Abstract
Aims
Previous genetic studies have shown that a C/T polymorphism at position −889 of the IL1A promoter, specifically allele 2 (−889T), increases the risk for development of several inflammation-related disorders, such as periodontitis, osteomyelitis, toxoplasmic retinochoroiditis, contact dermatitis, as well as neurodegenerative conditions such as Alzheimer’s disease. We sought to detemine the differential abilities of C- and T- containing versions of the −889 sequence to bind nuclear proteins from microglia.
Main methods
Microglial cells were subjected to inflammatory activation prior to the harvest of nuclear proteins. Electrophoretic mobility shift assays (EMSA) were performed using oligonucleotide probes representing 25 base pairs surrounding the IL1A −889 polymorphism. Antibodies reactive against transcription factors were used to identify the specific proteins involved in complexes with DNA.
Key findings
EMSA revealed multiple differences in DNA-binding profiles when microglial nuclear extracts were incubated with the polymorphic probes. The allele-2 probe formed specific complexes that were not detected with the allele-1 (−889C) probe, and vice versa. Formation of allele-2 nucleoprotein complexes was increased in activated microglia. Antibody supershift analysis indicated that multiple Jun-family members but not Fos-family proteins contributed to the LPS-activated allele-2 EMSA complexes. LPS-activation of allele-2 EMSA complexes could be blocked by the specific c-Jun N-terminal kinase (JNK) inhibitor SP600125.
Significance
These results suggest that the −889 polymorphism creates differential interactions with transcription factors that could lead to differential expression rates under proinflammatory conditions.
Keywords: Activator Protein-1, Alzheimer’s disease, c-Jun, Cytokine, c-Fos, Inflammation, Interleukin-1, Jun N-terminal kinase, Microglia, Transcription Factor
Introduction
Along with other genetic variants in the complex interleukin-1 locus of the human genome, polymorphisms at the −889 position in the gene (IL1A) for interleukin-1α have been associated with differential risk for inflammation-related diseases such as periodontitis (Kornman 2006, Kornman et al. 1997), Alzheimer’s disease (AD) (Combarros et al. 2003, Grimaldi et al. 2000, Nicoll et al. 2000, Rainero et al. 2004), osteomyelitis (Tsezou et al. 2008), toxoplasmic retinochoroiditis (Cordeiro et al. 2008), and irritant contact dermatitis (de Jongh et al. 2008). They may also differentially segregate in polycystic ovary syndrome (Kolbus et al. 2007) and multiple myeloma (Abazis-Stamboulieh et al. 2007).
Multiple research groups and two meta analyses (Combarros et al. 2003, Rainero et al. 2004) have reported that a C/T polymorphism at position −889 of the IL1A promoter influences susceptibility for AD. Homozygosity for the −889T genotype (allele 2) is associated with an approximately three-fold elevation in AD risk. The mechanism responsible for linkage between IL1A and AD or other conditions is unknown. The location of the polymorphism in the IL1A promoter region suggests that regulation of IL1A transcription may be affected. Dominici et al. (2002) reported that transient promoter-luciferase activity of IL1A −889 allele 2 was significantly (23%) higher than allele 1 in pancreatic carcinoma cells. They also found that LPS-stimulated IL-1α protein levels increased more rapidly in peripheral blood mononuclear cells (PBMCs) from homozygous TT (allele 2) individuals than homozygous CC (allele 1) individuals, with intermediate results from heterozygous CT individuals. Wei et al. (2007) demonstrated increased transient promoter-luciferase activity of allele 2 in human astroglial cells. The allelic imbalance was markedly enhanced by inflammatory activation of the cells with amyloid β-peptide (Aβ) or lipopolysaccharide (LPS); it was diminished by the anti-inflammatory agents salicylate and lovastatin. These data indicate that the IL1A −889 polymorphism has an impact on transcriptional regulatory mechanisms. In this report, we describe our efforts to characterize regulatory transcription factors binding to the IL1A −889 polymorphic locus.
