Abstract
Hepatic stellate cell (HSC) activation is a pivotal event in initiation and progression of hepatic fibrosis and a major contributor to collagen deposition driven by transforming growth factor beta (TGFβ). microRNAs (miRs), small non-coding RNAs modulating mRNA and protein expression, have emerged as key regulatory molecules in chronic liver disease. We investigated differentially expressed miRs in quiescent and activated HSCs to identify novel regulators of profibrotic TGFβ signaling. miR microarray analysis was performed on quiescent and activated rat HSCs. Members of the miR-17-92 cluster (19a, 19b, 92a) were significantly down-regulated in activated HSCs. Since miR 19b showed the highest fold-change of the cluster members, activated HSCs were transfected with miR 19b mimic or negative control and TGFβ signaling and HSC activation assessed. miR 19b expression was determined in fibrotic rat and human liver specimens. miR 19b mimic negatively regulated TGFβ signaling components demonstrated by decreased TGFβ receptor II (TGFβRII) and SMAD3 expression. Computational prediction of miR 19b binding to the 3’UTR of TGFβRII was validated by luciferase reporter assay. Inhibition of TGFβ signaling by miR 19b was confirmed by decreased expression of type I collagen and by blocking TGFβ-induced expression of α1(I) and α2(I) procollagen mRNAs. miR 19b blunted the activated HSC phenotype by morphological assessment and decreased αSMA expression. Additionally, miR 19b expression was markedly diminished in fibrotic rat liver compared to normal liver; similarly, miR 19b expression was markedly down-regulated in fibrotic compared to normal human livers.
CONCLUSIONS
miR 19b is a novel regulator of TGFβ signaling in HSCs suggesting a potential therapeutic approach for hepatic fibrosis.
Keywords: Transforming growth factor β, fibrosis, miR 19b, biomarker, hepatic stellate cell
Fibrosis of the liver is characterized by excessive deposition of extracellular matrix (ECM) components, predominantly type I collagen. Disproportionate deposition of fibrillar collagens disrupts normal liver architecture and hepatic function, and if left untreated, progresses to cirrhosis. Cytokine signaling predominates during fibrogenesis initiating activation of resident immune and hepatic stellate cells (HSCs) promoting wound repair(1). Activated HSCs are the principal cell type promoting synthesis and deposition of ECM proteins in response to increased levels of circulating inflammatory signals derived from damaged parenchymal cells. These resident vitamin A storing cells are found within the perisinusoidal space of Disse in a quiescent state, but upon hepatic injury, HSCs transdifferentiate into myofibroblast-like cells marked by expression of smooth muscle-α actin (αSMA)(2), loss of retinyl ester stores and neural marker glial fibrillary acidic protein (GFAP)(2-3), and increased proliferation and contractility. Myofibroblastic HSCs respond to and secrete a variety of profibrogenic cytokines including connective tissue growth factor, tissue inhibitor of metalloproteinases and transforming growth factor-β (TGFβ). Of these, TGFβ is recognized as the most potent fibrogenic cytokine regulating HSC collagen production via autocrine and paracrine signaling(4). TGFβ signal transduction plays a critical role in establishment of the myofibroblast phenotype, as it directly up-regulates hallmarks of HSC activation propelling the disease state forward(5-7). Inhibition of TGFβ receptors (soluble TGFβRII and knockout models) and/or signaling components decreases HSC activation and dramatically blunts chronic hepatic wound-healing in experimental animal models(8-9). In addition to induction of TGFβ signaling, numerous morphological and gene expression profile changes are acquired during transdifferentiation(10).
microRNAs (miRNAs, miRs) are small non-coding RNAs which negatively regulate target gene expression through base pairing with 3’UTRs inducing mRNA cleavage or translational repression. With multiple and diverse targets, miRNAs exert control over key cellular developmental processes including differentiation and proliferation. Specific contribution of select miRNAs in hepatic disease development and progression has been described(11). Recent studies report the process of HSC transdifferentiation is governed by differential miRNA expression. Specifically, down-regulation of miRNAs that control fat accumulation and adipocyte programming and up-regulaton of miRNAs that promote sustained activation of the cell concurrent with increased proliferation and suppression of apoptotic responses are observed(12-14). Forced expression of miRs 150 and 194 in activated HSCs resulted in suppression of the fibrotic phenotype and inhibition of ECM production through downstream regulators of collagen expression(15). Additional studies by Ogawa et al. reported direct regulation of collagen synthesis via binding of miR 29b to the 3’UTR of collagen and transcriptional regulator SP1 in a human HSC line(16). While the field continues to advance, studies to date have lacked accurate miRNA profiling of the divergent HSC phenotypes in primary cells. Additionally, no studies have identified any miRs that have a global effect on profibrotic TGFβ signaling in the liver which could be more efficient than targeting a single gene.
