An Arg 132 mutation in isocitrate dehydrogenase 1 (IDH1) has been identified in gliomas, glioblastomas, and acute myeloid leukemia, but its physiological significance is not known. Here, Sasaki et al. have generated and characterized brain-specific Idh1 R132H conditional knock-in mice to explore the correlation of the IDH1 mutation with tumorigenesis. Their results suggest that D2HG accumulation as a result of this mutation produces multiple metabolic effects that preclude embryo viability and may contribute to tumor progression. For example, D2HG blocks prolyl-hydroxylation of collagen, causing a defect in collagen protein maturation. An endoplasmic reticulum (ER) stress response induced by the accumulation of immature collagens may account for the embryonic lethality of these mutants. Importantly, D2HG-mediated impairment of collagen maturation also led to basement membrane (BM) aberrations.
Keywords: metabolism, brain hemorrhage, oxidative stress, angiogenesis, basement membrane, collagen biosynthesis
Abstract
Isocitrate dehydrogenase-1 (IDH1) R132 mutations occur in glioma, but their physiological significance is unknown. Here we describe the generation and characterization of brain-specific Idh1 R132H conditional knock-in (KI) mice. Idh1 mutation results in hemorrhage and perinatal lethality. Surprisingly, intracellular reactive oxygen species (ROS) are attenuated in Idh1-KI brain cells despite an apparent increase in the NADP+/NADPH ratio. Idh1-KI cells also show high levels of D-2-hydroxyglutarate (D2HG) that are associated with inhibited prolyl-hydroxylation of hypoxia-inducible transcription factor-1α (Hif1α) and up-regulated Hif1α target gene transcription. Intriguingly, D2HG also blocks prolyl-hydroxylation of collagen, causing a defect in collagen protein maturation. An endoplasmic reticulum (ER) stress response induced by the accumulation of immature collagens may account for the embryonic lethality of these mutants. Importantly, D2HG-mediated impairment of collagen maturation also led to basement membrane (BM) aberrations that could play a part in glioma progression. Our study presents strong in vivo evidence that the D2HG produced by the mutant Idh1 enzyme is responsible for the above effects.
In 2008, a genome-wide mutational analysis identified somatic mutations of isocitrate dehydrogenase 1 (IDH1) in glioblastomas (GBMs; World Health Organization [WHO] grade IV glioma) (Parsons et al. 2008). Subsequent studies revealed mutations in either IDH1 or its mitochondrial counterpart, IDH2, in >70% of grade II–III gliomas and secondary GBMs and in ∼20% of acute myeloid leukemias (AMLs) (Mardis et al. 2009; Yan et al. 2009). IDH1 mutations are found most often in tumors of younger individuals and correlate with a better prognosis (Parsons et al. 2008; Sanson et al. 2009; Yan et al. 2009; Hartmann et al. 2010).
Mammalian IDH1 and IDH2 proteins form homodimers that catalyze the conversion of isocitrate to α-ketoglutarate (αKG; also known as 2-oxoglutarate) and concomitantly produce reduced nicotinamide adenine dinucleotide phosphate (NADPH) from NADP+. The most common IDH1 and IDH2 mutations affect a single amino acid residue: Arg 132 (R132) in IDH1, and R172 or R140 in IDH2 (Parsons et al. 2008; Mardis et al. 2009; Yan et al. 2009; Green and Beer 2010; Ward et al. 2010). Patients are generally heterozygous for these mutant alleles, and the mutated subunit is believed to have a dominant-negative effect (Zhao et al. 2009). The IDH1 R132 mutant protein loses its ability to generate αKG but gains abnormal catalytic activity that allows it to convert αKG to D-2-hydroxyglutarate [D2HG; also known as R(−)-2-hydroxyglutarate] (Dang et al. 2009; Gross et al. 2010; Ward et al. 2010). Cells expressing IDH1 R132 mutant proteins have decreased NADPH not only due to the loss of the isocitrate–αKG reaction, but also because NADPH is consumed (at least in vitro) during the abnormal conversion of αKG to D2HG. Because NADPH is a key component of cellular anti-oxidation systems, the possibility exists that IDH1/2 mutations may decrease intracellular NADPH and also increase intracellular reactive oxygen species (ROS). Failed control of intracellular ROS has been associated with not only cellular senescence and apoptosis, but also tumorigenesis (Finkel and Holbrook 2000). However, there is, as yet, no direct evidence in vivo that IDH1/2 mutations influence intracellular ROS and promote tumor development.
D2HG and αKG are structurally identical, except that the C2 carbonyl group in αKG is replaced by a hydroxyl group in D2HG. D2HG may thus compete with αKG and inhibit various αKG-dependent enzymes, including the ten-eleven translocation (TET) family of 5-methylcytosine hydroxylases and the Jumonji-C domain-containing histone demethylases (JHDMs) (Chowdhury et al. 2011; Xu et al. 2011). In normal cells, these enzymes mediate epigenetic changes, such as modifications to cytosine nucleotides and histone proteins, that maintain cellular homeostasis. In cancer cells, these patterns of epigenetic modification are altered and constitute a key hallmark of tumorigenesis. It has been proposed that D2HG-mediated inhibition of the activities of TET hydroxylases and JHDMs may contribute to tumors associated with IDH1/2 mutations (Figueroa et al. 2010; Noushmehr et al. 2010).
Another group of αKG-dependent enzymes includes the prolyl hydroxylases. Important substrates of prolyl hydroxylase domain-containing proteins (PHDs; also known as EGLNs) and collagen prolyl 4-hydroxylases are the hypoxia-inducible transcription factors 1/2 α (HIF1/2α) and various collagen proteins, respectively. In the presence of oxygen, PHDs carry out prolyl-hydroxylation of HIF1/2α proteins, which then interact with the von Hippel-Lindau protein (pVHL)-ubiquitin E3 ligase and undergo ubiquitination and proteasomal degradation (Rankin and Giaccia 2008). HIF target gene expression is thus suppressed. Conversely, PHDs are inhibited under low-oxygen conditions or in the presence of succinate or fumarate (Isaacs et al. 2005; Selak et al. 2005). HIF1/2α are stabilized, resulting in enhanced expression of HIF target genes that drive angiogenesis, glucose metabolism, and other signaling pathways critical for tumor growth. With respect to the effects of D2HG on PHD regulation, two in vitro studies recently examined this issue but yielded conflicting results (Xu et al. 2011; Koivunen et al. 2012).
Although much of the available evidence implies that IDH1/2 mutation correlates well with tumorigenesis, most of these findings were obtained using correlative clinical data and gene overexpression systems; appropriate genetically engineered mouse models have yet to be characterized. To clearly define the effects of IDH1 mutation in vivo, we generated brain-specific Idh1 R132H knock-in (Idh-KI) mice. Brain-specific Idh1-KI mice show brain hemorrhage accompanied by high D2HG levels but decreased ROS. HIF1α proteins are stabilized due to impaired prolyl-hydroxylation, and HIF target gene expression is up-regulated. Importantly, collagen maturation is aberrant, perturbing basement membrane (BM) formation and triggering an endoplasmic reticulum (ER) stress response. We suggest that these defects are attributable to D2HG produced by the mutant Idh1 enzyme and that these metabolic disruptions are responsible for the embryonic lethality, brain hemorrhage, and potentially tumorigenic alterations to the BM that we observed in our mutant mice.
