markers from mouse showed expression difference in human, while glial and vascular marker expression was relatively conserved. With additional genes dropping out due to weak expression in mouse and undetectable in human (although not scored as “different”) as well as 25 newly identified human marker genes, consequently a total of 165 genes were designated as human cortical markers (Table S2), and among these 38% had human-mouse difference in expression pattern or level.
Looking at functional classes of genes, which overlap with the cortical markers, several showed high cross-species expression variation (Figure 4C). These include secreted protein (48%), extracellular matrix (50%), cell adhesion (36%), and peptide ligand (31%) genes, which are interestingly all involved in intercellular communication. Overall, cortical marker genes accounted for 68% of the total 199 genes having cross-species difference. Of the remaining genes with difference, some also have non-uniform expression patterns. Therefore, the small proportion of genes exhibiting differential expression in the visual cortex between human and mouse contains predominantly genes showing unique and non-widespread expression patterns.
We examined a group of genes hypothesized to be under positive selection or human lineage-specific (HLS) genes that have increased copy number along the human lineage (Popesco et al., 2006; Sikela, 2006). Nearly all of these genes, including those only present in human, were found to be expressed in human cortex, mostly in widespread pattern with variable expression levels (Figure S2 and Table S2). Among the genes present in both species, substantially more genes showed increased expression in human than decreased expression (number of genes 16:4, see Figure S2B for examples). These results indicate that the HLS genes may have more significant roles in the function of human brain.
To extend the human-mouse comparison to a multimodal association area, we compared human area 21 with the caudal part of mouse temporal association cortex (TEa), likely to be the closest homolog of area 21. Because TEa can only be unambiguously identified in mouse coronal sections, only 611 genes that have mouse coronal data in Allen Mouse Brain Atlas were included in this analysis. Gene expression again showed a high degree of concordance between the two temporal cortical areas, similar to the visual areas (Table S2). Among the 611 genes, expression difference was seen in 159 genes (26%) between human and mouse visual cortices and in 149 genes (24%) between human and mouse temporal cortices. The majority (126) of these genes showed expression difference in both types of comparison, with only a small number of genes showing unique difference in one cortical area (5% visual area only, 4% temporal area only) (Figure 4D). In addition, similar to the above cortical region comparison in human, a small degree of expression difference was observed between mouse TEa and primary visual cortex (V1) (6%), as well as between TEa and the adjacent mouse auditory cortex A1/A2 (3%). These results suggest that primary and association cortical areas have a similar degree of cross-species gene expression differences.
Distinct molecular signatures of cortical cell types
Despite the relatively large proportion of genes showing cross-species expression difference at either levels or patterns, many of the marker genes labeled similar cell types in mouse and human, supporting their functional conservation (Figure 5 and Figure S3). These include 56 laminar markers, 61 putative interneuron markers, 12 putative astrocyte markers, 10 oligodendrocyte markers, and 3 vascular markers (Table S2). (It should be noted that “putative” means the identities of the labeled cells cannot be ascertained purely by the scattered ISH patterns. We cross-referenced previous studies (Cahoy et al., 2008) to assign some of the new interneuron or astrocyte marker genes.) These conserved markers offer useful tools for labeling and studying specific cell populations.