Material and Methods
Materials
Complementary single-stranded oligonucleotides containing the the −889 polymorphic site of the IL1A promoter (bases −901 to −877 relative to the transcription start site) were obtained from Invitrogen (Carlsbad CA). Oligos were resuspended in TEN (10 mM Tris-HCl pH 7.4, 1 mM EDTA, 50 mM NaCl) and heated to 55 °C for 10 min prior to quantitation by spectrophotometry. To anneal equimolar amounts of complementary oligonucleotides, each oligonucleotide mixture was first denatured by heating to 80 °C for 10 min in an aluminum heat block, and then left in the block as it cooled slowly to room temperature after switching off power. Double-stranded AP1, CREB, and NFκB oligonucleotides were obtained from Santa Cruz Biotechnology (Santa Cruz CA). Antibodies specific to AP1 family proteins used in supershift experiments were obtained from Santa Cruz: c-Fos (K-25) sc-253 X (“pan-Fos”), c-Jun (D) sc-44 X (“pan-Jun”), c-Jun (H-79) sc-1694 X, Jun B (N-17) sc-46 X, Jun D (329) sc-74 X. All supershift antibodies were supplied as 1 µg/µl solutions. SP600125 was from EMD/Millipore (Billerica MA), which reports its IC50 as 40 nM for JNKs-1 and -2 and 90 nM for JNK-3.
Cell cultures
The BV2 cell line (American Type Culture Collection, Rockville MD) was maintained in minimal essential medium with Earle's salts (MEM), supplemented to 5% with fetal bovine serum (FBS). For passaging, these cells were dislodged by a short incubation Hank’s balanced salt solution (HBSS) lacking divalent cations and containing 0.5 mM EDTA (no trypsin). For experimental treatments and preparation of nuclear extracts, BV2 cells (0.3 × 106) were plated into 60-mm culture dishes in 4 ml MEM + 5% FBS, and allowed to grow for 2 days.
Rat primary mixed glial cultures were established from the cerebral cortex of Sprague-Dawley rats at postnatal day 1, as described previously (Barger et al. 2007). For use in experiments, microglia were gently washed from the surface of the astrocyte monolayer, and replated into 60-mm dishes in MEM + 10% FBS (1.2 × 106 cells per dish).
For treatments with LPS, growth medium was replaced with serum-free MEM 2 h (BV2) or 4 h (primary microglia) prior to LPS application. Cultures were activated with 300 ng/ml LPS for 4 h, unless noted otherwise. Activation was confirmed by testing the culture medium for nitrite accumulation by Griess reaction (Barger et al., 2007). The JNK inhibitor SP600125 was dissolved in dimethyl sulfoxide (DMSO), and used at a final concentration of 30 µM (0.3% DMSO). Inhibitor was added to cells 15 min prior to LPS treatment.
Electrophoretic mobility shift assay (EMSA)
For nuclear extraction, all steps were performed on ice or under refrigeration at 4 °C. Culture dishes were placed on ice and washed once with 10 mM ice-cold phosphate-buffered saline (PBS) and once with ice-cold lysis buffer (10 mM HEPES, 10 mM KCl, and 1.5 mM MgCl2, pH 7.9). Cells were lysed by scraping from the plate in lysis buffer containing 0.5% Nonidet P-40, 0.5 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml leupeptin, and 10 µg/ml aprotinin. Protease inhibitors were added to all buffers immediately before use. The lysate was microcentrifuged (12,000 × g, 5 min, 4 °C) to collect nuclei, which were washed with lysis buffer containing protease inhibitors but no detergent. Nuclear proteins were extracted by suspending the nuclei in ice-cold extraction buffer (420 mM NaCl, 20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM β-mercaptoethanol, 0.5 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin) for 10 min on ice. The extracted nuclei were collected by centrifugation (12,000 × g, 5 min, 4 °C), and the supernatant was transferred to another tube. To avoid prolonged high-salt exposure, the extracts were diluted immediately with 1.5 volumes of dilution buffer (50 mM KCl, 20 mM HEPES, pH 7.9, 0.2 mM EDTA, 20% glycerol, 0.5 mM β-mercaptoethanol, 0.2 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin). Aliquots were removed for quantification of nuclear protein by BCA (Pierce) so that the remainder of the sample could be frozen immediately at −80 °C. DNA-binding activity decreased dramatically if the samples were frozen a second time. Most binding reactions were performed within one week of extraction.