Herein, we report a set of differentially expressed miRNAs in quiescent (freshly isolated) vs activated HSCs, among which, miRNA 19b (miR 19b) directly inhibited fibrotic TGFβ signaling. Specifically, we validated computational prediction of miR 19b binding to the 3’UTR of TGFβRII by luciferase reporter assay. miR 19b mimic significantly decreased expression of TGFβRII as well as downstream target gene collagen, with additional suppression of HSC activation (αSMA) and concurrent increases in quiescent characteristics. In vitro findings translated to in vivo studies with decreased levels of miR 19b evident in fibrotic rat and human liver tissue compared to normal controls. These results identify miR 19b as a novel regulator of TGFβ signaling in HSC-mediated fibrogenesis and suggest a potential therapeutic approach for treating hepatic fibrosis.
Materials and Methods
miRNA Isolation, Purification and Microarray
Total RNA was isolated from samples using Trizol Reagent (Invitrogen, Carlsbad, CA) per manufacturer's instructions. RNA integrity was verified by an Agilent 2100 Bioanalyzer profile (Agilent Technologies Inc., Santa Clara, CA). miRNA purification and microarray details are described in the Supplementary Materials and Methods.
Primary Cell Isolation, Culture and Imaging
Male Sprague Dawley rats (> 500g; Charles River Laboratories, Wilmington, MA) were used for these studies. All experiments were reviewed and approved by Carolinas Medical Center Institutional Animal Care and Use Committee. Primary rat HSCs were isolated by pronase/collagenase perfusion digestion followed by subsequent density gradient centrifugation as previously reported(17). Cell purity (>95%) and viability were confirmed by autofluorescence and trypan blue staining. HSCs were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. Culture medium was replaced every 48 hours unless otherwise described and cells incubated at 37°C with 5% CO2. To document morphological changes, representative images were captured using an Olympus IX71 microscope (Olympus America Inc., Center Valley, PA). Primary rat hepatocytes were isolated and cultured as previously described(18).
Quantitative Real-Time Polymerase Chain Reaction and Immunoblotting
qRT-PCR reactions and immunoblotting details are described in the Supplementary Materials and Methods.
Transient Transfection
Activated HSCs (day 6) were transfected with mature miR 19b mimic and negative control probes (scramble, SCR) (Dharmacon, Inc.; Lafayette, CO) using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) according to manufacturer's instructions. Transient transfection details are described in the Supplementary Materials and Methods.
Immunocytochemistry and In Situ Hybridization
Prior to transfection culture-activated HSCs were seeded onto glass coverslips. Cells were transfected as described and fixed with 4% paraformaldehyde and stained with anti αSMA antibody (Millipore, Billerica, MA) as previously described(19). Liver tissues were obtained from the following fibrotic models: bile duct-ligation (BDL)/sham [Schrum lab, unpublished data], and ethanol/lipopolysaccharide(20). Sections (6μm) were cut from paraffin embedded tissues (RNase free). In situ hybridization was performed using mercury LNA™ detection probes, 5’-DIG and 3’-DIG labeled miR 19b according to manufacturer's instructions (Exiqon, Woburn, MA). To determine successful HSC activation in liver injury models, paired immunohistochemistry staining for quiescent and activated markers was performed (Supplementary Fig. S1).
Human Tissue Samples
Human fibrotic (Metavir fibrosis score of 3 or 4) liver biopsy samples (n=21) were obtained from the Liver-Biliary-Pancreatic Program Repository at Carolinas Medical Center (Charlotte, NC). The biopsies were being performed for clinical investigations. The consent was for a small portion of the biopsies (1-4 mg wet weight) to be stored for research studies. Samples were obtained under a protocol approved by the Institutional Review Board of CMC. All subjects provided written informed consent. Normal controls (n=7) were obtained from the Liver Tissue Cell Distribution Center (LTCDS) specimen bank (Minneapolis, MN).
Statistical Analysis
Data are presented as mean ± SE as determined from at least three independent experiments unless otherwise stated. Statistical analyses were performed using one way analysis of variance or Student's t-test or Mann-Whitney rank sum test where appropriate. p < 0.05 was considered significant, and denoted by *.
Results
miRNA Profiling in Quiescent and Activated Hepatic Stellate Cells
A total of 55 significantly differentially expressed miRs were identified by array analyses of quiescent (freshly isolated) and activated (day 14 of culture) HSCs (Fig. 1A). Validation of previously described miR expression levels was obtained, with miRs 16, 29a/b/c, 150 and 194 all significantly down-regulated during culture activation (Fig. 1A,B). These experiments also identified ~20 differentially expressed miRs not previously reported in published array data available at the time of manuscript preparation (data not shown). Analysis of differentially expressed miRs revealed members of the miR-17-92 cluster (19a, 19b, 92a) were significantly down-regulated in the profibrotic activated phenotype (Fig. 1B). Based upon previous literature and in silico analyses (TargetScan and miRanda prediction databases), which predict putative seed match sites for miR 19b in the 3’UTR of TGFβRII, this miR was selected for further analysis. qRT-PCR confirmed array data, verifying a significant decrease of miR 19b in activated compared to quiescent HSCs (Fig. 1C). Expression profile of miR 19b and predicted target mRNA TGFβRII were followed over 14 days in culture and a significant inverse relationship was observed (Fig. 1D) with a dramatic decrease seen in expression of the miR from quiescence to day 3 and a significant up-regulation of TGFβRII.