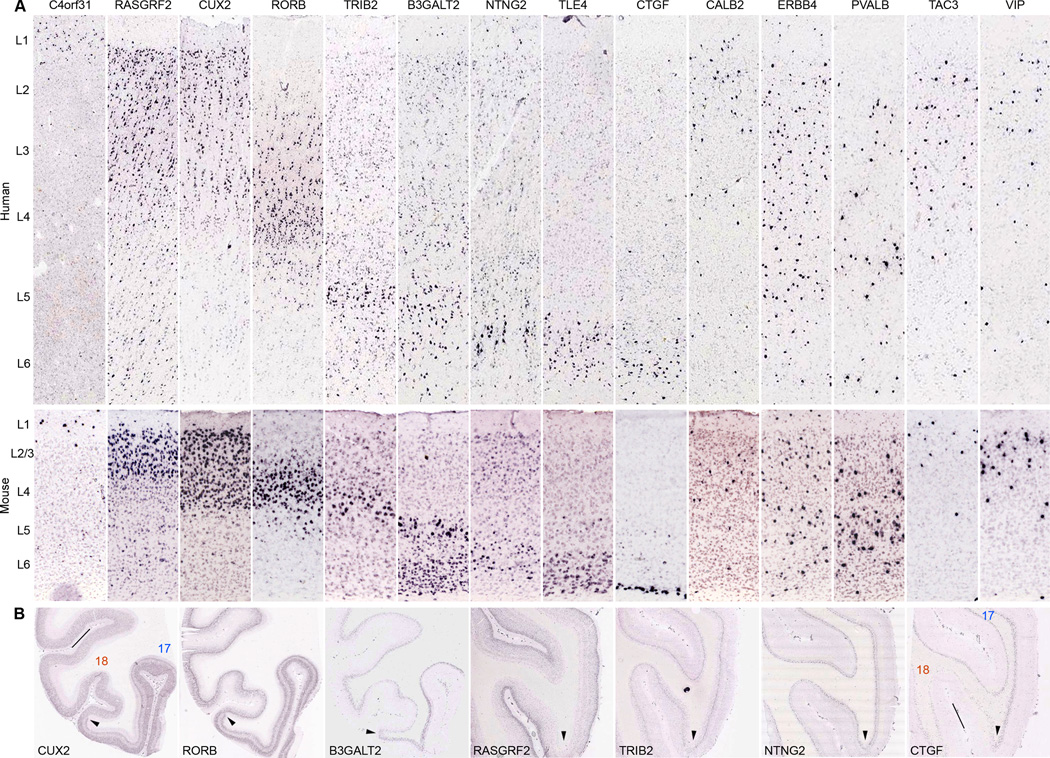
Figure 5.
Cell type marker genes conserved between human and mouse. (A) Examples of genes with specific or enriched expression in different cortical layers or different interneuron populations. Corresponding layers of human and mouse are marked on left. Enriched expression patterns: C4orf31, layer 1; RASGRF2, layers 2/3; CUX2, layers 2/3/4; RORB, layer 4; TRIB2, layer 5A; B3GALT2, layers 5/6; NTNG2, layer 6; TLE4, layer 6; CTGF, layer 6B. Putative interneurons: CALB2, ERBB4, PVALB, TAC3, VIP. See also Figure S3. (B) Laminar delineation seen in human visual cortical sections under low magnification. For direct comparisons, sections for CUX2, RORB and B3GALT2 were from a common tissue block, and those for RASGRF2, TRIB2, NTNG2 and CTGF were from another tissue block. Arrowheads point to the borders between areas 17 and 18. Lines indicate pial surface within the sulci.
On the other hand, a similarly large number of genes were found to label specific cell types uniquely in one species (Table S2). The combination of common and unique marker genes thus provides a distinct molecular signature for each cell type in each species. Below we present a detailed analysis of gene expression in different cortical neuronal cell types.
Layer-specific excitatory neuronal cell types
Cortical excitatory neurons in different layers are believed to have differential input/output connections and play distinct roles in the cortical circuits (Douglas and Martin, 2004). Layer 4 neurons are the main targets of thalamocortical inputs. Layer 5 and 6 neurons mainly send output projections to various subcortical and contralateral regions. Layer 2 and 3 neurons mainly mediate intracortical connections both locally and distantly. Layers 2 and 3 in human and other primates are greatly expanded compared to mouse, suggesting the occurrence of a substantially increased intracortical connectivity in human (Marin-Padilla, 1992). As shown in Figure 6, clustering of layer-specific marker genes in human and mouse visual cortex demonstrates that each layer can be identified by a set of common genes and a set of species-unique genes. In addition, two prominent features emerged. First, a large set of genes with specific expression in mouse layer 5 (36 genes, 51% of all mouse layer 5 expressing markers, some shown by the orange bars in Figure 6) have greatly diminished or no specific expression in human layer 5 (see examples in Figure 7A, as well as BCL6, HCN1 and RELN in Figure 4). Second, another set of genes (indicated by the green bar in Figure 6, as well as several non-marker genes such as KCNC3, NEFH and VAMP1) have greatly enhanced expression in the large pyramidal neurons in human layer 3 (mainly in association areas 18 and 21, see examples in Figure 7B, as well as KCNC3, VAMP1 and CRYM in Figures 2–4). Interestingly, some of these genes (linked by green lines in Figure 6) exhibited a shift of expression from preferentially layer 5 in mouse to preferentially layer 3 in human (see NEFH, SCN4B and SYT2 in Figure 7B). Being mostly ion channels (e.g., SCN4B and KCNC3), neurofilament (e.g., NEFH), adhesion (e.g., MFGE8), secreted (e.g., LGALS1), and synaptic (e.g., SYT2 and VAMP1) genes, we hypothesize that these genes label and support a unique set of human layer 3 pyramidal neurons that may have human (or primate) specific long-range intracortical projections.