For DNA binding reactions, 5 µg of nuclear extract was diluted in binding buffer (10 mM Tris-HCl, pH 7.4, 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.1% Nonidet P-40, 1 mM β-mercaptoethanol, 50 µg/ml poly[dI:dC]). After a 15-min incubation on ice, a 20-min room temperature incubation was performed in the presence of 30–50 fmol 32P-labeled probes (1–2 × 105 cpm). In cases where multiple probes were compared, specific activities were equilibrated by addition of unlabeled oligonucleotide to those having higher specific activities after labeling. Protein/DNA complexes were resolved by nondenaturing, 0.25X TBE/6% PAGE. Gels were dried and exposed to autoradiographic film. For competition assays, the indicated unlabeled oligonucleotides were added in 10-, 30-, and 100-fold molar excess over the amount of radiolabeled probe. Antibodies and unlabeled oligonucleotides were added to the binding reaction prior to the 15-min incubation on ice.
Results
EMSA was performed using nuclear extracts from BV2 microglia to determine if the −889C/T polymorphism confers differential transcription factor binding activities. Nuclear proteins from BV2 microglia formed different complexes with probes corresponding to each of the IL1A −889 allelic sequences. The two major allele-1 complexes (C1 and C2, Figure 1) exhibited higher gel mobility than the major allele-2 complexes (T1 and T2). Neither allele-1 complex was induced by LPS, while both allele-2 complexes appeared to respond to LPS with peak activity after 4 h of stimulation.
Figure 1.
Nuclear proteins from BV2 microglia form different complexes with allele-1 versus allele-2 probes. Duplicate cultures of BV2 microglia were treated with 300 ng/ml LPS for the times indicated (hours). Nuclear extract from each culture was incubated with [32P]-labeled IL1A −889 oligonucleotide probes representing each allele. Protein-DNA complexes were resolved by nondenaturing PAGE. Results are representative of 2 experiments.
Competition assays were performed to determine which transcription factors were responsible for the LPS-activatable allele-2 EMSA complexes. LPS activity is often mediated by inflammatory transcription factors such as AP1 and NFκB. Binding to the labeled allele-2 probe was effectively blocked by 100-fold molar excess of unlabeled competitor DNA of the same sequence (Figure 2). By contrast, unlabeled competitor containing canonical AP1 or CREB sequences were able to completely obliterate the T1 and T2 complexes at just a 10-fold molar excess. Neither NFκB nor IL1A −889 allele-1 sequences were able to compete for binding to the allele-2 probe.
Figure 2.
Sequence specificity of IL1A −889 allele-2 EMSA complexes. Nuclear extracts from LPS-activated BV2 microglia were analyzed by EMSA with radiolabeled IL1A −889 allele-2 probe alone (−) or in the presence of 10-, 30-, or 100-fold molar excess of the unlabeled competitor oligonucleotides indicated. Results are representative of 3 experiments.