Results
Brain-specific Idh1 R132 mutation results in perinatal lethality
Previous in vitro studies have indicated that the mutant protein encoded by IDH1 R132 gains abnormal catalytic activity that allows it to produce D2HG (Dang et al. 2009; Gross et al. 2010; Ward et al. 2010). To determine whether this effect occurs in vivo, we crossed Idh1LSL/wt mice (Sasaki et al. 2012) with Nestin-Cre transgenic mice (Tronche et al. 1999) to generate Nes-Idh1R132H/wt (Nes-KI) mice and the corresponding control Nes-Idh1wt/wt (Nes-WT) animals. Under the control of Nestin, Cre is expressed as early as embryonic day 10.5 (E10.5) in neural stem cells (NSCs), and nearly complete recombination in all CNS cells is achieved by E12.5 (Graus-Porta et al. 2001). We found that Nes-KI mice were born at the expected Mendelian ratio but died shortly after birth (Table 1). Gross examination of whole brains from postnatal day 0 (P0) Nes-KI and Nes-WT mice revealed that expression of the mutant Idh1 protein driven by the Nestin promoter resulted in massive hemorrhage within the cerebral hemispheres and cerebellum (Fig. 1A). Histological analysis confirmed the lack of cerebral development and the presence of large cavities in Nes-KI brains at P0 (Fig. 1B). Even at E15.5, significant cavitation and hemorrhage were already present in Nes-KI embryos (Supplemental Fig. 1A,B), and the laminar nature of the cortex was disturbed (Fig. 1C–E). Cellular proliferation in Nes-KI brains at E15.5 was virtually absent, as determined by BrdU incorporation (Fig. 1F–H). Massive cell death throughout these brains was evidenced by the presence of an increased number of cells positive for cleaved caspase-3, a marker of apoptosis (Fig. 1I–K). Notably, there were no obvious differences between Nes-KI and Nes-WT embryos in NSC distribution at E12.5 (Supplemental Fig. 1C–H).
Table 1.
Genotypes of live-born mice and embryos from Nestin-Cre × Idh1LSL/wt crosses

Figure 1.
Histological characterization of brains of Nes-Idh1KI/wt (Nes-KI) mice. (A) Dorsal and ventral views of whole brains dissected from Nes-WT (control) or Nes-KI mice at the newborn stage (P0) or at E16.5 or E14.5. Hemorrhage (white arrows) was observed in Nes-KI embryos starting at E14.5. Bars, 2 mm. (B) Sagittal sections of brains of Nes-WT and Nes-KI P0 embryos that were immunostained to detect BrdU. Bars, 2 mm. (C–K) Sagittal sections of the brains of one Nes-WT (C,F,I) and two Nes-KI E15.5 embryos (D,E,G,H,J,K) that were stained with H&E (C–E) or immunostained to detect BrdU (F–H) or cleaved caspase-3 (I–K). Black arrowheads indicate brain cells positive for BrdU or cleaved caspase-3. Small black arrows indicate regions of hemorrhage within the brain. Bars, 100 μm. For A–K, results are representative of at least three embryos/group.
To extend our results to another brain-specific promoter, we crossed Idh1LSL/wt mice with GFAP-Cre transgenic mice (Kwon et al. 2001) to generate GFAP-KI mutants and the corresponding GFAP-WT control mice. Previous reports on GFAP-Cre transgenic mice have indicated that Cre is expressed starting at E14.5 but only in the astrocytes of these animals and not in their neurons (Bajenaru et al. 2002). GFAP-KI embryos did not show any significant abnormalities until E15.5, after which ∼60% of embryos exhibited brain hemorrhage (Supplemental Fig. 2A,B). Interestingly, GFAP-KI embryos of the same litter demonstrated variable symptoms (Supplemental Fig. 2B), and, unlike Nes-KI mice, 8.3% of GFAP-KI mice survived until adulthood (Supplemental Table 1). This generally milder phenotype of GFAP-KI embryos may be partly due to the variable mosaic pattern of Cre expression documented among GFAP-Cre mice (see below).
D2HG accumulates to high levels in Nes-KI brains
D2HG accumulates in glioma and AML patients whose malignancies bear IDH1 or IDH2 mutations and inhibits αKG-dependent dioxygenase activity by competing with αKG (Dang et al. 2009; Gross et al. 2010; Ward et al. 2010; Chowdhury et al. 2011; Xu et al. 2011). When we measured αKG and D2HG levels in Nes-KI brain cells, αKG was modestly decreased in the mutant compared with the Nes-WT control, as expected (Fig. 2A). Strikingly, huge amounts of D2HG were detected in the brain cells of both Nes-KI and GFAP-KI mice compared with controls (Fig. 2B; Supplemental Fig. 3A). Significantly, GFAP-KI mice with brain hemorrhage also showed high levels of D2HG, whereas GFAP-KI mice without hemorrhage did not (Supplemental Fig. 3B). Thus, D2HG levels correlate closely with the hemorrhage phenotype.
Figure 2.
Altered αKG and D2HG levels correlate with expression of Idh1 R132 protein in mouse brains. (A) Brains of Nes-WT (n = 4) and Nes-KI (n = 6) E14.5 embryos were dissociated to generate single-cell suspensions, and intracellular αKG levels were determined in extracts of 2 × 106 brain cells. (B) Brain tissue extracts from Nes-WT (n = 4) and Nes-KI (n = 5) E13.5 embryos were assayed by LS-MS to detect D2HG. For A and B, results are the mean ± SEM of triplicates; P-values, unpaired Student's t-test.