Figure 6.
Species-differential molecular signatures of cortical layers. Mouse (left panel) and human (right panel) marker genes with selective expression in specific cortical layer(s) are clustered based on layers. Layers are marked at the top of each panel. WM, white matter. Layer(s) with predominant or secondary expression for each gene is labeled in dark or light blue, respectively. Names of the marker genes appearing in both mouse and human panels are shown in black, and names of those unique to mouse or human are shown in red. For each layer, the common set of mouse and human genes labeling the same layer are linked by black lines. The genes labeling different layers between mouse and human are linked by red lines, except that the genes with a switch from predominantly layer 5 or layers 5–6 expression in mouse to predominantly layer 3 or layers 2–3 expression in human are linked by green lines. The cluster of human genes showing a human-specific layer 3 expression pattern is indicated by a green vertical bar to the right of the human panel. The clusters of mouse genes showing mouse-specific layer 5 expression are indicated by the orange vertical bars to the left of the mouse panel. See also Figure S4.
Figure 7.
Downregulation of layer 5 and upregulation of layer 3 gene expression in the human cortex. (A) Representative genes with layer 5 specific or enriched expression in mouse but not human visual cortex. Expression changes include absence of layer 5 (or 5/6) expression in human (C20orf103, DEPDC6, MYL4), change from layer 5 enriched in mouse to scattered or widespread in human (NRIP3, PARM1), and much sparser layer 5 expression in human (VAT1L). (B) Representative genes with enhanced layer 3 expression in human associational cortex. The sparse (SNCG) or layer 5 enriched expression (NEFH, SCN4B, SYT2) in mouse area V2 (left panels) is in striking contrast with the specific (SCN4B, SYT2) or prominent (NEFH, SNCG) expression in layer 3 of human area 18 (middle panels, enlarged view; right panels, lower magnification showing the continuous band of layer 3 expression throughout the area). Corresponding layers of human (H) and mouse (M) are marked on left.
Cortical interneurons
Different types of cortical interneurons are traditionally marked by a set of calcium binding proteins (e.g., PVALB, CALB1, CALB2), neuropeptides (e.g., CCK, CORT, CRH, NPY, PDYN, PNOC, SST, TAC1, TAC3, VIP) or other genes (e.g., NOS1, RELN, TH), which are partially correlated with the morphological, connectional and physiological properties of interneuron subtypes in rodent and primate studies (Ascoli et al., 2008; Markram et al., 2004; Zaitsev et al., 2009). Genes involved in GABA synthesis or transport are also pan-interneuron markers (e.g., GAD1, GAD2, SLC32A1, SLC6A1). Nearly all of these genes (except CCK) were found to have interneuron-specific expressions which were well-conserved between mouse and human (with the exception of CALB1, CORT, CRH, NPY, PDYN). Even the laminar pattern exhibited by some genes was conserved: CALB2, TAC3, and VIP were enriched in superficial layers in both mouse and human (Figure 5A), and TH labeled a sparse population of cells in layer 6 of both mouse and human. Of the differentially expressed genes, CORT, NPY, and PDYN all showed a dramatic reduction in the number and density of labeled cells in human (see Figure 4A for NPY and PDYN). In addition to these well-known interneuron markers, a number of new putative interneuron marker genes as well as genes with enriched expression in interneurons were identified, again some are common between human and mouse and others are differentially expressed (Table S2). These include subtypes of potassium channels (e.g., HCN1, KCNC1, KCNC2, KCNC3, KCNAB1) and glutamate receptors (e.g., GRIK1, GRIN3A), which were hypothesized to define electrophysiological properties of different interneuron types (Markram et al., 2004).