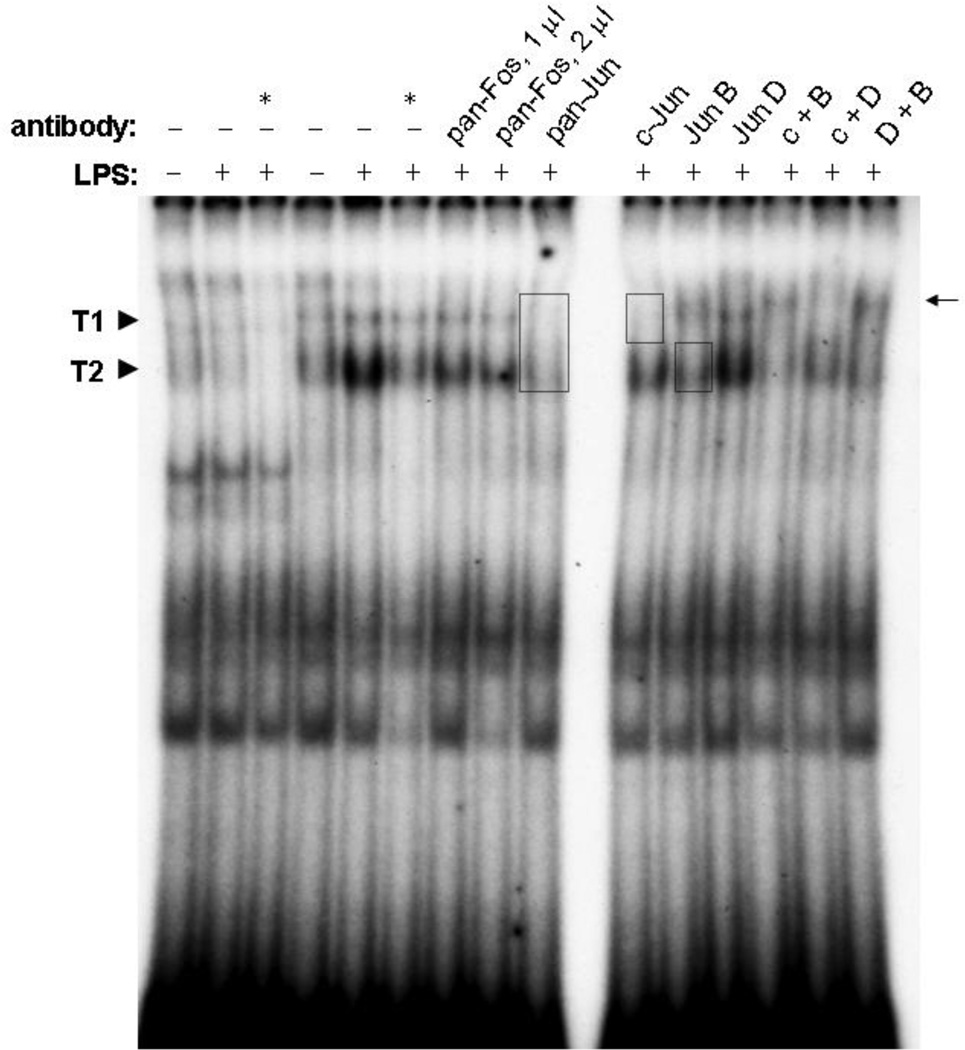
The acute sensitivity of T1 and T2 complexes to competition by the AP1 sequence suggested that the proteins responsible for these complexes might bear a relationship to AP1 transcription factors. These are a heterogeneous group of homo- and heterodimers of Jun and Fos family proteins. To determine which proteins may contribute to LPS-stimulated IL1A allele-2 complexes, we attempted to influence the EMSA complexes with antibodies specific to Jun and Fos family members. The anti-pan-Jun antibody, which recognizes all Jun proteins, completely obliterated complex T1 and reduced complex T2 to its basal level (Figure 3). The anti-pan-Fos antibody, which similarly recognizes all Fos proteins, had a much smaller effect on complex T2 and no effect on complex T1. Due to the success of anti-pan-Jun, antibodies to individual Jun proteins were tested. An antibody specific to c-Jun obliterated complex T1 and slightly diminished complex T2. An antibody against Jun B did not affect complex T1, but “supershifted” some of complex T2 to a position (Figure 3, arrow) slightly above complex T1. The supershifted band can be more easily seen in the lane where anti-c-Jun and anti-Jun B were used together (c + B), due to the absence of complex T1. An antibody specific to Jun D did not appear to affect either complex T1 or T2. These supershift results were confirmed in nuclear extracts from primary microglia (Figure 4). None of the antibodies had a detectible effect on IL1A −889 allele-1 complexes (not shown).
Figure 3.
EMSA supershift analysis. BV2 microglia were treated with (+) or without (−) 300 ng/ml LPS for 4 h. Nuclear extracts in lanes 1–3 were probed with [32P]-labeled IL1A −889 allele-1 probe, while extracts in the remaining lanes were probed with allele-2 probe. Reactions designated by an asterisk (*) included the same unlabeled oligonucleotide at 100-fold molar excess. The nuclear extracts were combined with the indicated antibodies alone or in combination (e.g., “c+B” indicates 1 µl c-Jun antibody combined with 1 µl Jun B antibody). Unless otherwise indicated, the antibodies were used a 1 µl; all were combined with extracts prior to the addition of probe DNA. The boxes indicate complexes that were diminished by the presence of antibody, and the arrow indicates an apparent supershifted band in the presence of the Jun B antibody. Results are representative of 3 experiments.
Figure 4.
Confirmation in primary microglia. Primary microglia were treated with (+) or without (−) 100 ng/ml LPS for 4 h. Nuclear extracts were probed with [32P]-labeled IL1A −889 allele-2 probe. The nuclear extracts were combined with the indicated antibodies prior to the addition of probe DNA. Complexes that appear similar to T1 and T2 identified in BV2 cells are marked (left), and the arrow (right) indicates an apparent supershifted band in the presence of the Jun B antibody. Results are representative of 4 experiments.