ROS levels are reduced in Nes-KI brains
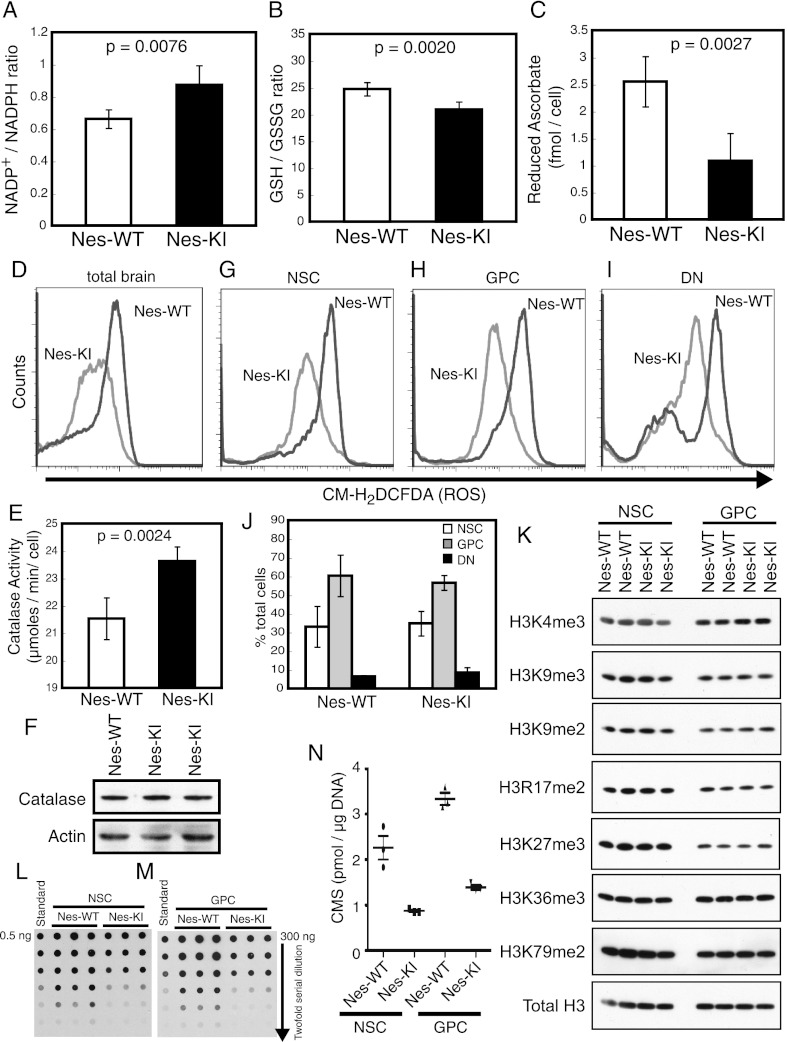
To elucidate the mechanisms underlying the brain hemorrhage observed in Idh1-KI mice, we first investigated the regulation of ROS in the mutant brain cells. Since the mutated IDH1 protein loses catalytic activity for the normal αKG- and NADPH-generating reaction and gains abnormal enzymatic reactivity that produces D2HG and consumes NADPH, it is plausible that cells expressing the mutated IDH1 enzyme would show an increase in their intracellular NADP+/NADPH ratio. As NADPH is a cofactor in the recycling of anti-oxidant molecules such as glutathione (GSH) and ascorbate, it has also been assumed that such an increase in the NADP+/NADPH ratio would lead to a corresponding increase in intracellular ROS. Indeed, total brain cells from E14.5 Nes-KI embryos showed a slight but significant increase in their intracellular NADP+/NADPH ratio compared with Nes-WT controls (Fig. 3A), consistent with a loss of conventional NADPH production and enhanced NADPH consumption. In addition, intracellular levels of reduced GSH and ascorbate were decreased in Nes-KI cells (Fig. 3B,C), implying that the observed reduction in NADPH attenuates the recycling of GSH and ascorbate. However, to our surprise, intracellular ROS levels measured using the CM-H2DCFDA fluorescent probe were not elevated in total Nes-KI brain cells but instead were dramatically decreased (Fig. 3D). One possible explanation for this reduction of intracellular ROS may be due to enhanced catalase activity, but not protein expression, detected in the mutant cells (Fig. 3E,F).
Figure 3.
Altered intracellular ROS, increased NADP+/NADPH ratio, and epigenetic modifications in Nes-KI mice. (A) The effect of the mutant Idh1 enzyme on NADP+/NADPH homeostasis was evaluated in brain cells of Nes-WT (n = 6) and Nes-KI (n = 3) E14.5 embryos. The intracellular NADP+/NADPH ratio was determined as described in the Materials and Methods. (B) The GSH/GSSG ratio was determined to evaluate GST utilization in the brain cells of Nes-WT (n = 5) and Nes-KI (n = 5) E14.5 embryos. (C) The amount of reduced ascorbate was determined to evaluate ascorbate utilization in the brain cells of Nes-WT (n = 5) and Nes-KI (n = 4) E14.5 embryos. (D) ROS production by total brain cells of Nes-WT and Nes-KI E14.5 embryos was measured by flow cytometry using the fluorescent dye CM-H2DCFDA. (E) Catalase activity was evaluated in the brain cells of Nes-WT (n = 5) and Nes-KI (n = 4) E14.5 embryos. (F) Immunoblot of catalase protein in brain extracts from the indicated Nes-WT and Nes-KI embryos. Actin was used as a loading control. (G–I) ROS production by NSCs (prominin+) (G), GPCs (prominin−A2B5+) (H), and other brain cell types (DNs; prominin−A2B5−) (I) from Nes-WT and Nes-KI E13.5 embryos was measured by flow cytometry using the fluorescent dye CM-H2DCFDA. (J) Proportions of NSCs, GPCs, and DNs among total brain cells from the embryos in G–I. No alterations due to Idh1 R132 mutation were observed. (K) Acid extracts of brains from Nes-WT and Nes-KI E14.5 embryos were immunoblotted to detect levels of the indicated H3-methylated histones. Total H3 was used as a loading control. (L–N) Genomic DNA (300 ng) of NSCs (L) and GPCs (M) isolated from the brains of Nes-WT (n = 3) and Nes-KI (n = 3) E14.5 embryos was serially diluted and subjected to cytosine 5-methylenesulfonate (CMS) dot blotting to quantify 5hmC levels (Ko et al. 2010). A synthetic oligonucleotide (0.5 ng) containing a known amount of CMS was serially diluted and used as the standard. Data are representative of two independent experiments. Results of L and M are summarized graphically in N. (Horizontal lines) Median values (n = 3). For A–C and E, results are the mean ± SEM of triplicates; P-values, unpaired Student's t-test. For D, F, G–J, results are representative of three independent trials. For K–N, results are representative of two independent trials.
We then examined intracellular ROS levels in various subpopulations of embryonic brain cells. Moderate ROS accumulation in NSCs reportedly promotes their proliferation and differentiation (Le Belle et al. 2011), so we wondered whether a drop in intracellular ROS would alter NSC numbers and perhaps account for the brain hemorrhage observed in Idh1-KI mice. For our analysis, we monitored the expression of Prominin-1 (CD133 in humans) as a marker of NSCs, and A2B5 as a marker of glial-restricted precursor cells (GPCs). The double-negative (DN) subpopulation that lacks both of these markers includes neural-restricted precursor cells, terminally differentiated neurons and glial cells, and other cell types such as endothelial cells. We found that, compared with Nes-WT controls, ROS levels were reduced in the Prominin-1+ (NSC), Prominin-1−A2B5+ (GPC), and Prominin-1−A2B5− (DN) cell populations in Nes-KI brains (Fig. 3G–I). However, the proportions of these three subsets among total Nes-KI brain cells were normal and consistent with the pattern of Nestin expression in Nes-KI brains (Fig. 3J; Supplemental Fig. 1D,G). Taken together, these results indicate that the increased cell death and reduced proliferation in the mutant brains cannot be due to ROS accumulation and suggest that the Idh1 R132 mutation does not influence NSC proliferation or differentiation, at least during the earliest stages of embryonic brain development.
In addition to its effects on ROS levels, D2HG reportedly increases histone methylation because it inhibits the JHDMs required for cell differentiation (Lu et al. 2012). However, we found no differences in histone H3 methylation between the E14.5 control and Nes-KI NSCs or GPCs (Fig. 3K). On the other hand, Nes-KI NSCs and GPCs both showed reduced levels of 5-hydroxymethylcytosine (5hmC) compared with controls (Fig. 3L–N), presumably due to D2HG-mediated inhibition of TET hydroxylases. Precisely how such altered epigenetic modifications could contribute to the phenotypes of Idh1-KI mice remains to be clarified.