Fig. 1.
Differential miRNA expression in quiescent vs activated HSCs. (A) Microarray analysis for miRNA was performed with RNA extracts from primary quiescent (n=7) and activated (n=6) HSCs. Hierarchical cluster analysis of significantly differentially expressed miRs: bright green, underexpression; gray, no change; bright red, overexpression. (B) Listing of differentially expressed miRs in the array analysis including p value and fold change. (C) Rat HSCs were harvested and miR 19b expression was determined in quiescent (n=7) and activated (day 14, n=6) HSCs as assessed by qRT-PCR. (D) qRT-PCR analysis of miR 19b and TGFβRII expression levels over days in culture as normalized to 4.5S rRNA and β-actin, respectively (n=3). Data are presented as mean ± SE. *Differs from quiescent, p<0.05.
miR 19b Negatively Regulates Profibrotic TGFβ Signaling
Activated HSCs were transfected with miR 19b (19b) or a miRNA mimic negative control (scramble, SCR) and following 24 or 48 h of transfection, RNA and protein were analyzed. Preliminary studies validated the SCR sequence did not significantly affect TGFβRII expression or invariant control β-actin compared to mock transfection or untransfected cells (Lipofectamine 2000 alone) (data not shown). Additionally, effective transfection of primary cells was verified by qRT-PCR and consistent concentration dependent increases were observed in miR 19b relative to 4.5S rRNA expression (Supplementary Fig. S2). TGFβRII mRNA levels at both 24 and 48 h post-transfection were significantly decreased compared to SCR control, with greatest reduction seen when transfected with 75 nM 19b (Fig. 2A,B). Similar results were observed in human HSCs (LX-2) (Supplementary Fig. S3). Protein expression of the receptor was also significantly blunted by forced expression of miR 19b (Fig. 2C).
Fig. 2.
miR 19b negatively regulates TGFβRII expression. Day 6 HSCs were transiently transfected with miR 19b mimic (25-75 nM) or miR mimic negative control (SCR), and TGFβRII gene expression was measured by qRT-PCR at (A) 24 h and (B) 48 h (n=4). Expression was normalized to β-actin. (C) Representative immunoblot of TGFβRII protein expression 48 h post-transfection (miR 19b, 75 nM) (n=3). β-actin was used as an invariant control. (D) SMAD gene expression in activated HSCs (day 6) transfected with mature miR 19b for 24 and 48 h was determined by qRT-PCR (n=3). (E) Representative immunoblot of phosphorylated SMAD3 (p-SMAD3). β-actin was used as an invariant control. Data are presented as mean ± SE. *Differs from SCR, p<0.05.
Fibrotic TGFβ signaling propagates through the SMAD family of transcriptional activators, and like TGFβRII, SMAD2 and SMAD3 are up-regulated following fibrotic liver injury(21). Down-regulation of TGFβ signaling can impact expression of downstream SMAD3 and SMAD7(22-23). While SMAD2/3 3’UTRs do not harbor putative miR 19b binding sites (as predicted by TargetScan and miRanda), mRNA expression of SMAD3 is significantly down-regulated after 48 h of miR 19b transfection (Fig. 2D). miR 19b is also predicted to bind to the 3’UTR of Co-SMAD4, but no significant changes were observed in SMAD4 mRNA expression following transfections (Supplementary Fig. S4). More importantly, to determine whether downstream TGFβ signaling was impacted by disrupting TGFβRII, phosphorylation of SMAD3 was assessed. Compared to SCR, cells transfected with miR 19b showed a marked decrease in p-SMAD3 (Fig. 2E). Computational prediction of miR 19b binding to the 3’UTR of TGFβRII was validated by luciferase reporter assay using LX-2 cells (Fig. 3). These cells were selected to achieve higher transfection efficiency than primary rat HSCs. Addition of miR 19b mimic induced a 50-60% reduction in luciferase activity compared to controls.
Fig. 3.

miR 19b directly inhibits TGFβRII. Inhibition of firefly luciferase activities of pEZX-TGFβRII reporter by miR 19b mimic. Human HSCs (LX-2) were co-transfected with 4.8 μg of pEZX-TGFβRII reporter plasmid or empty vector and 75 nM miR 19b mimic or negative miR mimic control (SCR) using Lipofectamine 2000 (n=3). Data are presented as mean ± SE. *Differs from TGFβRII/SCR and CTRL/19b, p<0.05.
miR 19b Decreases Expression of TGFβ Target Genes
Effects of increasing miR 19b on downstream TGFβ signaling target procollagen mRNA and protein were measured. Forced expression of miR 19b dampened mRNA expression of both procollagen Col α1(I) and Col α2(I), with more significant effects observed on the transcription of Col α2(I) (Fig. 4A,B). Translation of the fibrillar collagen is also markedly decreased after 48 h of miR 19b treatment as denoted by decreased intracellular protein expression (Fig. 4C), confirming negative regulation of TGFβRII signaling by miR 19b as both procollagen 3’UTRs lack predicted binding sites (TargetScan). Additionally, functional secretion of this protein is disrupted by miR 19b as determined by immunoblot utilizing proteins concentrated from harvested culture medium (48 h) (Supplementary Fig. S5).