One component of the activation of AP1 trans-acting factors is phosphorylation of their Jun-family components by Jun N-terminal kinase (JNK). The potential role of JNK in protein binding to IL1A −889 sequences was tested by application of a specific inhibitor of JNK: SP600125. The elevation in intensity of the T2 complex evoked by LPS was blocked by a pretreatment with SP600125 (Figure 5). However, the basal level of complex T2 in unstimulated cells was unaffected. Complex T1 was reduced by SP600125, not only in LPS-stimulated microglia, but in untimulated cells as well. DMSO (the vehicle for SP600125) had no effect on these complexes.
Figure 5.
JNK inhibitor SP600125 reduces LPS-induced IL1A allele-2 complex T2. Triplicate cultures of BV2 microglia were treated with 30 µM SP600125 or 0.3% DMSO (vehicle control) for 15 min prior to and throughout a 2-h treatment with 300 ng/ml LPS. For all lanes, BV2 nuclear extracts were incubated with the IL1A −889 allele-2 probe. Results are representative of 2 experiments.
Discussion
Our data demonstrate striking differences in the nucleoprotein (EMSA) complexes formed by probing microglial nuclear extracts with IL1A promoter oligonucleotides differing only at the −889 position. Further, only complexes formed with the allele-2 probe were increased by treatment of the cells with LPS. This suggests that the increased risk for disease imparted by IL1A −889 allele 2 may be rooted in inflammatory hyper-responsiveness. Competition EMSA revealed that the protein factors that contribute to allele-2 complexes prefer canonical CREB and AP1 binding sites to the allele-2 probe. Antibodies to c-Jun and Jun B were able to eradicate complex T1 and supershift a significant portion of complex T2, respectively, identifying these AP1 subunits as components of the LPS-activated allele-2 complexes. The specific JNK inhibitor SP600125 significantly reduced the formation of LPS-activated allele-2 complexes, indicating that phosphorylation of c-Jun, Jun B, and/or other unidentified binding partners potentiates formation of allele-2 complexes following activation of microglia with LPS.
Inhibition of allele-2 complex T1 (containing c-Jun) by SP600125 in the absence of LPS stimulation suggests that JNK was active at some basal level in the unstimulated cells, a situation which may or may not be duplicated in vivo. Allele-2 complex T2, which was more strikingly activated by LPS, and contains Jun B, was not inhibited by SP600125 in the absence of LPS. Together with the inability of the Jun-B antibody to supershift all of complex T2, this may indicate that other JNK-independent allele-2 complexes migrate at the same position as the Jun-B complexes. Even so, it is clear that different complexes form with allele-1 and allele-2 probes and that these differences are exaggerated in nuclear extracts from LPS-activated microglia, partially through the action of JNK.
While Jun B has a reputation as a negative regulator of transcription, it has also been reported that positive transactivation by Jun B is both promoter-specific and dependent on phosphorylation by JNK (Li et al. 1999). At least one report indicates higher promoter activity from allele-2 versus allele-1 promoter-reporter constructs (Wei et al. 2007). Our observed inhibition of LPS-activated allele-2 complex T2 by the JNK inhibitor SP600125 raises the possibility that LPS could activate transcription of the IL1A promoter in microglia through a Jun B complex, similar to activation of peroxiredoxin-I transcription by LPS in mouse macrophage cells (Bast et al. 2010). IL-4 transcription in T-cell differentiation is also dependent on phosphorylation of Jun B by JNK (Li et al. 1999).
As a proinflammatory cytokine, IL-1α plays a key position in the “cytokine cycle” hypothesis of Alzheimer disease (AD) (Griffin 2006). IL-1 induces each of the substrates of the most prominent neuropathological features of AD and AD with Lewy body disease (AD/LBD), e.g., Aβ plaques (βAPP), NFTs (phosphorylated tau) (Mrak and Griffin 2005), and Lewy bodies (Griffin et al. 2006b). Injurious events such as head trauma or stroke are associated with an elevated risk for AD and with the induction of IL-1 and other inflammation-related gene products in the brain. Such circumstances typically produce injured and dysfunctional neurons which overexpress the β-amyloid precursor protein (βAPP), promoting the deposition of Aβ (Roberts et al. 1991). Both Aβ and secreted forms of βAPP (sAPP) activate microglia to express elevated levels of IL-1 (Barger and Harmon 1997), perhaps explaining the link to IL-1 elevation in head trauma (Griffin et al. 1994). IL-1 may further damage neurons by promoting the overexpression and phosphorylation of the microtubule-associated protein tau (Griffin et al. 2006b), the requisite component of neurofibrillary tangles in AD. IL-1 also reduces levels of acetylcholine (Li et al. 2000) and participates in an age-related decline in synaptic plasticity (Griffin et al. 2006a). Thus, IL-1 is envisioned as creating a positive feedback for βAPP expression and other sequelae that amplify the cytokine cycle and progression of AD.