D2HG stabilizes Hif1α proteins in Nes-KI mice
Xu et al. (2011) previously demonstrated that D2HG blocks the hydroxylation of proline residues in HIF1α proteins, but Koivunen et al. (2012) recently presented evidence suggesting that D2HG can activate PHDs. To resolve this controversy, we assessed Hif1α expression in our Nes-KI mutants. We detected higher levels of HIF1α protein in Nes-KI embryos compared with controls (Fig. 4A), but the same was not true for Hif1a mRNA (Fig. 4B,C, left). Moreover, the increased stabilization of Hif1α protein was observed even in E14.5 Nes-KI brains showing little or no hemorrhage. When we examined the mRNA expression of Hif1α target genes in Nes-KI embryos, we found that transcription of the vascular endothelial growth factor (Vegf) gene was preferentially up-regulated in E13.5–E16.5 Nes-KI brains compared with Nes-WT controls (Fig. 4B,C). Transcription levels of the Hif1α target genes glucose transporter-1 (Glut-1) and phosphoglycerate kinase 1 (Pgk1) were also moderately up-regulated at E15.5–E16.5 (Fig. 4C). On the other hand, the expression of angiopoietin-1 (Ang1), which is not a Hif1α target gene, was unaffected. Thus, mutation of Idh1 is associated with stabilized Hif1α and up-regulated Hif1α-dependent transcription. We therefore concur with Xu et al. (2011) that the D2HG associated with the Idh1 mutation blocks PHD-mediated modification of HIF1α protein and thus promotes its transactivation activity.
Figure 4.
Idh1 R132 stabilizes Hif1α protein and induces Hif1α target gene expression. (A) Brain tissue extracts from the indicated Nes-WT and Nes-KI embryos were immunoblotted to detect Hif1α protein levels. Actin was used as a loading control. Results are representative of three trials. (B,C) qRT–PCR analysis of Hif1a, Vegf, Glut-1, Pgk1, and Ang1 mRNA levels in Nes-WT and Nes-KI embryos at E13.5–E14.5 (B) or E15.5–E16.5 (C) (n = 7 per group). Data are expressed as the mean fold difference between Nes-WT (set to 1) and Nes-KI values after normalization to 18S rRNA. Results are the mean ± SEM of triplicates; P-values, unpaired Student's t-test. (NS) Not significant.
Collagen maturation is impaired in Nes-KI mice
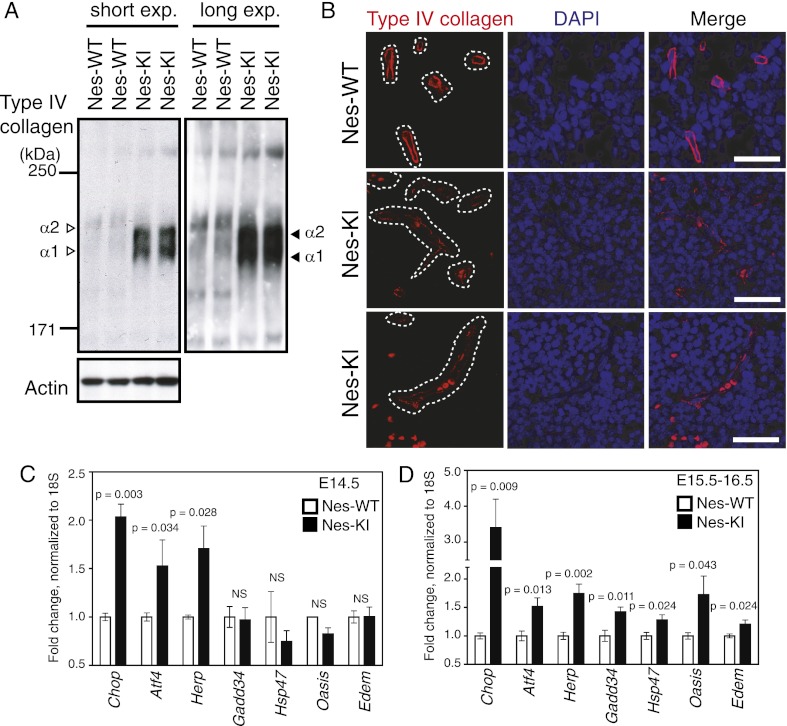
The impaired proline hydroxylation of Hif1α in Nes-KI brains prompted us to explore whether other processes involving prolyl hydroxylases were affected in our mutants. Prolines in collagen proteins are hydroxylated by collagen prolyl-4-hydroxylases I, II, and III (C-P4H I–III), and lysines in collagen proteins are hydroxylated by the procollagen-lysine 2-oxoglutarate 5-dioxygenases 1, 2, and 3 (Plod1–3) (Kivirikko and Myllylä 1985; Myllyharju and Kivirikko 2004). Increased amounts of hydroxyproline (Hyp) and hydroxylysine (Hyl) residues in a mature collagen protein reinforce its helical structure and promote its thermal stability (Kivirikko and Myllylä 1985; Myllyharju and Kivirikko 2004). Type IV collagen is a key BM component and promotes interactions between astrocytes and endothelial cells (Kalluri 2003). We hypothesized that the D2HG accumulation in Nes-KI mice might attenuate collagen modification by αKG-dependent enzymes and thus disrupt type IV collagen maturation, leading to BM breakdown. To investigate this possibility, we first analyzed levels of soluble collagen in Nes-KI and Nes-WT brains, since an increased proportion of soluble collagen indicates a lack of maturation of this protein. We recovered the fraction of cell extract proteins soluble in Triton X-100 and performed immunoblotting to detect type IV collagen. Soluble type IV collagens α1 and α2 were dramatically increased in tissues of Nes-KI mutants compared with Nes-WT controls (Fig. 5A). However, when we evaluated type IV collagen mRNAs in the brains of these embryos, four out of six Col4a mRNAs were expressed at comparable levels in mutant and control cells (Supplemental Fig. 4A). Thus, the increase in soluble type IV collagen proteins in Nes-KI brain cells is most likely due to the accumulation of immature, nonhelical forms of these molecules. Consistent with this hypothesis, the migration in a 5% SDS gel of the band representing the type IV collagen protein in the Nes-KI mutant was faster than the band in the Nes-WT extract (Fig. 5A), implying that Hyl-mediated glycosylation was impaired. Similar phenotypes have been observed in tissues of mice deficient for P4ha1 (the α subunit of C-P4H I) or Plod3 (Rautavuoma et al. 2004; Holster et al. 2007). Thus, we propose that the excess D2HG present due to the activity of the mutated Idh1 enzyme impairs Hyp- and Hyl-mediated modification of collagen proteins, preventing their maturation.
Figure 5.
D2HG impairs collagen maturation and BM formation and induces an ER stress response. (A) Triton X-100-soluble proteins were prepared from the brains of E14.5 Nes-WT and Nes-KI embryos, and equal amounts of protein were subjected to 5% SDS-PAGE under reducing conditions. The indicated proteins were detected by immunoblotting. The difference in the size of type IV collagens α1 and α2 between Nes-WT and Nes-KI extracts reflects the reduced Hyl content of the collagen produced in mutant cells. Results are representative of four trials. (Unfilled arrowheads) α1/α2 type IV collagen bands in Nes-WT extracts; (filled arrowheads) immature α1/α2 type IV collagen bands in Nes-KI extracts. (B) Immunofluorescence analysis of type IV collagen (red) in the brains from one Nes-WT and two Nes-KI E15.5 embryos. Nuclei were counterstained with DAPI (blue). (White dotted lines) Outlines of blood vessel structures. Bars, 25 μm. (C,D) Total RNA was isolated from the brains of Nes-WT (n = 5) and Nes-KI (n = 5) embryos at E14.5 (C) or E15.5–16.5 (D), and mRNA levels of the indicated ER stress-responsive genes were analyzed by qRT–PCR. Data are expressed as the mean fold difference between Nes-WT (set to 1) and Nes-KI values after normalization to 18S rRNA. Results are the mean ± SEM of triplicates; P-values, unpaired Student's t-test.