Fig. 4.
miR 19b exerts inhibitory effects on TGFβ target gene collagen. Day 6 HSCs were transiently transfected with miR 19b mimic (25-75 nM) or miR mimic negative control (SCR) and Col α1(I) (A) and Col α2(I) (B) gene expression was assessed by qRT-PCR at 24 h (n=4). Expression was normalized to β-actin. (C) 48 h post-transfection cells were harvested and immunoblot performed on whole cell lysates for type I collagen expression. β-actin was used as an invariant control (n=3). Data are presented as mean ± SE. *Differs from SCR, p<0.05.
miR 19b Inhibits Paracrine TGFβ Signals
Recombinant TGFβ was added to day 6 culture-activated HSCs transfected with miR 19b mimic and levels of procollagen mRNA determined. After 48 h Col α1(I) and α2(I) mRNA expression was decreased even in the presence of exogenous TGFβ as compared to respective control (Fig. 5), indicating a powerful role for miR 19b in the inflammatory hepatic microenvironment. Additionally, as TGFβRII has been shown to modulate TGFβ expression(24), miR 19b suppressed TGFβ1 expression as compared to control (Supplementary Fig. S6A,B).
Fig. 5.
miR 19b inhibits paracrine TGFβ signals. Day 6 activated HSCs were transfected with or without miR 19b mimic or miR mimic negative control (SCR) (75 nM), and following standard 6 h incubation, transfection medium was removed and fresh culture medium devoid of antibiotic was added containing 5 ng/mL of recombinant human TGFβ (TGFβ) for a period of 48 h. Cells were harvested and mRNA levels of Col α1(I) and Col α2(I) were assessed by qRT-PCR (n=3). Data are presented as mean ± SE. *Compared to SCR alone, #compared to SCR, TGFβ, p<0.05.
HSC Transdifferentiation is Regulated by miR 19b
Forced expression of miR 19b blunted the day 6 culture-activated HSC phenotype as denoted by shrunken cytoplasm, decreased polygonal shape and increased spindle shaped cellular protrusions (characteristic of the quiescent phenotype) (Supplementary Fig. S7). Morphological changes indicative of suppression of the activated phenotype correlated with levels of αSMA mRNA, which were significantly decreased after 48 h of transfection (Fig. 6A). Immunoctyochemical analysis of αSMA protein (as a marker of activated HSCs) expression corroborated the visible reduction in activated phenotype as visualized by markedly reduced red fluorescence as well as by disorganization and disorientation of actin fibers (Fig. 6B). Further, miR 19b restored GFAP expression, a marker of quiescent HSCs (Fig. 6C).
Fig. 6.
HSC transdifferentiation is regulated by miR 19b. (A) Activated HSCs (day 6) were transfected with negative control (SCR) or miR 19b (75 nM) and αSMA gene expression was assessed by qRT-PCR at 24 h and 48 h (n=4). (B) Representative images (40X) of fluorescent immunocytochemical analysis of αSMA expression (red) in activated HSCs following 48 h of transfection with SCR or miR 19b mimic (75 nM); DAPI staining (blue) was used to indicate cell nuclei (n=3). (C) Day 6 HSCs were transiently transfected with miR 19b mimic or SCR (75 nM) and GFAP expression was assessed by qRT-PCR at 48 h (n=4). Expression was normalized to β-actin. Data are presented as mean ± SE. *Differs from SCR, p<0.05.
miR 19b is Decreased Following Fibrotic Liver Injury in vivo
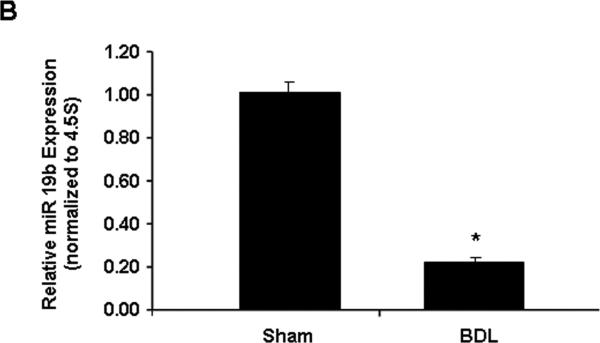
Next, we assessed whether decreased miR 19b also occurs in vivo (or in intact animals), in a rat model of hepatic fibrosis. Tissue sections from sham operated control and BDL rats were subjected to in situ hybridization and qRT-PCR experiments to assess expression of miR 19b. miR 19b was markedly decreased in fibrotic liver tissue compared to controls (Fig. 7A,B). miR 19b specific staining (red fluorescence, Fig. 7A; blue chromagen, Supplementary Fig. S8) in control tissue appears outside of the parenchymal cells and higher magnification inspection is indicative of perisinusoidal (HSC-specific location) expression. Supporting in situ hybridization data, low expression of miR 19b, comparable to that of activated HSCs, were observed in primary rat hepatocytes as compared to quiescent HSCs (Supplementary Fig. S9). To confirm initial observation of HSC specific expression, co-localization of miR 19b (red fluorescence) and quiescent HSC specific marker (GFAP, green fluorescence) was performed. Merged images (Fig. 7C) obtained from single channel images (Supplementary Fig. S10) showed high intensity yellow fluorescence in Sham tissue indicating miR 19b expression in quiescent HSCs. As expected, decreased yellow fluorescence was observed in BDL tissue (Fig. 7C). Interestingly, the decrease of miR 19b observed in hepatic injury does not appear to be stimulus-specific, as another rat model of liver injury/fibrosis (ethanol/lipopolysacchardide, ELPS(20)) also showed decreased hepatic miR 19b levels (Supplementary Fig. S11), strengthening the conserved importance of decreased miR 19b in hepatic fibrosis.