Conclusion
We find that proinflammatory activation of microglial cells promotes binding of Jun-family transcription factors to one version (allele 2) of polymorphic sequence in the promoter of the IL1A gene. This DNA sequence is associated with an elevated risk for inflammation-related disorders, including Alzheimer’s disease. The induction appears to be dependent upon JNK kinase(s). As the Jun-family transcription factors identified are stimulators of transcription, these findings are consistent with a prior study linking allele 2 to stronger promoter activity and add to the evidence that elevated expression of IL-1α contributes to the enhanced risk of disease in allele-2 carriers.
Table 1.
EMSA Oligonucleotide Sequences
| IL1A −889 allele 1 | 5’-AACCAGGCAACACCATTGAAGGCTC-3’ |
| IL1A −889 allele 2 | 5’-AACCAGGCAACATCATTGAAGGCTC-3’ |
| AP1 consensus | 5’-CGCTTGATGAGTCAGCCGGAA-3’ |
| CREB consensus | 5’-AGAGATTGCCTGACGTCAGAGAGCTAG-3’ |
| NF-κB consensus | 5’-AGTTGAGGGGACTTTCCCAGGC-3’ |
The sense strand of EMSA oligos used for competition is depicted above. The −889 polymorphic bases in the allele-1 and allele-2 oligonucleotides are in bold type. The putative binding site for each oligonucleotide’s target protein is underlined.
Acknowledgments
This work was supported by the National Institute on Aging (P01AG012411).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abazis-Stamboulieh D, Oikonomou P, Papadoulis N, Panayiotidis P, Vrakidou E, Tsezou A. Association of interleukin-1A, interleukin-1B and interleukin-1 receptor antagonist gene polymorphisms with multiple myeloma. Leuk Lymphoma. 2007;48:2196–2203. doi: 10.1080/10428190701615892. [DOI] [PubMed] [Google Scholar]
- Barger SW, Goodwin ME, Porter MM, Beggs ML. Glutamate release from activated microglia requires the oxidative burst and lipid peroxidation. J Neurochem. 2007;101:1205–1213. doi: 10.1111/j.1471-4159.2007.04487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger SW, Harmon AD. Microglial activation by secreted Alzheimer amyloid precursor protein and modulation by apolipoprotein E. Nature. 1997;388:878–881. doi: 10.1038/42257. [DOI] [PubMed] [Google Scholar]
- Bast A, Fischer K, Erttmann SF, Walther R. Induction of peroxiredoxin I gene expression by LPS involves the Src/PI3K/JNK signalling pathway. Biochim Biophys Acta. 2010;1799:402–410. doi: 10.1016/j.bbagrm.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Combarros O, Llorca J, Sanchez-Guerra M, Infante J, Berciano J. Age-dependent association between interleukin-1A (-889) genetic polymorphism and sporadic Alzheimer's disease. A meta-analysis. J Neurol. 2003;250:987–989. doi: 10.1007/s00415-003-1136-7. [DOI] [PubMed] [Google Scholar]
- Cordeiro CA, Moreira PR, Costa GC, Dutra WO, Campos WR, Orefice F, Teixeira AL. Interleukin-1 gene polymorphisms and toxoplasmic retinochoroiditis. Mol Vis. 2008;14:1845–1849. [PMC free article] [PubMed] [Google Scholar]
- de Jongh CM, John SM, Bruynzeel DP, Calkoen F, van Dijk FJ, Khrenova L, Rustemeyer T, Verberk MM, Kezic S. Cytokine gene polymorphisms and susceptibility to chronic irritant contact dermatitis. Contact Dermatitis. 2008;58:269–277. doi: 10.1111/j.1600-0536.2008.01317.x. [DOI] [PubMed] [Google Scholar]
- Dominici R, Cattaneo M, Malferrari G, Archi D, Mariani C, Grimaldi LM, Biunno I. Cloning and functional analysis of the allelic polymorphism in the transcription regulatory region of interleukin-1 alpha. Immunogenetics. 2002;54:82–86. doi: 10.1007/s00251-002-0445-9. [DOI] [PubMed] [Google Scholar]