To confirm the physiological significance of these in vitro findings, we performed immunofluorescence staining of type IV collagen to analyze its distribution between blood vessels and perivascular astrocytes. Intense, continuous staining of blood vessel type IV collagen was present in Nes-WT E15.5 brains, whereas only faint, dispersed staining was seen in the mutant (Fig. 5B). These data further support our hypothesis that D2HG attenuates type IV collagen maturation and thereby impairs BM structure and function.
Defective collagen maturation in Nes-KI mice is associated with an ER stress response
C-P4H I–III and Plod1–3 are localized in the ER. When the activities of these enzymes are suppressed, newly synthesized collagens are not properly modified, leading to the accumulation of unfolded proteins in the ER (Marutani et al. 2004; Rautavuoma et al. 2004; Holster et al. 2007). Because the presence of these abnormal proteins in bulk triggers an ER stress response that leads to apoptosis (Ron and Walter 2007), we hypothesized that such a response might be induced by the impaired collagen maturation in Nes-KI mutants. To test this idea, we used quantitative RT–PCR (qRT–PCR) to examine the mRNA levels of several ER stress-responsive genes in the brains of Nes-KI and Nes-WT embryos at E14.5–E16.5. Specifically, we assayed ER stress-responsive transcription factors (Atf4 and Chop), ER chaperones (Hsp47, Herp, Edem1, Grp78/Bip, Grp94, and p58IPK), an ER stress sensor protein (Oasis), and other ER stress-related proteins (Gadd34, PDI, and Erol1). Of these 12 genes, three (Chop, Atf4, and Herp) were significantly up-regulated in E14.5 Nes-KI brains, whereas seven (Chop, Atf4, Hsp47, Herp, Edem1, Oasis, and Gadd34) were up-regulated by E15.5–E16.5 (Fig. 5C,D; Supplemental Fig. 4B). This elevation of ER stress-responsive gene expression is most likely responsible for the increased apoptosis observed in Nes-KI brains.
GFAP-KI mice do not develop glioma
To determine whether the Idh1 R132H mutation on its own can drive glioma formation, we monitored the health and longevity of our live-born GFAP-KI mice (8.3% of the total) (Supplemental Table 1). Surviving GFAP-KI mice showed a markedly shorter life span than GFAP-WT mice (Supplemental Fig. 5A) without observable glioma formation. These results suggest that, in the brain, expression of the mutated Idh1 enzyme alone is insufficient for gliomagenesis. Alternatively, glioma may be formed at a later time point, but this may be complicated by the reduced life span of the mutant mice.
Intriguingly, many surviving GFAP-KI mice exhibited splenomegaly (39%) and/or liver tumors (21%) or other abnormalities (Supplemental Fig. 5B–D). GFAP-KI mice with splenomegaly showed a modest increase in total white blood cells and reticulocytes and a decrease in red blood cells but no alteration to platelet numbers (Supplemental Fig. 5E). To determine the cause of these non-brain-related phenotypes, we verified the expression pattern of GFAP-Cre with Rosa26-LSL-TdTomato reporter mice (Madisen et al. 2010). GFAP-Cre-mediated expression of TdTomato protein was observed not only in the brains, but also in a wide variety of other tissues. In most mutants, Cre was expressed in a mosaic pattern unique to each individual, but some mutants showed high Cre expression throughout their whole bodies (Supplemental Fig. 6). Thus, it is most likely that the non-brain phenotypes in GFAP-KI mice arose due to leaky expression of Cre in hematopoietic cells. It has been previously shown that Tet2 knockout mice also exhibit defects in hematopoietic stem cell homeostasis but show no brain abnormalities (Ko et al. 2011; Li et al. 2011; Moran-Crusio et al. 2011; Quivoron et al. 2011). This similarity in phenotypes between GFAP-KI mice and Tet2 knockout mice suggests both that hematopoietic cells are probably more sensitive to D2HG than brain cells and that these phenotypes may partly depend on D2HG-mediated inhibition of Tet2. We also examined the phenotypes of mice in which the Idh1-KI mutation is driven by Cre under the control of the myeloid lineage-specific promoter LysM (Sasaki et al. 2012). These animals have hematopoietic phenotypes similar, but not identical, to those of GFAP-KI mice, presumably reflecting a different Cre expression pattern under the two different promoters in various hematopoietic lineages.
An obvious way to enhance the chance of glioma formation in GFAP-KI mice was to cross these animals to mutants with a deletion of the tumor suppressor p53. However, due to the leaky expression of GFAP-Cre, both control GFAP-WT/p53flox/flox mice and GFAP-KI/p53flox/flox double-mutant mice developed the same broad spectrum of tumors (Supplemental Fig. 7). It remains a formidable challenge to generate a live Idh1-KI mouse that expresses mutant Idh1 protein only in the brain, can be crossed into a tumor-prone background, and can thus be used to identify the other factors that combine with altered Idh1 activity to drive glioma formation.
Discussion
In this study, we provide the first in vivo evidence that the aberrant metabolite D2HG associated with the Idh1 R132H mutation causes a failure in mouse embryonic brain development. Our major findings (illustrated in Fig. 6A) are as follows: (1) Brain-specific expression of the mutant form of Idh1 causes brain hemorrhage. (2) Intracellular ROS levels are dramatically reduced in Idh1-KI brain cells, which also show an increased NADP+/NADPH ratio and catalase activity. (3) D2HG is associated with Hif1α stabilization that up-regulates the expression of Vegf and other Hif1α target genes. (4) D2HG impairs collagen maturation, disrupting BM structure and triggering an ER stress response.
Figure 6.
The Idh1 R132 mutant protein profoundly alters the microenvironment supporting the interaction between astrocytes and endothelial cells, resulting in brain hemorrhage. (A) Relationship between Nes-Cre expression, D2HG production, and mutant mouse phenotypes. Schematic time line indicates Nes-Cre expression, with D2HG production shown in red. Arrows indicate the overall outcome for a given phenotype. (B) Model postulating how the Idh1 R132 mutant protein could promote brain hemorrhage. In a wild-type (IDH1-WT) astrocyte, type IV procollagen proteins in the ER are modified at their Pro and Lys residues, which allows the assembly of mature macromolecular collagen molecules that are secreted into the extracellular space and contribute to BM formation. An intact BM is crucial for sustaining proper interaction between astrocytes and endothelial cells. Routine degradation of HIF1/2α mediated by the actions of PHD and VHL maintains the intracellular homeostasis of wild-type astrocytes. In contrast, in astrocytes expressing the IDH1 R132 mutant protein (IDH1 Mut), we propose that intracellular homeostasis is perturbed by the large amounts of D2HG produced by the abnormal reaction catalyzed by IDH1 R132. D2HG inhibits collagen maturation, resulting in the formation of a fragile BM. Elevated D2HG also triggers an ER stress response, causing intrinsic cell death, and stabilizes HIF1/2α, leading to increased VEGF that drives aberrant blood vessel formation. These imbalances alter the microenvironment surrounding astrocytes and endothelial cells and affect the interaction between them, resulting in brain hemorrhage.