Fig. 7.
Down-regulation of miR 19b in a rodent model of hepatic fibrosis. (A) Representative immunofluorescent images of sham (normal) and BDL (fibrotic) tissue sections following in situ hybridization with double DIG labeled LNA miR 19b probes. miR 19b expression levels are marked by red fluorescence. Zonal damage (Z1, Z2, Z3) is indicated. (B) RNA was harvested from sham (n=6) and BDL (n=6) tissues and miR 19b expression was determined by qRT-PCR. (C) Co-localization of miR 19b (red fluorescence) and quiescent HSC specific marker (GFAP, green fluorescence) was performed. Merged immunofluorescent images (yellow fluorescence indicates quiescent HSCs expressing miR 19b) of sham and BDL tissue sections. Data are presented as mean ± SE. *Differs from sham, p<0.05.
Human Hepatic Fibrosis is Associated with Altered miR 19b Expression
To determine if miR 19b expression is also affected in human hepatic fibrosis, total RNA was isolated from fibrotic (Metavir fibrosis score of 3 or 4 due to varying etiologies including PBC, HCC and HCV) and normal control livers. qRT-PCR was used to determine relative expression levels of miR 19b. As observed in the rodent fibrotic injury models, levels of miR 19b were also significantly decreased by approximately 80% in human patients with fibrotic livers (Fig. 8A). Recent studies have shown inverse correlations between tissue and plasma miR levels(25-26). miR 19b levels were assessed in the sera of fibrotic patients, and when directly compared to pair-matched tissue levels, a clear inverse relationship was observed (Fig. 8B).
Fig. 8.
miR 19b is a putative biomarker for human hepatic fibrosis. RNA was harvested from normal (n=7) and fibrotic (n=21) human liver tissue (A) and sera (B; 6 pair-matched fibrotic liver and sera samples) and miR 19b expression was assessed by qRT-PCR. Expression was normalized to SNORD44 (A) and total cDNA (B). Data are presented as mean ± SE. *Differs from control, p<0.05.
Discussion
This study provides the first evidence that miR 19b has a functional role in rat and human liver fibrosis. Mechanistically, miR 19b acts as a novel inhibitor of fibrotic TGFβ signaling in the HSC and holds clinical promise as a therapeutic molecule and/or biomarker for fibrosis. Significant down-regulation of miR 19b was observed in activated HSCs as well as in rodent models of fibrosis and in human disease. Forced expression of mature miR 19b in activated HSCs significantly reduced expression of TGFβRII through direct binding to the 3’UTR, which is vital for efficient activation of downstream profibrotic gene expression. Although miR 19b levels were increased substantially at lower mimic concentrations (Supplementary Fig. S2), it is not surprising that higher concentrations were necessary to have a physiological effect as other factors may also modulate TGFβ signaling. Additionally, since observations were made in a fibrotic phenotype, higher expression of miR 19b (compared to quiescent) is likely needed to suppress TGFβRII expression. Expression of procollagen mRNAs and secreted type I collagen were also markedly reduced by miR 19b, even in the presence of exogenous TGFβ. As firmly established in the literature, disruption of TGFβ signaling impedes HSC activation and fibrosis as evidenced by altered expression of transdifferentiation markers (i.e. decreased aSMA and increased GFAP)(2, 8-9, 27). miR 19b mediated-down-regulation of TGFβRII impeded HSC activation and produced reversion to a more quiescent phenotype, and since αSMA and GFAP do not harbor putative miR 19b binding sites, this effect is likely attributed to disruption of profibrotic TGFβ signaling.