Cortical subplate and white matter neurons
There are generally two types of subplate neurons in the adult cortex. The first is the so-called layer 6B neurons, which are mostly excitatory (Ayoub and Kostovic, 2009; Hoerder-Suabedissen et al., 2009). Layer 6B is present in both human and mouse cortex and was found to be labeled by a common set of genes (i.e., ADRA2A, CTGF, NR4A2, Figure 5A and Figure 6). In addition, another set of genes were found to be expressed specifically in mouse layer 6B only (Figure 6). The second type is only present in human white matter (WM) and is known as interstitial neurons (Suarez-Sola et al., 2009). These neurons are mostly GABAergic but also heterogeneous. Indeed, we found that all the above-mentioned interneuron marker genes were sparsely expressed in WM with variable densities. NPY, NOS1, SST, as well as pan-interneuron genes SLC6A1 and SLC6A12 had more prominent expression in WM than other interneuron markers (Figure S4). In particular, NPY-positive and NOS1-positive cells were more strongly expressed in WM than in cortical layers. On the other hand, we also found very significant expression of the glutamatergic neuronal marker SLC17A7 as well as the layer 6B marker CTGF in WM (Figure S4). These findings indicate the heterogeneous nature of WM interstitial neurons, and the identified genes could help to differentiate potential subtypes.
Discussion
A variety of expression patterns is seen among the genes, ranging from highly localized or very sparse to widespread and dense. Despite the expression pattern heterogeneity among the genes, each gene’s expression pattern is remarkably well-conserved across the two cortical regions, among different individual subjects, and between human and mouse, for the majority of genes. This finding is consistent with previous microarray gene expression profiling studies (Caceres et al., 2003; Gu and Gu, 2003; Johnson et al., 2009; Miller et al., 2010; Oldham et al., 2008), extending the conclusion further by providing cellular level details. However, it should be noted that this study provides qualitative, not quantitative, assessment, due to the small number of specimens used for each gene and the difficulty in quantifying anatomical data in such a large, anatomically irregular dataset. Therefore, small, quantitative changes could have been missed in the current analysis, although the overall trends strongly favor consistency. Importantly, the majority of the genes carrying out essential neural functions and the genes implicated in brain diseases have well-conserved expression in all aspects of comparison. Also, the identification of genes with unique expression patterns that are conserved in mouse and human, especially the cortical cell-type markers, provides a form of validation for the functional relevance of these marker-defined cell types, and offers a means to track these cell types and investigate variations in different cortical regions or under various mutant or disease conditions.
A small percentage of genes did show substantial expression difference among human individuals (5%) or between human and mouse (21%). Considering that only two cortical regions were analyzed, extrapolating to the whole brain, it is possible that a larger portion of genes would exhibit localized expression difference somewhere in the brain. Interestingly, most of the genes showing cross-species expression difference in this study have distinct, non-widespread expression patterns, suggesting that genes localized to discrete cell populations also tend to be more susceptible to changes in gene expression control or regulation. This could indicate a lesser selection pressure on these genes, or that distinct, subtle changes may be opted for in species divergence than global changes.
We investigated in detail gene expression patterns in all major cortical neuronal cell types. A large panel of genes was found to have selective expression in one or more cell types. These include both designated cell type “markers” as well as other genes with more complex expression patterns but nonetheless displaying preference for certain cell types. Each neuronal cell type can be identified by a unique set of genes, composed of those with similar expression patterns in human and mouse and those with species-specific patterns. This suggests that the same cell types can have both similar and distinct properties or functions between human and mouse, and the distinct gene profiles can provide an informative means to elucidate such similarity or difference.
Specifically, the WM interstitial neurons are a primate-specific population and have been implicated in psychiatric diseases such as schizophrenia (Suárez-Solá et al., 2009), although their function remains poorly understood. Here we show that the WM neurons are composed of glutamatergic, GABAergic and peptidergic neurons and express a diversity of genes. Further investigation of these genes could help to identify different subtypes and lead to understanding of their properties.
The GABAergic interneurons within the cortical layers have huge diversity in structure and function in both mouse and human (Ascoli et al., 2008; Markram et al., 2004). Again we identified well-known as well as novel interneuron markers and other genes with common or differential expression patterns in both species. Furthermore, a major cross-species distinction during development is that, whereas rodent cortical interneurons are almost exclusively generated in the ganglionic eminence (GE) and migrate into cortex, primate cortical interneurons are generated both in the GE at earlier stage and in the ventricular and subventricular zones (VZ and SVZ) at later stage (Petanjek et al., 2009). In human, GE-originated interneurons preferentially express NPY, NOS1, and SST, whereas VZ/SVZ-originated interneurons preferentially express CALB2. This correlates well with our finding that NPY, NOS1, and SST have an enriched presence in the WM, and CALB2 is enriched in superficial layers. Further, although our analysis is not quantitative, higher-density expression of CALB2 can be seen in human cortex, especially area 21, pointing to potentially increased function of CALB2-positive interneurons (e.g., the double bouquet cells) in human.