Idh1 mutation and ROS regulation
The conversion of intracellular NADPH to NADP+ supplies vital reducing power for the generation of anti-oxidant molecules such as reduced GSH and ascorbic acid (Finkel and Holbrook 2000). As expected, we detected an increase in the NADP+/NADPH ratio and a decrease in GSH and reduced ascorbic acid in Idh1-KI cells. However, although an elevated NADP+/NADPH ratio has been assumed to induce ROS accumulation, we found dramatically lower ROS levels and increased catalase activity in Idh1-KI cells. Thus, excessive oxidative stress is not the cause of the brain hemorrhage in Idh1-KI mice. The normal Idh1 protein is localized in both the cytosol and the peroxisome, where catalase resides. Peroxisomes are organelles with essential metabolic functions that affect the generation and decomposition of ROS (Geisbrecht and Gould 1999). It is possible that aberrant metabolism in the peroxisomes of Idh1-KI mice results in the observed dysregulation of anti-oxidants. In addition to changes in catalase activation, alterations in the level of NADPH oxidases, which are enzymes that generate superoxide from oxygen and NADPH (Ying 2008), may also contribute to the observed phenotype, although it is technically challenging to experimentally interrogate this idea.
Idh1 mutation and brain hemorrhage
Brain hemorrhage is an often fatal complication of premature birth, vascular malformations, and brain disorders. The mechanisms that cause brain hemorrhage may be specific to this organ because of the close connection between brain endothelial cells and perivascular astrocytes (Ballabh et al. 2004). Two lines of evidence support our view that D2HG accumulation is directly linked to brain hemorrhage.
Up-regulated HIF target gene expression
In normal mice, angiogenesis is regulated by a balance of endogenous proangiogenic and anti-angiogenic factors such that a breakdown in this balance leads to abnormal angiogenesis and hemorrhage (Carmeliet and Jain 2000). One important proangiogenic factor is Vegf, which is an endothelial cell mitogen that activates these cells to release proteases, proliferate, migrate, and form new vascular structures (Ferrara et al. 2003). In our study, high D2HG levels in vivo correlated with Hif1α stabilization and increased expression of several HIF target genes, including Vegf (Fig. 4). Because numerous reports have indicated that Vegf overproduction causes blood vessel hyperformation and sometimes hemorrhage (Cheng et al. 1997; Belteki et al. 2005; Jung et al. 2006), we propose that the excess Vegf present in the brains of Nes-KI and GFAP-KI mice contributed to their hemorrhage. It remains possible that the Vegf overproduction that we observed is not due as much to the effects of D2HG on Hif1α as to the stabilization of HIF proteins caused by hypoxia induced by the abnormal blood supply present in Idh1-KI brains. This issue remains to be resolved.
Impaired collagen maturation
Among the 28 members of the collagen superfamily, type IV collagen is a major component of the BM. Six different type IV collagen α chains (α1–6) have been identified that all have the same basic domain configuration: an N-terminal 7S domain, a middle triple-helical domain with a characteristic Gly-X-Y motif, and a C-terminal globular noncollagenous (NC1) domain (Kalluri 2003). Studies of P4ha1- or Plod3-deficient mice have shown that type IV collagen maturation is easily derailed by impaired prolyl-/lysyl-hydroxylation and that the phenotypes of these mutants are highly similar to that of Col4a1/2-deficient mice (Pöschl et al. 2004; Rautavuoma et al. 2004; Holster et al. 2007). Importantly, mutation of Col4a1 is one of the risk factors for brain hemorrhage in both humans and mice (Gould et al. 2005).
In our study, the accumulation of D2HG in Idh1-KI mice inhibited the prolyl-/lysyl-hydroxylation of collagen proteins and thus their maturation, resulting in the formation of a BM of increased fragility. The accumulation of these unfolded collagen proteins in the ER of mutant brain cells could have triggered their ER stress response, which in turn may have induced their apoptosis. Such a scenario would be consistent with our observation of widespread cleaved caspase-3 in mutant brains. Similarly, the decreased NADPH in Idh1-KI brain cells may be responsible for their impaired recycling of ascorbic acid (Fig. 3C), important because ascorbate is a cofactor for prolyl-/lysyl-hydroxylation (Kivirikko and Myllylä 1985; Myllyharju and Kivirikko 2004). This lack of ascorbate recycling may also have contributed to the abnormal collagen maturation in our Idh1-KI mutants. We therefore propose that the brain hemorrhage arising in Idh1-KI mice is due to D2HG-induced changes resulting in (1) Vegf overproduction, (2) increased BM fragility, and (3) enhanced ER stress-mediated cell death. This model is illustrated in Figure 6B.
Idh1 mutation and tumorigenesis
Due to the embryonic lethality of Nes-KI mice and the leakiness of Cre expression in GFAP-KI mice, we cannot state unequivocally that Idh1 mutations cause brain tumors in vivo. However, none of our live-born GFAP-KI mice developed spontaneous brain tumors, implying that genetic alterations in addition to Idh1 mutation are required for tumor initiation in mice. Nevertheless, our findings clearly demonstrate that Idh1 mutation induces cellular and microenvironmental changes that have been linked to tumor promotion. Although ROS possess mutagenic/oncogenic potential, reduced ROS levels have been previously shown to promote tumor growth and progression (at least in certain contexts) (Finkel and Holbrook 2000). BM breakdown may therefore contribute to tumor growth and progression (Tate and Aghi 2009). In addition, D2HG is associated with increased Hif1α activity and Vegf expression, which can facilitate tumor angiogenesis. Notably, glioma patients with IDH1 mutations that are treated with VEGF(R)-blocking agents such as bevacizumab and sunitinib malate exhibit prolonged survival and delayed tumor recurrence compared with patients treated with cetuximab, a monoclonal antibody that blocks the epidermal growth factor receptor (EGFR) (Lv et al. 2011).
Our findings are consistent with a previous human study in which mutations in the coding sequences of genes encoding collagens and other extracellular matrix-related proteins were identified in 13 out of 21 GBM tumor samples (Supplemental Table 2; Parsons et al. 2008). A different study of 91 GBM tumor samples discovered additional mutations in exons of four collagen genes (The Cancer Genome Atlas Research Network 2008). Most recently, 152 mutations of 28 collagen genes were identified in 316 ovarian tumors (The Cancer Genome Atlas Research Network 2011). Importantly, GBM patients with BM-related gene mutations rarely carry IDH1 mutations (Supplemental Table 2). Moreover, IDH1 mutations occur in chondromas (Amary et al. 2011a,b; Pansuriya et al. 2011), and these neoplasms show alterations to collagen distribution upon progression to malignancy (Ueda et al. 1990). These data collectively indicate that normal collagen maturation is important for suppressing cancer and suggest that IDH1 mutation leading to D2HG production severely impairs collagen functions in glioma cells such that tumor progression is facilitated.