Although pathologies of chronic hepatic disease (ALD, HCV, HCC) are variable, TGFβ-mediated fibrosis is an underlying commonality(28). TGFβ is the most potent stimulus for HSC-mediated fibrogenesis(28-29) as it plays a critical role in initiation of the transdifferentiation process. In addition to paracrine sources of the cytokine, TGFβ synthesis is markedly increased by the HSC as a result of activation, further perpetuating the fibrotic phenotype. Inhibition of TGFβ receptors, specifically TGFβRII, abrogates the signaling pathway and HSC activation(5-7). These findings have established HSC-mediated TGFβ signaling as a pivotal mechanism in hepatic fibrogenesis and disruption of HSC activation and collagen deposition via inhibition of TGFβ signaling as a mechanism to ameliorate and/or reverse fibrosis.
miRs have emerged as key regulatory molecules in chronic liver disease, including hepatic fibrosis(30-32). Array profiling studies report differential miR expression in normal vs. fibrotic liver tissue in a variety of rodent injury models including BDL and carbon tetrachloride (CCl4) (33). miRs 150, 187, 194 and 207 were significantly down-regulated in HSCs isolated from BDL animals compared to sham controls, while let7 family members were significantly up-regulated. Recently we have seen evidence that these small non-coding RNAs modulate fibrogenesis and HSC activation(11). Overexpression of miRs 150 and 194 in human HSCs (LX-2) resulted in inhibition of proliferation as well as decreases in type I collagen and αSMA(15).
miR profiling in human and murine liver fibrosis, and additional published in vitro manipulation studies, have highlighted a role for the miR 29 family in fibrosis via regulation of collagen expression(16, 33). While fibrosis underlies most chronic liver diseases, including HCV and ALD, when expression of miR 29 was examined in human samples, Roderburg et al. found expression of the miR highly variable amongst patients with viral vs alcohol-induced fibrosis, indicating the role of miR 29 in fibrosis may be stimulus specific(33). Here we report that miR 19b levels are down-regulated in two experimental animal models of hepatic fibrosis (BDL, ELPS), and these results were confirmed in fibrotic human patients despite variable underlying etiologies (HBV, HCV, HCC, PBC), supporting a highly conserved role of this miR in fibrosis. Moreover, success in identifying target genes of dysregulated miRs in liver disease has been limited. Herein, we report that miR 19b binds directly to the 3’UTR of TGFβRII inhibiting fibrotic HSC activation.
miR expression patterns are organ and tissue specific, making systemic miR targeting problematic. However, recent reports have shown that miRs derived from the miR-17-92 cluster (including miR 19b) directly regulate TGFβ signaling in non-liver cell types, including neuroblastoma cells(34) and colonocytes(35). Additionally, miR 19b levels are down-regulated in fibrosis and ECM remodeling of other tissue/organs (pulmonary, cardiac)(36-37), indicating a highly conserved role of miR 19b in TGFβ-mediated fibrogenesis. While transcriptional regulation of miR17-92 cluster is mostly unknown, studies have reported that transcription factors p53, NFκB and E2Fs can modulate expression of this cluster(38-39). Additionally, several miRs are post-transcriptionally regulated by TGFβ and nuclear accumulation of SMAD proteins(38, 40). As levels of TGFβ are known to increase as a result of hepatic injury promoting fibrogenesis, it is important to note this cytokine may affect disease pathology through HSC-mediated actions as well as through affecting global miR processing/expression.
Currently there are no FDA-approved treatments for fibrosis. As the field of miR research is rapidly developing, pioneering advances have emphasized critical changes in miR expression profiles during development of fibrosis which regulate wound-healing transcripts. While acknowledged that therapeutic modulation of single miRs in vivo has aimed to inhibit expression via antisense oligos/antagomirs, miR over-expression strategies are also ongoing and hold great promise to restore delicate genetic programs vital to normal organ function. Adenoviral delivery of miR-17-92 cluster inhibited HCV replication in cell culture propogated HCV(41). Additionally, recent reports in HCC demonstrate miR 26a administration is capable of repressing tumorigenesis without significant systemic effects(42). While we cannot definitively demonstrate loss of miR 19b expression is a cause or consequence of fibrosis, our data indicate that over-expression of miR 19b may be a useful therapeutic agent for TGFβ-mediated fibrosis.
In addition to therapeutic applications, miR 19b may also serve as an accurate biomarker of liver fibrosis and/or HSC activation. Several studies in settings of chronic liver disease with underlying fibrosis have shown strong correlations between specific miR expression patterns and responses to drug treatments as well as disease progression/prognostic outcome(11). Plasma levels of miR 122 are elevated in both HBV and HCV patients as well as in models of alcohol and drug-induced liver damage reinforcing a role for miRs as biomarkers(26). Recent studies have also shown inverse correlations between tissue and plasma miR levels(25-26). In fact, miR 19b was up-regulated 4.3-fold in serum of individuals with cirrhotic livers compared to normal controls(43) suggesting a potentially non-invasive route for diagnosing hepatic fibrosis. In support of existing literature(43), despite the small sample size, higher expression of miR 19b was evident in the sera compared to pair-matched tissue of fibrotic patients. Future studies are needed to carefully monitor plasma miR 19b expression in relation to tissue from healthy individuals compared to varying fibrotic stages and etiologies.