A surprising finding is the dramatic shift of cortical layer-specific gene expression patterns. More than half of the genes with robust layer 5-specific expression patterns in mouse have diminished or no specific expression in human layer 5 neurons. This may serve as an example that one should be cautious in translating mouse studies of cell types into human situations. At the same time, increased expression in superficial layers, especially in the large layer 3 pyramidal neurons throughout areas 18 and 21, is seen in a set of genes that include major neurofilament, synaptic and ion channel genes. This result is consistent with the notion that, with the great expansion of cortical surface and the thickening of layers 2–3 in the human cortex, intracortical connections linking distant cortical areas may be critically contributing to the more advanced cortical function in human. And it suggests that these large layer 3 neurons may be a main neuronal type mediating such corticocortical connections, consistent with tracing studies in monkeys (Hof et al., 1995).
In conclusion, a large number of genes related to neural functions were profiled in an unprecedented scale with cellular resolution in two cortical regions of human brains, revealing a wide range of distinct expression patterns in different cell types or populations. Each gene’s unique expression pattern was remarkably conserved across individual human brains, supporting the notion that despite substantial structural and functional variability among individuals, the basic cellular composition and gene expression profiles of the human brain are fundamentally similar, variations being likely reflected in more subtle ways. This large dataset also enabled a detailed comparison of gene expression between human and mouse cerebral cortex, revealing cross-species conservation and divergence of gene expression at anatomical and cell type levels. These results support the use of mouse as a good model system for the understanding of human brain function, while pointing out important differences in the cellular organization between mouse and human brains and the differential functions individual genes may play in each species.
Experimental Procedures
Tissue processing and in situ gene expression profiling
The process, equipment, and workflow for generation of gene expression data in human cortex closely follows that described for generation of the Allen Mouse Brain Atlas (Lein et al., 2007) with some adaptation to manage specific challenges posed by working with human tissue (see “In situ hybridization in the Allen Human Brain Atlas” white paper as well as the Supplemental Experimental Procedures accompanying this paper for detailed methodology).
Postmortem human tissue
Frozen postmortem tissue samples from adult men and women subjects at least 20 years of age were provided by the brain tissue collection of the Section on Neuropathology of the Clinical Disorders Branch, GCAP, IRP, National Institute of Mental Health, NIH, Bethesda, MD. Tissue was also obtained from the University of Miami Brain Endowment Bank, University of Miami Miller School of Medicine, Miami, FL. Specimens from the NIMH collection were processed and characterized as previously described (Lipska et al., 2006). Subjects selected for this study had normal neuropathological examination results and no known history of neuropsychiatric disease. Cases with evidence of drug use, drug overdose, or poisoning, or with suicide as cause of death were excluded.
Probe design and synthesis
Digoxigenin-labeled riboprobes were designed and synthesized following previously described methods (Lein et al., 2007) with some modification. Briefly, probes were designed to be between 400–1000 bp in length (optimally > 600 bp) and to contain no more than 200 bp with > 90% homology to non-target transcripts. In addition, to allow comparability of mouse and human gene expression datasets, each human probe was designed to have > 50% overlap with the existing Allen Mouse Brain Atlas probe when the mouse and human genes were orthologous.
Laser capture microdissection and RT-qPCR
After tissue was validated, 20 µm tissue sections were collected onto PEN (polyethylene napththalate) membrane slides for laser capture microdissection (LCM) or standard glass slides for ISH and thionin-based Nissl staining, respectively. LCM was performed using a Leica LMD6000 system on predefined layer 4C of area 17 for RNA isolation. Equal amount (100 ng) of total RNA was used to make cDNA. Real-time qPCR was conducted with a gene-specific primer pair as well as a positive control primer pair of GAPDH, using the Roche LightCycler 480 system and SYBR Green PCR Master Mix (Roche). Difference in number of cycles needed to reach a threshold level of fluorescence with gene-specific primers as compared with GAPDH primers (ΔCp) was used as measure of relative mRNA abundance.
Highlight.