Another factor associated with tumorigenesis in general is altered epigenetic modification, and dramatic epigenetic changes are observed in tumor cells in glioma and AML patients with IDH1/2 mutations (Figueroa et al. 2010; Noushmehr et al. 2010). Although we did not detect any significant changes in histone H3 methylation of proteins in very early stage Nes-KI embryos, 5hmC was greatly reduced in both NSCs and GPCs of these mutants (Fig. 3K–N). We therefore speculate that a key tumorigenic aspect of IDH1 mutation is D2HG-mediated alteration of epigenetic modification that also favors tumor promotion.
In conclusion, our in vivo study has demonstrated that brain-specific mutation of Idh1 is embryonic-lethal in mice due to brain hemorrhage. In brain cells isolated from these mutants, D2HG produced by the activity of the abnormal Idh1 enzyme may function as an oncometabolite that induces HIF target gene transactivation, disrupts collagen maturation, and impairs BM structure. Since GFAP-Cre-driven Idh1 mutant mice are not suitable for the glioma model, future studies are needed to establish the tumorigenic roles of Idh1 mutation by using inducible Cre systems.
Materials and methods
Mice
Idh1 (Sasaki et al. 2012), Nestin-Cre (Tronche et al. 1999), GFAP-Cre (Kwon et al. 2001), Rosa-LSL-TdTomato (Madisen et al. 2010), and p53flox (Jonkers et al. 2001) mice have been described previously. All animals were treated in accordance with the NIH Guide for Care and Use of Laboratory Animals as approved by the Ontario Cancer Institute Animal Care Committee.
Mouse tissue preparation
For total brain cell preparations, brains were dissected from mouse embryos and dissociated to create single-cell suspensions via enzymatic degradation using a neural tissue dissociation kit (Miltenyi Biotec) according to the manufacturer's protocol. Briefly, whole embryonic brains were minced, a prewarmed enzyme mix was added to the tissue pieces, and the mixture was incubated for 15 min at 37°C. The tissue was further mechanically dissociated by trituration, incubated for 20 min at 37°C, and passed through a 70-μm cell strainer. To isolate NSCs, GPCs, and DN cells, we applied the extracts to MACS prominin-1- or A2B5-coupled microbeads (Miltenyi Biotec). After two washes in PBS(−), cells were processed for various assays as described below.
Metabolite determinations
For D2HG measurement, metabolites were extracted using 80% aqueous methanol as previously described (Lu et al. 2006). Briefly, 30–50 mg of embryonic brain was immersed in 80% methanol at −80°C and homogenized. Extracts were centrifuged at 13,000 rpm for 15 min at 4°C, and supernatants were dried at room temperature and stored at −80°C. Extracts were subjected to ion-paired reverse-phase LC coupled to negative mode electrospray triple-quadrupole mass spectrometry using multiple reaction monitoring. Integrated elution peaks were compared with metabolite standard curves for absolute quantification. For intracellular ROS measurement, cells (1 × 105) were incubated with 100 μM CM-H2-DCFDA (Invitrogen) for 30 min at 37°C. Data were collected on a FACSCalibur flow cytometer (Becton Dickinson). For NADP+/NADPH ratios and αKG measurements, cells (2 × 106) were processed using the Fluoro NADP/NADPH detection kit (Cell Technology) or the α-Ketoglutarate Assay kit (BioVision), respectively. For GSH/GSSG ratios, 2 × 106 (for GSH) and 1 × 106 (for GSSG) cells were lysed by two freeze/thaw cycles and assessed using the Bioxytech GSH/GSSG-412 kit (OxisResearch). For ascorbic acid, cells (4 × 105) were evaluated using the EnzyChrom Ascorbic Acid Assay kit (BioAssay Systems). For catalase activity, cells (1 × 106) were assessed using the Catalase Assay kit (Sigma-Aldrich). For all kits, the manufacturers' protocols were followed.
qRT–PCR
Total RNA was extracted and purified using the RNeasy kit (Qiagen) according to the manufacturer's protocol. Purified RNA was treated with DNaseI and reverse-transcribed using the iScript cDNA Synthesis kit (Bio-Rad). cDNAs were subjected to qRT–PCR using SYBR Green PCR Master Mix (Applied Biosystems). Primers used for qRT–PCR are listed in Supplemental Table 3.
Immunoblotting
For detection of most proteins, brain cell lysates were prepared in lysis buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% [v/v] 2-mercaptoethanol, 0.1% [v/v] Triton X-100, 1× protease inhibitor cocktail [Roche]), electrophoretically separated on denaturing SDS polyacrylamide gels, and transferred to a PVDF membrane (Roche). For histone protein detection, E14.5 NSCs and GPCs were suspended in extraction buffer (PBS[–] containing 0.5% [v/v] Triton X100, 1× protease inhibitor cocktail [Roche]) for 10 min on ice. HCl was added to 0.2 M followed by incubation overnight at 4°C. After centrifugation to recover the supernatant, proteins were diluted in 1× NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and electrophoresed as above. For collagen protein detection, E14.5–E15.5 whole embryonic brains were homogenized in homogenization buffer (20 mM Tris-HCl at pH 7.4, 0.2 M NaCl, 0.1 M glycine, 0.1% [v/v] Triton X-100, 50 μM DTT, 1× protease inhibitor cocktail [Roche]). Proteins were separated on a 5% Tris-glycine SDS-PAGE gel and transferred to a PVDF membrane.
Proteins fixed on PVDF membranes were visualized using antibodies to type IV collagen (600-401-106, Rockland), HIF1α (10006421, Cayman Chemical), H3K4me3 (ab1012, Abcam), H3K9me3 (ab8898, Abcam), H3K9me2 (ab1220, Abcam), H3R17me2 (8284, Abcam), H3K36me3 (ab9050, Abcam), histone H3 (ab10799, Abcam), H3K27me3 (07-449, Millipore), H3K79me2 (9757, Cell Signaling Technologies), catalase (ab1877, Abcam), or β-actin (A2066, Sigma-Aldrich). Primary antibodies were detected using HRP-conjugated secondary antibodies to mouse IgG or rabbit IgG (GE Healthcare). Immunocomplexes were visualized using ECL + reagents (GE Healthcare).
Quantitative analysis of 5hmC
Detection of 5hmC was performed as previously described (Ko et al. 2010). Briefly, genomic DNA was treated with sodium bisulfite using the EpiTect bisulfite kit (Qiagen). DNA samples were denatured, and twofold serial dilutions were spotted onto a nitrocellulose membrane in an assembled Bio-Dot apparatus (Bio-Rad). The membrane was incubated with anti-cytosine 5-methylenesulfonate antibody. After incubating with HRP-conjugated anti-rabbit IgG secondary antibody, the membrane was visualized by enhanced chemoluminescence.
Histology
Tissue samples were fixed in 10% buffered formalin or 4% paraformaldehyde (PFA) at 4°C and embedded in paraffin. Sections (5 μm) were stained with Harris hematoxylin and eosin according to standard protocols. For cell proliferation assays, pregnant females were injected intraperitoneally with 0.1 mg BrdU (Roche) per gram body weight 45 min before dissection to isolate embryos. Paraffin sections of fixed embryos were processed using the BrdU In Situ Detection kit (Boehringer Mannheim) to detect BrdU. For immunostaining, sections were dewaxed, rehydrated, and microwaved in 10 mM sodium citrate. Slides for peroxidase reactions were treated with 3% hydrogen peroxide in PBS to kill endogenous peroxidase activity. Slides were incubated overnight at 4°C in primary antibody, washed, and incubated in secondary antibody for 1 h at room temperature. Slides for peroxidase reactions were treated with ABC reagent (Vector Laboratories) for 1 h and washed in PBS. Primary antibodies recognizing cleaved caspase-3 (9661, Cell Signaling) or type IV collagen (600-401-106, Rockland) were visualized using secondary antibodies as above. Microscopic observations and images were acquired using a Leica DM6000 or Leica DMI6000 instrument.