- Griffin R, Nally R, Nolan Y, McCartney Y, Linden J, Lynch MA. The age-related attenuation in long-term potentiation is associated with microglial activation. J Neurochem. 2006a;99:1263–1272. doi: 10.1111/j.1471-4159.2006.04165.x. [DOI] [PubMed] [Google Scholar]
- Griffin WS. Inflammation and neurodegenerative diseases. Am J Clin Nutr. 2006;83:470S–474S. doi: 10.1093/ajcn/83.2.470S. [DOI] [PubMed] [Google Scholar]
- Griffin WS, Liu L, Li Y, Mrak RE, Barger SW. Interleukin-1 mediates Alzheimer and Lewy body pathologies. J Neuroinflammation. 2006b;3:5. doi: 10.1186/1742-2094-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin WS, Sheng JG, Gentleman SM, Graham DI, Mrak RE, Roberts GW. Microglial interleukin-1a expression in human head injury: correlations with neuronal and neuritic b-amyloid precursor protein expression. Neurosci Lett. 1994;176:133–136. doi: 10.1016/0304-3940(94)90066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi LM, Casadei VM, Ferri C, Veglia F, Licastro F, Annoni G, Biunno I, De Bellis G, Sorbi S, Mariani C, Canal N, Griffin WS, Franceschi M. Association of early-onset Alzheimer's disease with an interleukin- 1alpha gene polymorphism. Ann Neurol. 2000;47:361–365. [PubMed] [Google Scholar]
- Kolbus A, Walch K, Nagele F, Wenzl R, Unfried G, Huber JC. Interleukin-1 alpha but not interleukin-1 beta gene polymorphism is associated with polycystic ovary syndrome. J Reprod Immunol. 2007;73:188–193. doi: 10.1016/j.jri.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Kornman KS. Interleukin 1 genetics, inflammatory mechanisms, and nutrigenetic opportunities to modulate diseases of aging. Am J Clin Nutr. 2006;83:475S–483S. doi: 10.1093/ajcn/83.2.475S. [DOI] [PubMed] [Google Scholar]
- Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, Wilson TG, Jr, Higginbottom FL, Duff GW. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–77. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu L, Kang J, Sheng JG, Barger SW, Mrak RE, Griffin WST. Neuronal-glial interactions mediated by interleukin-1 enhance neuronal acetylcholinesterase activity and mRNA expression. J Neurosci. 2000;20:149–155. doi: 10.1523/JNEUROSCI.20-01-00149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak RE, Griffin WS. Glia and their cytokines in progression of neurodegeneration. Neurobiol Aging. 2005;26:349–354. doi: 10.1016/j.neurobiolaging.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Rainero I, Bo M, Ferrero M, Valfre W, Vaula G, Pinessi L. Association between the interleukin- 1alpha gene and Alzheimer's disease: a meta-analysis. Neurobiol Aging. 2004;25:1293–1298. doi: 10.1016/j.neurobiolaging.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Roberts GW, Gentleman SM, Lynch A, Graham DI. bA4 amyloid protein deposition in brain after head injury. Lancet. 1991;338:1422–1423. doi: 10.1016/0140-6736(91)92724-g. [DOI] [PubMed] [Google Scholar]
- Tsezou A, Poultsides L, Kostopoulou F, Zintzaras E, Satra M, Kitsiou-Tzeli S, Malizos KN. Influence of interleukin 1alpha (IL-1alpha), IL-4, and IL-6 polymorphisms on genetic susceptibility to chronic osteomyelitis. Clin Vaccine Immunol. 2008;15:1888–1890. doi: 10.1128/CVI.00209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Chen X, Fontanilla C, Zhao L, Liang Z, Dodel R, Hampel H, Farlow M, Du Y. C/T conversion alters interleukin-1A promoter function in a human astrocyte cell line. Life Sci. 2007;80:1152–1156. doi: 10.1016/j.lfs.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]