HCV represents the major cause of hepatic fibrosis on a global scale and is the most frequent indication of liver transplantation(44); however, recurrent hepatitis C occurs in 80% of patients by 3 year post-transplant(45) and up to 20% advance to bridging fibrosis or cirrhosis within 2 years(46) due to TGFβ signaling and HSC activation (47-48). Antiviral therapy with interferon and ribavirin in transplant recipients is only 10-30% effective, and therapy may not be well-tolerated. Identifying liver transplant recipients at greatest risk for rapid development of fibrosis from recurrent hepatitis C would target those recipients most urgently in need of antiviral therapy and defer treatment to those at less risk for disease progression(48). In support of our findings, miR 19b levels were significantly higher in HCV responder vs non-responder patient populations, underscoring the importance of this specific miR. Examining expression levels of miR 19b in HCV transplant patients could lead to development of a reliable marker to identify rapid progressors of fibrosis.
Overall these systematic studies indicate miR 19b is a novel regulator of fibrotic TGFβ signaling and indicates the loss of miR 19b following HSC activation perpetuates the fibrotic response. Restored miR 19b expression in activated HSCs indicated this miR may be a possible therapy for the treatment or reversion of fibrosis, and patient data indicates this powerful miR may prove to be an accurate biomarker for the fibrotic condition. These studies provide novel insight into the global regulation of a key signaling pathway which promotes hepatic fibrosis, and more importantly, provides a new avenue to be explored for translational research.
Supplementary Material
Acknowledgments
Grant Support: NIH 014891 (LWS) and institutional funds from CMC
Abbreviations
- ALD
alcoholic liver disease
- BDL
bile duct-ligated
- ECM
extracellular matrix
- GFAP
glial fibrillary acidic protein
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HSC
hepatic stellate cell
- IHC
immunohistochemistry
- miRNA
microRNA
- PBC
primary biliary cirrhosis
- qRT-PCR
quantitative real time polymerase chain reaction
- αSMA
smooth muscle α-actin
- TGFβ
transforming growth factor beta
References
- 1.Saile B, Ramadori G. Inflammation, damage repair and liver fibrosis--role of cytokines and different cell types. Z Gastroenterol. 2007;45:77–86. doi: 10.1055/s-2006-927395. [DOI] [PubMed] [Google Scholar]
- 2.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neubauer K, Knittel T, Aurisch S, Fellmer P, Ramadori G. Glial fibrillary acidic protein--a cell type specific marker for Ito cells in vivo and in vitro. J Hepatol. 1996;24:719–730. doi: 10.1016/s0168-8278(96)80269-8. [DOI] [PubMed] [Google Scholar]
- 4.Dooley S, Delvoux B, Streckert M, Bonzel L, Stopa M, ten Dijke P, Gressner AM. Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 2001;502:4–10. doi: 10.1016/s0014-5793(01)02656-4. [DOI] [PubMed] [Google Scholar]
- 5.Kanzler S, Meyer E, Lohse AW, Schirmacher P, Henninger J, Galle PR, Blessing M. Hepatocellular expression of a dominant-negative mutant TGF-beta type II receptor accelerates chemically induced hepatocarcinogenesis. Oncogene. 2001;20:5015–5024. doi: 10.1038/sj.onc.1204544. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T, Sakata R, Ueno T, Sata M, Ueno H. Inhibition of transforming growth factor beta prevents progression of liver fibrosis and enhances hepatocyte regeneration in dimethylnitrosamine-treated rats. Hepatology. 2000;32:247–255. doi: 10.1053/jhep.2000.9109. [DOI] [PubMed] [Google Scholar]
- 7.Ueno H, Sakamoto T, Nakamura T, Qi Z, Astuchi N, Takeshita A, Shimizu K, et al. A soluble transforming growth factor beta receptor expressed in muscle prevents liver fibrogenesis and dysfunction in rats. Hum Gene Ther. 2000;11:33–42. doi: 10.1089/10430340050016139. [DOI] [PubMed] [Google Scholar]
- 8.Dooley S, Delvoux B, Lahme B, Mangasser-Stephan K, Gressner AM. Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology. 2000;31:1094–1106. doi: 10.1053/he.2000.6126. [DOI] [PubMed] [Google Scholar]
- 9.George J, Roulot D, Koteliansky VE, Bissell DM. In vivo inhibition of rat stellate cell activation by soluble transforming growth factor beta type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci U S A. 1999;96:12719–12724. doi: 10.1073/pnas.96.22.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakner AM, Moore CC, Gulledge AA, Schrum LW. Daily genetic profiling indicates JAK/STAT signaling promotes early hepatic stellate cell transdifferentiation. World J Gastroenterol. 2010;16:5047–5056. doi: 10.3748/wjg.v16.i40.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakner AM, Bonkovsky HL, Schrum LW. microRNAs: Fad or future of liver disease. World J Gastroenterol. 2011;17:2536–2542. doi: 10.3748/wjg.v17.i20.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo CJ, Pan Q, Jiang B, Chen GY, Li DG. Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis. 2009;14:1331–1340. doi: 10.1007/s10495-009-0401-3. [DOI] [PubMed] [Google Scholar]