Acknowledgments
We are grateful to M. Kawazu, H. Kuwata, A. Suzuki, L. Clarke, D.V.D. Kooy, E. Bogdanovic, N. Sabha, and A. Guha for helpful discussions; V.R. Fantin, S. Schalm, A. Shahinian, G. Duncan, and J. Silvester for expert technical support and insightful comments; M. Saunders for scientific editing; and Irene Ng for administrative help. This work was supported by grants to T.W.M. from the Canadian Institutes of Health Research, and by grants to A.R. from the U.S. National Institutes of Health, the Leukemia and Lymphoma Society, and the California Institute for Regenerative Medicine. M.S., A.W., J.H., and A.Y.T. generated mutant mice. M.S., M.I., A.J.E., I.S.H., I.I.C.C., B.S., M.K., A.R., K.E.Y., and S.M.S. conducted experiments. M.S., M.I., A.J.E., R.A.C., C.B.K., I.S.H., S.M., I.I.C.C., T.U., S.I., K.Y., and T.W.M. discussed the hypothesis and interpreted the data. M.S. and T.W.M. wrote the manuscript. K.E.Y. and S.M.S. disclose financial interests as employees of Agios Pharmaceuticals. T.W.M. owns stock options in Agios Pharmaceuticals. The authors declare that no other conflicts of interest exist.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.198200.112.
References
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. 2011a. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 [DOI] [PubMed] [Google Scholar]
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. 2011b. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet 43: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Bajenaru ML, Zhu Y, Hedrick NM, Donahoe J, Parada LF, Gutmann DH 2002. Astrocyte-specific inactivation of the neurofibromatosis 1 gene (NF1) is insufficient for astrocytoma formation. Mol Cell Biol 22: 5100–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabh P, Braun A, Nedergaard M 2004. The blood-brain barrier: An overview: Structure, regulation, and clinical implications. Neurobiol Dis 16: 1–13 [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A 2005. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res 33: e51 doi: 10.1093/nar/gki559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK 2000. Angiogenesis in cancer and other diseases. Nature 407: 249–257 [DOI] [PubMed] [Google Scholar]
- Cheng SY, Nagane M, Huang HS, Cavenee WK 1997. Intracerebral tumor-associated hemorrhage caused by overexpression of the vascular endothelial growth factor isoforms VEGF121 and VEGF165 but not VEGF189. Proc Natl Acad Sci 94: 2081–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, LeCouter J 2003. The biology of VEGF and its receptors. Nat Med 9: 669–676 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Gould SJ 1999. The human PICD gene encodes a cytoplasmic and peroxisomal NADP+-dependent isocitrate dehydrogenase. J Biol Chem 274: 30527–30533 [DOI] [PubMed] [Google Scholar]
- Gould DB, Phalan FC, Breedveld GJ, van Mil SE, Smith RS, Schimenti JC, Aguglia U, van der Knaap MS, Heutink P, John SW 2005. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science 308: 1167–1171 [DOI] [PubMed] [Google Scholar]
- Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny JC, Müller U 2001. β1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31: 367–379 [DOI] [PubMed] [Google Scholar]
- Green A, Beer P 2010. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med 362: 369–370 [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. 2010. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, et al. 2010. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: Implications for classification of gliomas. Acta Neuropathol 120: 707–718 [DOI] [PubMed] [Google Scholar]
- Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, Myllyharju J 2007. Loss of assembly of the main basement membrane collagen, type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem 282: 2512–2519 [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. 2005. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell 8: 143–153 [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 [DOI] [PubMed] [Google Scholar]
- Jung S, Moon KS, Jung TY, Kim IY, Lee YH, Rhu HH, Sun HS, Jeong YI, Kim KK, Kang SS 2006. Possible pathophysiological role of vascular endothelial growth factor (VEGF) and matrix metalloproteinases (MMPs) in metastatic brain tumor-associated intracerebral hemorrhage. J Neurooncol 76: 257–263 [DOI] [PubMed] [Google Scholar]
- Kalluri R 2003. Basement membranes: Structure, assembly and role in tumour angiogenesis. Nat Rev Cancer 3: 422–433 [DOI] [PubMed] [Google Scholar]
- Kivirikko KI, Myllylä R 1985. Post-translational processing of procollagens. Ann N Y Acad Sci 460: 187–201 [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A 2011. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci 108: 14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ 2001. Pten regulates neuronal soma size: A mouse model of Lhermitte-Duclos disease. Nat Genet 29: 404–411 [DOI] [PubMed] [Google Scholar]
- Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI 2011. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell 8: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M 2011. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Kimball E, Rabinowitz JD 2006. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J Am Soc Mass Spectrom 17: 37–50 [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Teugels E, Sadones J, Quartier E, Huylebrouck M, Du Four S, Le Mercier M, De Witte O, Salmon I, Michotte A, et al. 2011. Correlation between IDH1 gene mutation status and survival of patients treated for recurrent glioma. Anticancer Res 31: 4457–4463 [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani T, Yamamoto A, Nagai N, Kubota H, Nagata K 2004. Accumulation of type IV collagen in dilated ER leads to apoptosis in Hsp47-knockout mouse embryos via induction of CHOP. J Cell Sci 117: 5913–5922 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI 2004. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 20: 33–43 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansuriya TC, van Eijk R, d'Adamo P, van Ruler MA, Kuijjer ML, Oosting J, Cleton-Jansen AM, van Oosterwijk JG, Verbeke SL, Meijer D, et al. 2011. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet 43: 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöschl E, Schlötzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U 2004. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 131: 1619–1628 [DOI] [PubMed] [Google Scholar]
- Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38 [DOI] [PubMed] [Google Scholar]
- Rankin EB, Giaccia AJ 2008. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ 15: 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautavuoma K, Takaluoma K, Sormunen R, Myllyharju J, Kivirikko KI, Soininen R 2004. Premature aggregation of type IV collagen and early lethality in lysyl hydroxylase 3 null mice. Proc Natl Acad Sci 101: 14120–14125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, et al. 2009. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27: 4150–4154 [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. 2012. Idh1(R132H) mutation increases murine hematopoietic progenitors and alters epigenetics. Nature doi: 10.1038/nature11323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E 2005. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7: 77–85 [DOI] [PubMed] [Google Scholar]
- Tate MC, Aghi MK 2009. Biology of angiogenesis and invasion in glioma. Neurotherapeutics 6: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 23: 99–103 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Oda Y, Tsuchiya H, Tomita K, Nakanishi I 1990. Immunohistological study on collagenous proteins of benign and malignant human cartilaginous tumours of bone. Virchows Arch A Pathol Anat Histopathol 417: 291–297 [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W 2008. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid Redox Signal 10: 179–206 [DOI] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. 2009. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]