First online database profiling ~1000 genes at cellular resolution in human cortex
Diverse expression patterns delineate regional boundaries and distinct cell types
While 5% genes vary among human individuals, 21% differ between human and mouse
Species-specific molecular signatures for major cortical cell types are identified
Supplementary Material
Acknowledgements
We are grateful for the technical support and expertise of the Atlas Production Team, led by Paul Wohnoutka, and the Information Technology Team, led by Chinh Dang, at the Allen Institute, without which the work would have not been accomplished. We thank Drs. Deborah Mash and Margaret Basile of the University of Miami Brain Endowment Bank for providing tissue. This work was funded by the Allen Institute for Brain Science. The authors wish to thank the Allen Institute founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interests.
References
- Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl. Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, Fairen A, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub AE, Kostovic I. New horizons for the subplate zone and its pioneering neurons. Cereb. Cortex. 2009;19:1705–1707. doi: 10.1093/cercor/bhp025. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat. Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA. Neuronal circuits of the neocortex. Annu. Rev. Neurosci. 2004;27:419–451. doi: 10.1146/annurev.neuro.27.070203.144152. [DOI] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gilbert SL, Dobyns WB, Lahn BT. Genetic links between brain development and brain evolution. Nat. Rev. Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- Gu J, Gu X. Induced gene expression in human brain after the split from chimpanzee. Trends Genet. 2003;19:63–65. doi: 10.1016/s0168-9525(02)00040-9. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Wang WZ, Lee S, Davies KE, Goffinet AM, Rakic S, Parnavelas J, Reim K, Nicolic M, Paulsen O, et al. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb. Cortex. 2009;19:1738–1750. doi: 10.1093/cercor/bhn195. [DOI] [PubMed] [Google Scholar]
- Hof PR, Nimchinsky EA, Morrison JH. Neurochemical phenotype of corticocortical connections in the macaque monkey: quantitative analysis of a subset of neurofilament protein-immunoreactive projection neurons in frontal, parietal, temporal, and cingulate cortices. J. Comp. Neurol. 1995;362:109–133. doi: 10.1002/cne.903620107. [DOI] [PubMed] [Google Scholar]
- Hurd YL, Fagergren P. Human cocaine- and amphetamine-regulated transcript (CART) mRNA is highly expressed in limbic- and sensory-related brain regions. J. Comp. Neurol. 2000;425:583–598. doi: 10.1002/1096-9861(20001002)425:4<583::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol. Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M. Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. J. Comp. Neurol. 1992;321:223–240. doi: 10.1002/cne.903210205. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, et al. A four-dimensional probabilistic atlas of the human brain. J. Am. Med. Inform. Assoc. 2001;8:401–430. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mori S, Wakana S, Nagae-Poetscher LM, van Zijl PC. MRI Atlas of Human White Matter. Amsterdam, The Netherlands: Elsevier; 2005. [Google Scholar]
- Morris JA, Royall JJ, Bertagnolli D, Boe AF, Burnell JJ, Byrnes EJ, Copeland C, Desta T, Fischer SR, Goldy J, et al. Divergent and nonuniform gene expression patterns in mouse brain. Proc. Natl. Acad. Sci. USA. 2010;107:19049–19054. doi: 10.1073/pnas.1003732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Kostovic I, Esclapez M. Primate-specific origins and migration of cortical GABAergic neurons. Front. Neuroanat. 2009;3:26. doi: 10.3389/neuro.05.026.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Caceres M, Oldham MC, Geschwind DH. Human brain evolution: insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela JM. The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS Genet. 2006;2:e80. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J. Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Sola ML, Gonzalez-Delgado FJ, Pueyo-Morlans M, Medina-Bolivar OC, Hernandez-Acosta NC, Gonzalez-Gomez M, Meyer G. Neurons in the white matter of the adult human neocortex. Front. Neuroanat. 2009;3:7. doi: 10.3389/neuro.05.007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Mekel-Bobrov N, Lahn BT. Genetic basis of human brain evolution. Trends Neurosci. 2008;31:637–644. doi: 10.1016/j.tins.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat. Rev. Genet. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci. STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- Zaitsev AV, Povysheva NV, Gonzalez-Burgos G, Rotaru D, Fish KN, Krimer LS, Lewis DA. Interneuron diversity in layers 2–3 of monkey prefrontal cortex. Cereb. Cortex. 2009;19:1597–1615. doi: 10.1093/cercor/bhn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.