- 13.Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 15.Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J, et al. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol. 2010;298:G101–106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa T, Iizuka M, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. Suppression of type I collagen production by microRNA-29b in cultured human stellate cells. Biochem Biophys Res Commun. 2010;391:316–321. doi: 10.1016/j.bbrc.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 17.Lakner AM, Walling TL, McKillop IH, Schrum LW. Altered aquaporin expression and role in apoptosis during hepatic stellate cell activation. Liver Int. 2011;31:42–51. doi: 10.1111/j.1478-3231.2010.02356.x. [DOI] [PubMed] [Google Scholar]
- 18.Schrum LW, Black D, Iimuro Y, Rippe RA, Brenner DA, Behrns KE. c-Jun does not mediate hepatocyte apoptosis following NFkappaB inhibition and partial hepatectomy. J Surg Res. 2000;88:142–149. doi: 10.1006/jsre.1999.5784. [DOI] [PubMed] [Google Scholar]
- 19.Chung GG, Yoon HH, Zerkowski MP, Ghosh S, Thomas L, Harigopal M, Charette LA, et al. Vascular endothelial growth factor, FLT-1, and FLK-1 analysis in a pancreatic cancer tissue microarray. Cancer. 2006;106:1677–1684. doi: 10.1002/cncr.21783. [DOI] [PubMed] [Google Scholar]
- 20.Karaa A, Thompson KJ, McKillop IH, Clemens MG, Schrum LW. S-adenosyl-L-methionine attenuates oxidative stress and hepatic stellate cell activation in an ethanol-LPS-induced fibrotic rat model. Shock. 2008;30:197–205. doi: 10.1097/shk.0b013e318160f417. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura Y, Ninomiya H. Smad expression of hepatic stellate cells in liver cirrhosis in vivo and hepatic stellate cell line in vitro. Pathol Int. 2003;53:18–26. doi: 10.1046/j.1440-1827.2003.01431.x. [DOI] [PubMed] [Google Scholar]
- 22.Bauge C, Cauvard O, Leclercq S, Galera P, Boumediene K. Modulation of transforming growth factor beta signalling pathway genes by transforming growth factor beta in human osteoarthritic chondrocytes: involvement of Sp1 in both early and late response cells to transforming growth factor beta. Arthritis Res Ther. 2011;13:R23. doi: 10.1186/ar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov. 2004;3:1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 24.Miyazono K. Positive and negative regulation of TGF-beta signaling. J Cell Sci. 2000;113(Pt 7):1101–1109. doi: 10.1242/jcs.113.7.1101. [DOI] [PubMed] [Google Scholar]
- 25.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 27.Isono M, Soda M, Inoue A, Akiyoshi H, Sato K. Reverse transformation of hepatic myofibroblast-like cells by TGFbeta1/LAP. Biochem Biophys Res Commun. 2003;311:959–965. doi: 10.1016/j.bbrc.2003.10.093. [DOI] [PubMed] [Google Scholar]
- 28.Brenner DA. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361–368. [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons CJ, Takashima M, Rippe RA. Molecular mechanisms of hepatic fibrogenesis. J Gastroenterol Hepatol. 2007;22(Suppl 1):S79–84. doi: 10.1111/j.1440-1746.2006.04659.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009;15:1665–1672. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin X, Ye YF, Chen SH, Yu CH, Liu J, Li YM. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 2009;41:289–297. doi: 10.1016/j.dld.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 34.Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70:8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, Swinnen M, et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardie WD, Glasser SW, Hagood JS. Emerging concepts in the pathogenesis of lung fibrosis. Am J Pathol. 2009;175:3–16. doi: 10.2353/ajpath.2009.081170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Haurigot V, Zhou S, Luo G, Couto LB. Inhibition of hepatitis C virus replication using adeno-associated virus vector delivery of an exogenous anti-hepatitis C virus microRNA cluster. Hepatology. 2010;52:1877–1887. doi: 10.1002/hep.23908. [DOI] [PubMed] [Google Scholar]
- 42.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, Run W, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Martin E, Senzolo M, Gambato M, Germani G, Vitale A, Russo FR, Burra P. Fibrosis progression and the pros and cons of antiviral therapy for hepatitis C virus recurrence after liver transplantation: a review. Transplant Proc. 2010;42:2223–2225. doi: 10.1016/j.transproceed.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 45.Szabo G, Katz E, Bonkovsky HL. Management of recurrent hepatitis C after liver transplantation: a concise review. Am J Gastroenterol. 2000;95:2164–2170. doi: 10.1111/j.1572-0241.2000.02296.x. [DOI] [PubMed] [Google Scholar]
- 46.Berenguer M. Natural history of recurrent hepatitis C. Liver Transpl. 2002;8:S14–18. doi: 10.1053/jlts.2002.35781. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Ari Z, Pappo O, Druzd T, Sulkes J, Klein T, Samra Z, Gadba R, et al. Role of cytokine gene polymorphism and hepatic transforming growth factor beta1 expression in recurrent hepatitis C after liver transplantation. Cytokine. 2004;27:7–14. doi: 10.1016/j.cyto.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Russo MW, Firpi RJ, Nelson DR, Schoonhoven R, Shrestha R, Fried MW. Early hepatic stellate cell activation is associated with advanced fibrosis after liver transplantation in recipients with hepatitis C. Liver Transpl. 2005;11:1235–1241. doi: 10.1002/lt.20432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.